Looked at Life from Both Sides Now
Abstract
:1. From Up and Down, and Still Somehow
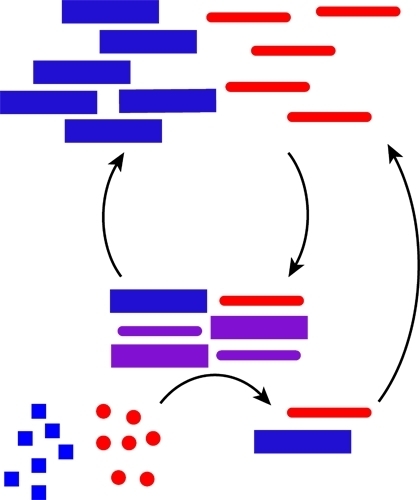
2. Biopolymer Diversity
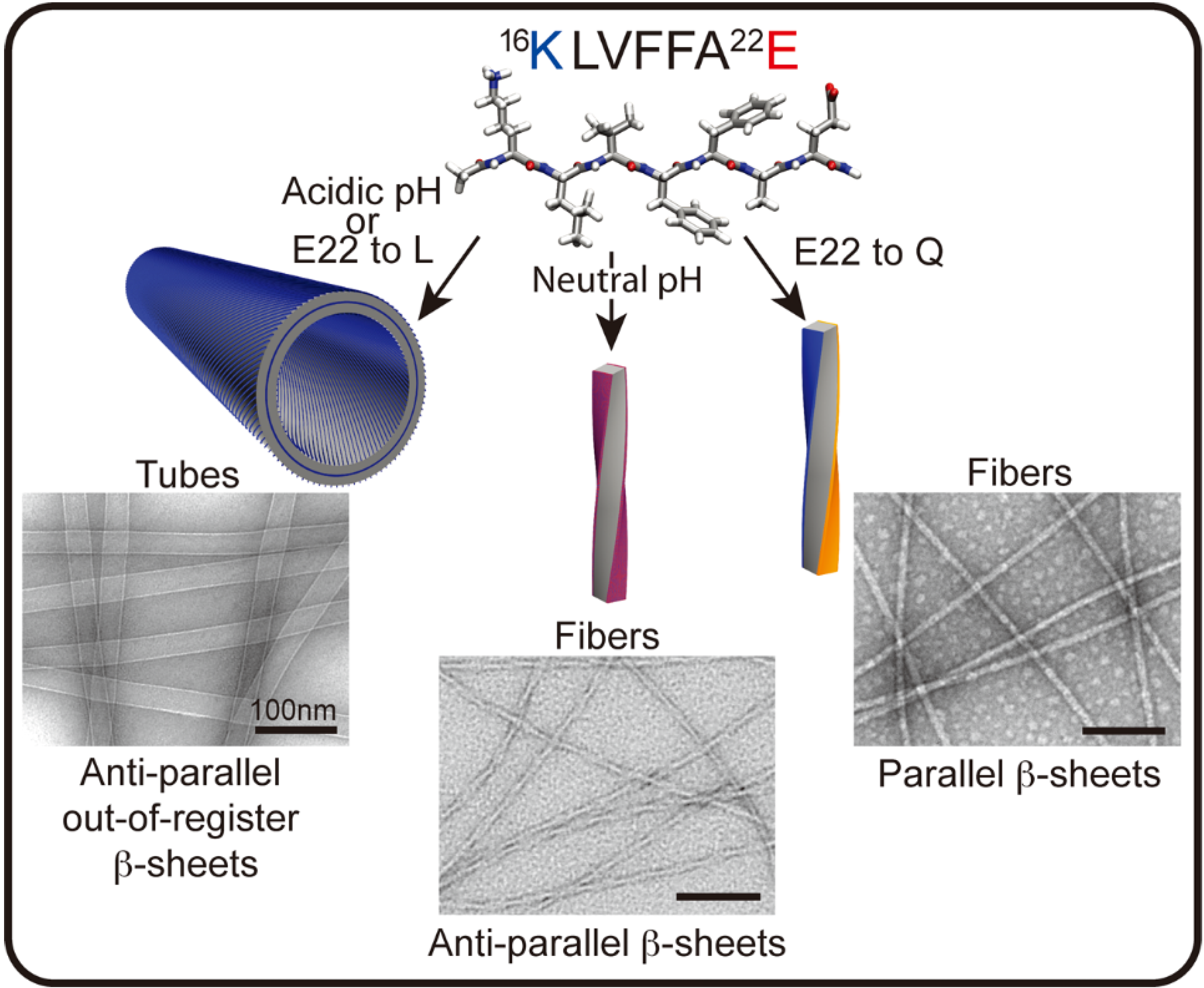
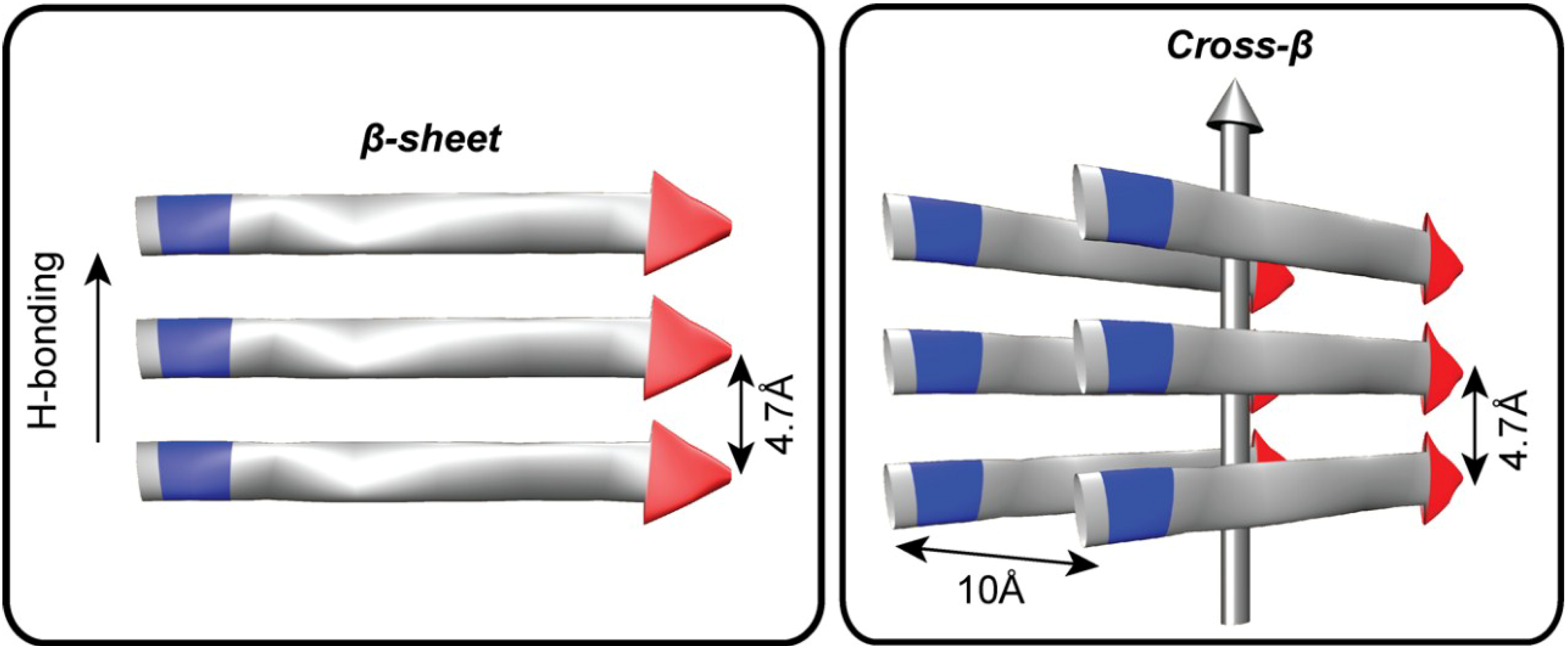
3. Functional Assemblies
4. From Both Sides Now
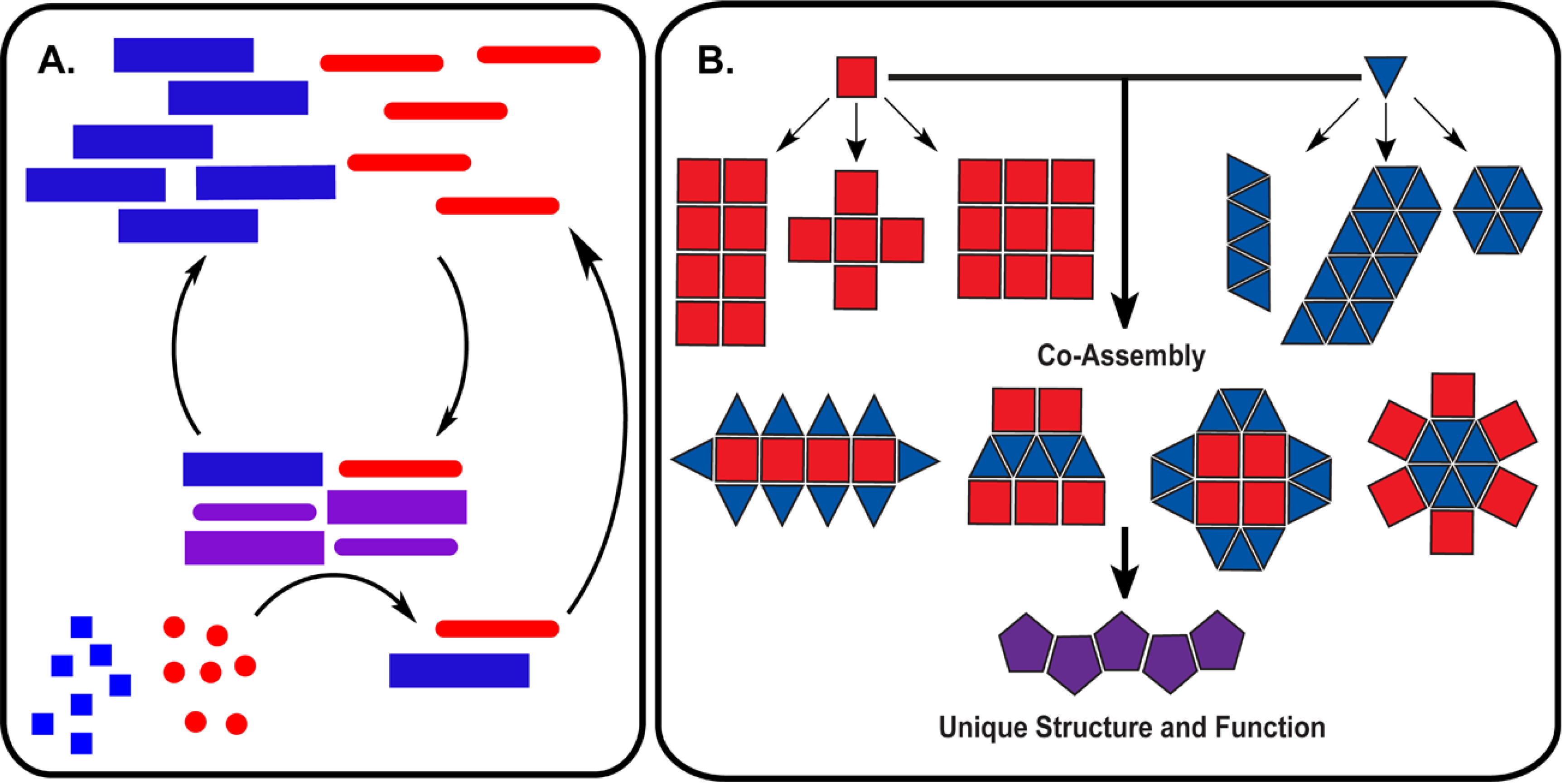
5. Conclusions: Towards Molecular Mutualisms
Acknowledgments
Conflicts of Interest
References
- Mann, S. Life as a nanoscale phenomenon. Angew. Chem. Int. Ed. 2008, 47, 5306–5320. [Google Scholar]
- Crick, F.H.C. The biological replication of macromolecules. Sym. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar]
- Woese, C.R. On the evolution of cells. Proc. Natl. Acad. Sci. USA 2002, 99, 8742–8747. [Google Scholar] [CrossRef]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of Ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Gesteland, R.F.; Cech, T.R.; Atkins, J.F. The RNA World, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1999. [Google Scholar]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Harish, A.; Caetano-Anolles, G. Ribosomal history reveals origins of modern protein synthesis. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Goodwin, J.T.; Mehta, A.K.; Lynn, D.G. Digital and analog chemical evolution. Acc. Chem. Res. 2012, 45, 2189–2199. [Google Scholar] [CrossRef]
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science 2005, 309, 1514–1518. [Google Scholar] [CrossRef]
- Bolton, D.C.; McKinley, M.P.; Prusiner, S.B. Identification of a protein that purifies with the scrapie prion. Science 1982, 218, 1309–1311. [Google Scholar] [CrossRef]
- Astbury, W.T.; Dickinson, S.; Bailey, K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 1935, 29, 2351–2360. [Google Scholar]
- Parker, K.D.; Rudall, K.M. Structure of the silk of chrysopa egg-stalks. Nature 1957, 179, 905–906. [Google Scholar] [CrossRef]
- Geddes, A.J.; Parker, K.D.; Atkins, E.D.T.; Beighton, E. “Cross-β” conformation in proteins. J. Mol. Biol. 1968, 32, 343–358. [Google Scholar] [CrossRef]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef]
- Tessier, P.M.; Lindquist, S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+]. Nat. Struct. Mol. Biol. 2009, 16, 598–605. [Google Scholar] [CrossRef] [Green Version]
- Inge-Vechtomov, S.G.; Zhouravleva, G.A.; Chernoff, Y.O. Biological roles of prion domains. Prion 2007, 1, 228–235. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Hamodrakas, S.J. Natural protective amyloids. Curr. Protein Pept. Sci. 2008, 9, 291–309. [Google Scholar]
- Barlow, D.E.; Dickinson, G.H.; Orihuela, B.; Kulp, J.L.; Rittschof, D.; Wahl, K.J. Characterization of the adhesive plaque of the barnacle Balanus amphitrite: Amyloid-like nanofibrils are a major component. Langmuir 2010, 26, 6549–6556. [Google Scholar] [CrossRef]
- Kenchington, W. The larval silk of Hypera spp. (Coleoptera: Curculionidae). A new example of the cross-β protein conformation in an insect silk. J. Insect Physiol. 1983, 29, 355–361. [Google Scholar] [CrossRef]
- Majumdar, A.; Cesario, W.C.; White-Grindley, E.; Jiang, H.; Ren, F.; Khan, M.R.; Li, L.; Choi, E.M.; Kannan, K.; Guo, F.; et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 2012, 148, 515–529. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006, 4. [Google Scholar] [CrossRef]
- Kemp, M. The Mona Lisa of modern science. Nature 2003, 421, 416–420. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structures of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Rich, A.; Zhang, S. Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003, 4, 566–572. [Google Scholar] [CrossRef]
- Hermann, T.; Westhof, E. Non-Watson-Crick base pairs in RNA-protein recognition. Chem. Biol. 1999, 6, R335–R343. [Google Scholar] [CrossRef]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010, 67, 43–62. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef]
- Saini, N.; Zhang, Y.; Usdin, K.; Lobachev, K.S. When secondary comes first—The importance of non-canonical DNA structures. Biochimie 2013, 95, 117–123. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Miyoshi, D.; Matsumura, S.; Li, W.; Sugimoto, N. Structural polymorphism of telomeric DNA regulated by pH and divalent cation. Nucleosides Nucleotides Nucl. Acids 2003, 22, 203–221. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar]
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Baral, A.; Kumar, P.; Pathak, R.; Chowdhury, S. Emerging trends in G-quadruplex biology—Role in epigenetic and evolutionary events. Mol. Biosyst. 2013, 9, 1568–1575. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Hiller, D.A.; Strobel, S.A. The chemical versatility of RNA. Phil. Trans. R. Soc. B 2011, 366, 2929–2935. [Google Scholar]
- Doudna, J.A.; Cech, T.R. The chemical repertoire of natural ribozymes. Nature 2002, 418, 222–228. [Google Scholar] [CrossRef]
- Ward, W.L.; Plakos, K.; DeRose, V.J. Nucleic acid catalysis: Metals, nucleobases, and other cofactors. Chem. Rev. 2014, 114, 4318–4342. [Google Scholar] [CrossRef]
- Silverman, S.K. Deoxyribozymes: DNA catalysts for bioorganic chemistry. Org. Biomol. Chem. 2004, 2, 2701–2706. [Google Scholar] [CrossRef]
- Dill, K.A.; MacCallum, J.L. The protein-folding problem, 50 years on. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef]
- Compiani, M.; Capriotti, E. Computational and theoretical methods for protein folding. Biochemistry 2013, 52, 8601–8624. [Google Scholar] [CrossRef]
- Vabulas, R.M.; Raychaudhuri, S.; Hayer-Hartl, M.; Hartl, F.U. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Chien, P.; Weissman, J.S.; DePace, A.H. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 2004, 73, 617–656. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Jucker, M.; Walker, L.C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013, 501, 45–51. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.W.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Watts, J.C.; Nguyen, H.O.; Hayashi, S.; DeArmond, S.J.; Prusiner, S.B. Conformational transformation and selection of synthetic prion strains. J. Mol. Biol. 2011, 413, 527–542. [Google Scholar] [CrossRef]
- Li, J.; Browning, S.; Mahal, S.P.; Oelschlegel, A.M.; Weissmann, C. Darwinian evolution of prions in cell culture. Science 2010, 327, 869–872. [Google Scholar] [CrossRef]
- Collinge, J.; Clarke, A.R. A general model of prion strains and their pathogenicity. Science 2007, 318, 930–936. [Google Scholar] [CrossRef]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef]
- Childers, W.S.; Mehta, A.K.; Bui, T.Q.; Liang, Y.; Lynn, D.G. Molecular Self-Assembly—Advances and Applications, 1st ed.; Li, A., Ed.; Pan Stanford Publishing Pte Ltd.: Singapore, Singapore, 2012; pp. 1–36. [Google Scholar]
- Childers, W.S.; Anthony, N.R.; Mehta, A.K.; Berland, K.M.; Lynn, D.G. Phase networks of cross-β peptide assemblies. Langmuir 2012, 28, 6386–6395. [Google Scholar] [CrossRef]
- Debeljuh, N.; Barrow, C.J.; Byrne, N. The impact of ionic liquids on amyloid fibrilization of Aβ16–22: Tuning the rate of fibrilization using a reverse Hofmeister strategy. Phys. Chem. Chem. Phys. 2011, 13, 16534–16536. [Google Scholar] [CrossRef]
- Mehta, A.K.; Lu, K.; Childers, W.S.; Liang, Y.; Dublin, S.N.; Gong, J.; Snyder, J.P.; Pingali, S.V.; Thiyagarajan, P.; Lynn, D.G. Facial symmetry in protein self-assembly. J. Am. Chem. Soc. 2008, 130, 9829–9835. [Google Scholar] [CrossRef]
- Balbach, J.J.; Ishii, Y.; Antzutkin, O.N.; Leapman, R.D.; Rizzo, N.W.; Dyda, F.; Reed, J.; Tycko, R. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer’s β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry 2000, 39, 13748–13759. [Google Scholar] [CrossRef]
- Lu, K.; Jacob, J.; Thiyagarajan, P.; Conticello, V.P.; Lynn, D.G. Exploiting amyloid fibril lamination for nanotube self-assembly. J. Am. Chem. Soc. 2003, 125, 6391–6393. [Google Scholar] [CrossRef]
- Dong, J.; Lu, K.; Lakdawala, A.; Mehta, A.K.; Lynn, D.G. Controlling amyloid growth in multiple dimensions. Amyloid 2006, 13, 206–215. [Google Scholar] [CrossRef]
- Liang, Y.; Pingali, S.V.; Jogalekar, A.S.; Snyder, J.P.; Thiyagarajan, P.; Lynn, D.G. Cross-strand pairing and amyloid assembly. Biochemistry 2008, 47, 10018–10026. [Google Scholar] [CrossRef]
- Childers, W.S.; Mehta, A.K.; Lu, K.; Lynn, D.G. Templating molecular arrays in amyloid’s cross-β grooves. J. Am. Chem. Soc. 2009, 131, 10165–10172. [Google Scholar] [CrossRef]
- Anthony, N.R.; Mehta, A.K.; Lynn, D.G.; Berland, K.M. Mapping amyloid-β(16–22) nucleation pathways using fluorescence lifetime imaging microscopy. Soft Matter 2014, 10, 4162–4172. [Google Scholar] [CrossRef]
- Liang, Y.; Lynn, D.G.; Berland, K.M. Direct observation of nucleation and growth in amyloid self-assembly. J. Am. Chem. Soc. 2010, 132, 6306–6308. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Dunkelberger, E.B.; Tran, H.Q.; Cheng, P.N.; Chiu, C.C.; Cao, P.; Raleigh, D.P.; de Pablo, J.J.; Nowick, J.S.; Zanni, M.T. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. USA 2013, 110, 19285–19290. [Google Scholar] [CrossRef]
- Liang, C.; Ni, R.; Smith, J.E.; Childer, W.S.; Mehta, A.K.; Lynn, D. Kinetic intermediates in amyloid assembly. J. Am. Chem. Soc. 2014, 136, 15146–15149. [Google Scholar] [CrossRef]
- Lin, D.; Luo, Y.; Wu, S.; Ma, Q.; Wei, G.; Yang, X. Investigation of the aggregation process of amyloid-β-(16–22) peptides and the dissolution of intermediate aggregates. Langmuir 2014, 30, 3170–3175. [Google Scholar] [CrossRef]
- Perween, S.; Chandanshive, B.; Kotamarthi, H.C.; Khushalani, D. Single amino acid based self-assembled structure. Soft Matter 2013, 9, 10141–10145. [Google Scholar] [CrossRef]
- Frederix, P.W.J.M.; Ulijn, R.V.; Hunt, N.T.; Tuttle, T. Virtual screening for dipeptide aggregation: Toward predictive tools for peptide self-assembly. J. Phys. Chem. Lett. 2011, 2, 2380–2384. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef]
- De Groot, N.S.; Parella, T.; Aviles, F.X.; Vendrell, J.; Ventura, S. Ile-Phe dipeptide self-assembly: Clues to amyloid formation. Biophys. J. 2007, 92, 1732–1741. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Formation of closed-cage nanostructures by self-assembly of aromatic dipeptides. Nano Lett. 2004, 4, 581–585. [Google Scholar] [CrossRef]
- Soldatov, D.V.; Moudrakovski, I.L.; Ripmeester, J.A. Dipeptides as microporous materials. Angew. Chem. Int. Ed. 2004, 43, 6308–6311. [Google Scholar] [CrossRef]
- Gorbitz, C.H. Nanotubes from hydrophobic dipeptides: Pore size regulation through side chain substitution. New J. Chem. 2003, 27, 1789–1793. [Google Scholar] [CrossRef]
- Gorbitz, C.H. Structures of dipeptides: The head-to-tail story. Acta Cryst. 2010, B66, 84–93. [Google Scholar] [CrossRef]
- Gorbitz, C.H. Microporous organic materials from hydrophobic dipeptides. Chem. Eur. J. 2007, 13, 1022–1031. [Google Scholar] [CrossRef]
- Flores, R.; Hernandez, C.; Martinez de Alba, A.E.; Daros, J.A.; di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Flores, R.; Serra, P.; Minoia, S.; di Serio, F.; Navarro, B. Viroids: From genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Sano, T.; Candresse, T.; Hammond, R.W.; Diener, T.O.; Owens, R.A. Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. USA 1992, 89, 10104–10108. [Google Scholar] [CrossRef]
- Daros, J.A.; Marcos, J.F.; Hernandez, C.; Flores, R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 1994, 91, 12813–12817. [Google Scholar] [CrossRef]
- Hutchins, C.J.; Rathjen, P.D.; Forster, A.C.; Symons, R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuan, R.; Elena, S.F. Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef]
- Buell, A.K.; Esbjorner, E.K.; Riss, P.J.; White, D.A.; Aigbirhio, F.I.; Toth, G.; Welland, M.E.; Dobson, C.M.; Knowles, T.P. Probing small molecule binding to amyloid fibrils. Phys. Chem. Chem. Phys. 2011, 13, 20044–20052. [Google Scholar] [CrossRef]
- Levine, H. Thioflavine T interactions with synthetic Alzheimers Disease β-amyloid peptides—detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef]
- Divry, P.; Florkin, M. The optic properties of amyloid. Comptes Rendus des Seances de la Societe de Biologie et de ses Filiales 1927, 97, 1808–1810. (In French) [Google Scholar]
- Buxbaum, J.N.; Linke, R.P. A molecular history of the amyloidoses. J. Mol. Biol. 2012, 421, 142–159. [Google Scholar] [CrossRef]
- Schutz, A.K.; Soragni, A.; Hornemann, S.; Aguzzi, A.; Ernst, M.; Bockmann, A.; Meier, B.H. The amyloid-Congo red interface at atomic resolution. Angew. Chem. Int. Ed. 2011, 50, 5956–5960. [Google Scholar] [CrossRef]
- Dos Reis, S.; Coulary-Salin, B.; Forge, V.; Lascu, I.; Begueret, J.; Saupe, S.J. The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J. Biol. Chem. 2002, 277, 5703–5706. [Google Scholar] [CrossRef]
- Bäcklund, F.G.; Wigenius, J.; Westerlund, F.; Inganäs, O.; Solin, N. Amyloid fibrils as dispersing agents for oligothiophenes: Control of photophysical properties through nanoscale templating and flow induced fibril alignment. J. Mater. Chem. C 2014, 2, 7811–7822. [Google Scholar] [CrossRef]
- Herland, A.; Bjork, P.; Hania, P.R.; Scheblykin, I.G.; Inganas, O. Alignment of a conjugated polymer onto amyloid-like protein fibrils. Small 2007, 3, 318–325. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Bio. 2007, 8, 479–490. [Google Scholar] [CrossRef]
- Saha, S.; Deep, S. Switch in the aggregation pathway of bovine serum albumin mediated by electrostatic interactions. J. Phys. Chem. B 2014, 118, 9155–9166. [Google Scholar]
- Gilbert, J.; Campanella, O.; Jones, O.G. Electrostatic stabilization of β-lactoglobulin fibrils at increased ph with cationic polymers. Biomacromolecules 2014, 15, 3119–3127. [Google Scholar] [CrossRef]
- Nielsen, S.B.; Yde, P.; Giehm, L.; Sundbye, S.; Christiansen, G.; Mathiesen, J.; Jensen, M.H.; Jensen, P.H.; Otzen, D.E. Multiple roles of heparin in the aggregation of p25alpha. J. Mol. Biol. 2012, 421, 601–615. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Kato, K.; Yanagisawa, K. Aβ polymerization through interaction with membrane gangliosides. Biochim. Biophys. Acta 2010, 1801, 868–877. [Google Scholar] [CrossRef]
- Li, S.; Sidorov, A.N.; Mehta, A.K.; Bisignano, A.J.; Das, D.; Childers, W.S.; Schuler, E.; Jiang, Z.; Orlando, T.M.; Berland, K.; et al. Neurofibrillar Tangle Surrogates: Histone H1 Binding to Patterned Phosphotyrosine Peptide Nanotubes. Biochemistry 2014, 53, 4225–4227. [Google Scholar] [CrossRef]
- Suk, J.Y.; Zhang, F.; Balch, W.E.; Linhardt, R.J.; Kelly, J.W. Heparin accelerates gelsolin amyloidogenesis. Biochemistry 2006, 45, 2234–2242. [Google Scholar]
- Terzi, E.; Holzemann, G.; Seelig, J. Interaction of Alzheimer β-amyloid peptide (1–40) with lipid membranes. Biochemistry 1997, 36, 14845–14852. [Google Scholar] [CrossRef]
- Illos, R.A.; Bisogno, F.R.; Clodic, G.; Bolbach, G.; Weissbuch, I.; Lahav, M. Oligopeptides and copeptides of homochiral sequence, via β-sheets, from mixtures of racemic alpha-amino acids, in a one-pot reaction in water; relevance to biochirogenesis. J. Am. Chem. Soc. 2008, 130, 8651–8659. [Google Scholar] [CrossRef]
- Rubinstein, I.; Eliash, R.; Bolbach, G.; Weissbuch, I.; Lahav, M. Racemic beta sheets in biochirogenesis. Angew. Chem. Int. Ed. 2007, 46, 3710–3713. [Google Scholar] [CrossRef]
- Wagner, N.; Rubinov, B.; Ashkenasy, G. β-Sheet-induced chirogenesis in polymerization of oligopeptides. Chemphyschem 2011, 12, 2771–2780. [Google Scholar] [CrossRef]
- Rubinov, B.; Wagner, N.; Rapaport, H.; Ashkenasy, G. Self-replicating amphiphilic β-sheet peptides. Angew. Chem. Int. Ed. 2009, 48, 6683–6686. [Google Scholar] [CrossRef]
- Takahashi, Y.; Mihara, H. Construction of a chemically and conformationally self-replicating system of amyloid-like fibrils. Bioorgan. Med. Chem. 2004, 12, 693–699. [Google Scholar]
- Rubinov, B.; Wagner, N.; Matmor, M.; Regev, O.; Ashkenasy, N.; Ashkenasy, G. Transient Fibril Structures Facilitating Nonenzymatic Self-Replication. ACS Nano 2012, 6, 7893–7901. [Google Scholar] [CrossRef]
- Weissbuch, I.; Illos, R.A.; Bolbacj, G.; Lahav, M. Racemic β-sheets as templates of relevance to the origin of homochirality of peptides: Lessons from crystal chemistry. Acc. Chem. Res. 2009, 42, 1128–1140. [Google Scholar] [CrossRef]
- Maury, C.P.J. Self-propagating β-sheet polypeptide structures as prebiotic informational molecular entities: The amyloid world. Orig. Life Evol. Biosph. 2009, 39, 141–150. [Google Scholar] [CrossRef]
- Childers, W.S.; Ni, R.; Mehta, A.K.; Lynn, D.G. Peptide membranes in chemical evolution. Curr. Opin. Chem Biol. 2009, 13, 652–659. [Google Scholar] [CrossRef]
- Brack, A.; Orgel, L.E. β Structures of alternating polypeptides and their prebiotic significance. Nature 1975, 256, 383–387. [Google Scholar] [CrossRef]
- Hoffman, M.M.; Khrapov, M.A.; Cox, J.C.; Yao, J.; Tong, L.; Ellington, A.D. AANT: The amino acid-nucleotide interaction database. Nucleic Acids Res. 2004, 32, D174–D181. [Google Scholar] [CrossRef]
- Jones, S.; van Heyningens, P.; Berman, H.M.; Thornton, J.M. Protein-DNA interactions: A structural analysis. J. Mol. Biol. 1999, 287, 877–896. [Google Scholar] [CrossRef]
- Nadassy, K.; Wodak, S.J.; Janin, J. Structural features of protein-nucleic acid recognition sites. Biochemistry 1999, 38, 1999–2017. [Google Scholar] [CrossRef]
- Wool, I.G. The structure and function of eukaryotic ribosomes. Annu. Rev. Biochem. 1979, 48, 719–754. [Google Scholar] [CrossRef]
- Esakova, O.; Krasilnikov, A.S. Of proteins and RNA: The RNase P/MRP family. RNA 2010, 16, 1725–1747. [Google Scholar] [CrossRef]
- Frank, D.N.; Pace, N. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998, 67, 153–180. [Google Scholar] [CrossRef]
- Woodson, S.A. RNA folding and ribosome assembly. Curr. Opin. Chem. Biol. 2008, 12, 667–673. [Google Scholar] [CrossRef]
- Macedo, B.; Millen, T.A.; Braga, C.A.; Gomes, M.P.; Ferreira, P.S.; Kraineva, J.; Winter, R.; Silva, J.L.; Cordeiro, Y. Nonspecific prion protein-nucleic acid interactions lead to different aggregates and cytotoxic species. Biochemistry 2012, 51, 5402–5413. [Google Scholar] [CrossRef]
- Supattapone, S. Biochemistry. What makes a prion infectious? Science 2010, 327, 1091–1092. [Google Scholar] [CrossRef]
- Geoghegan, J.C.; Valdes, P.A.; Orem, N.R.; Deleault, N.R.; Williamson, R.A.; Harris, B.T.; Supattapone, S. Selective incorporation of polyanionic molecules into hamster prions. J. Biol. Chem. 2007, 282, 36341–36353. [Google Scholar] [CrossRef]
- Deleault, N.R.; Geoghegan, J.C.; Nishina, K.; Kascsak, R.; Williamson, R.A.; Supattapone, S. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J. Biol. Chem. 2005, 280, 26873–26879. [Google Scholar] [CrossRef]
- Grossman, A.; Zeiler, B.; Sapirstein, V. Prion protein interactions with nucleic acid: Possible models for prion disease and prion function. Neurochem. Res. 2003, 28, 955–963. [Google Scholar] [CrossRef]
- Wang, M.; Law, M.; Duhamel, J.; Chen, P. Interaction of a self-assembling peptide with oligonucleotides: Complexation and aggregation. Biophys. J. 2007, 93, 2477–2490. [Google Scholar] [CrossRef]
- Braun, S.; Humphreys, C.; Fraser, E.; Brancale, A.; Bochtler, M.; Dale, T. Amyloid-associated nucleic acid hybridisation. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Turk, R.M.; Chumachenko, N.V.; Yarus, M. Multiple translational products from a five-nucleotide ribozyme. Proc. Natl. Acad. Sci. USA 2010, 107, 4585–4589. [Google Scholar] [CrossRef]
- Carny, O.; Gazit, E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005, 19, 1051–1055. [Google Scholar] [CrossRef]
- Carny, O.; Gazit, E. Creating prebiotic sanctuary: Self-assembling supramolecular peptide structures bind and stabilize RNA. Orig. Life Evol. Biosph. 2011, 41, 121–132. [Google Scholar] [CrossRef]
- Otto, S.; Furlan, R.L.; Sanders, J.K. Dynamic combinatorial libraries of macrocyclic disulfides in water. J. Am. Chem. Soc. 2000, 122, 12063–12064. [Google Scholar] [CrossRef]
- Carnell, J.; Waudby, C.A.; Belenguer, A.M.; Stuart, M.C.; Peyralans, J.J.-P.; Otto, S. Mechanosensitive self-replication driven by self-organization. Science 2010, 327, 1502–1506. [Google Scholar]
- Ivnitski, D.; Amit, M.; Rubinov, B.; Cohen-Luria, R.; Ashkenasy, N.; Ashkenasy, G. Introducing charge transfer functionality into prebiotically relevant β-sheet peptide fibrils. Chem. Commun. 2014, 50, 6733–6736. [Google Scholar] [CrossRef]
- Goodwin, J.T.; Lynn, D.G. Template-directed synthesis: Use of a reversible reaction. J. Am. Chem. Soc. 1992, 114, 9197–9198. [Google Scholar] [CrossRef]
- McCleskey, S.C.; Griffin, M.J.; Schneider, S.E.; McDevitt, J.T.; Anslyn, E.V. Differential receptors create patterns diagnostic for ATP and GTP. J. Am. Chem. Soc. 2003, 125, 1114–1115. [Google Scholar] [CrossRef]
- Schneider, S.E.; O’Neil, S.N.; Anslyn, E.V. Coupling rational design with libraries leads to the production of an ATP selective chemosensor. J. Am. Chem. Soc. 2000, 122, 542–543. [Google Scholar] [CrossRef]
- Butterfield, S.M.; Sweeney, M.M.; Waters, M.L. The recognition of nucleotides with model β-hairpin receptors: Investigation of critical contacts and nucleotide selectivity. J. Org. Chem. 2005, 70, 1105–1114. [Google Scholar] [CrossRef]
- Butterfield, S.M.; Waters, M.L. A designed β-hairpin peptide for molecular recognition of ATP in water. J. Am. Chem. Soc. 2003, 125, 9580–9581. [Google Scholar] [CrossRef]
- Norris, V.; Loutelier-Bourhis, C.; Thierry, A. How did metabolism and genetic replication get married? Orig. Life Evol. Biosph. 2012, 42, 487–495. [Google Scholar] [CrossRef]
- Root-Bernstein, R. Simultaneous origin of homochirality, the genetic code and its directionality. Bioessays 2007, 29, 689–698. [Google Scholar] [CrossRef]
- Hunding, A.; Kepes, F.; Lancet, D.; Minsky, A.; Norris, V.; Raine, D.; Sriram, K.; Root-Bernstein, R. Compositional complementarity and prebiotic ecology in the origin of life. Bioessays 2006, 28, 399–412. [Google Scholar] [CrossRef]
- Root-Bernstein, R.; Dillon, P.F. Molecular Complementarity I: The complementarity theory of the origin and evolution of life. J. Theor. Biol. 1997, 188, 447–479. [Google Scholar] [CrossRef]
- Root-Bernstein, R. A modular hierarchy-based theory of the chemical origins of life based on molecular complementarity. Acc. Chem. Res. 2012, 45, 2169–2177. [Google Scholar] [CrossRef]
- Dale, T. Protein and nucleic acid together: A mechanism for the emergence of biological selection. J. Theor. Biol. 2006, 240, 337–342. [Google Scholar] [CrossRef]
- Segre, D.; DBen-Eil, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar] [CrossRef]
- Flores Martinez, C.I. SETI in the light of cosmic convergent evolution. Acta Astronaut. 2014, 104, 341–349. [Google Scholar] [CrossRef]
- Chela-Flores, J. From systems chemistry to systems astrobiology: Life in the universe as an emergent phenomenon. Int. J. Astrobiol. 2013, 12, 8–16. [Google Scholar] [CrossRef]
- Goodwin, J.T.; Walker, S.I.; Amin, S.; Armbrust, G.; Burrows, C.J.; Lynn, D.G. Alternative Chemistries of Life: Empirical Approaches. Available online: http://chemistry.emory.edu/home/assets/alternativechem.pdf (accessed on 3 December 2014).
- Russell, M.J.; Barge, L.M.; Bhartia, R.; Bocanegra, D.; Bracher, P.J.; Branscomb, E.; Kidd, R.; McGlynn, S.; Meier, D.H.; Nitschke, W.; et al. The drive to life on wet and icy worlds. Astrobiology 2014, 14, 308–343. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Prebiotic systems chemistry: New perspectives for the origins of life. Chem. Rev. 2014, 114, 285–366. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, J.E.; Mowles, A.K.; Mehta, A.K.; Lynn, D.G. Looked at Life from Both Sides Now. Life 2014, 4, 887-902. https://doi.org/10.3390/life4040887
Smith JE, Mowles AK, Mehta AK, Lynn DG. Looked at Life from Both Sides Now. Life. 2014; 4(4):887-902. https://doi.org/10.3390/life4040887
Chicago/Turabian StyleSmith, Jillian E., Allisandra K. Mowles, Anil K. Mehta, and David G. Lynn. 2014. "Looked at Life from Both Sides Now" Life 4, no. 4: 887-902. https://doi.org/10.3390/life4040887
APA StyleSmith, J. E., Mowles, A. K., Mehta, A. K., & Lynn, D. G. (2014). Looked at Life from Both Sides Now. Life, 4(4), 887-902. https://doi.org/10.3390/life4040887