RNA Sociology: Group Behavioral Motifs of RNA Consortia
Abstract
:1. Introduction
2. RNA Structural Agents
2.1. Important Riboagents Act as RNA Stem Loop Groups
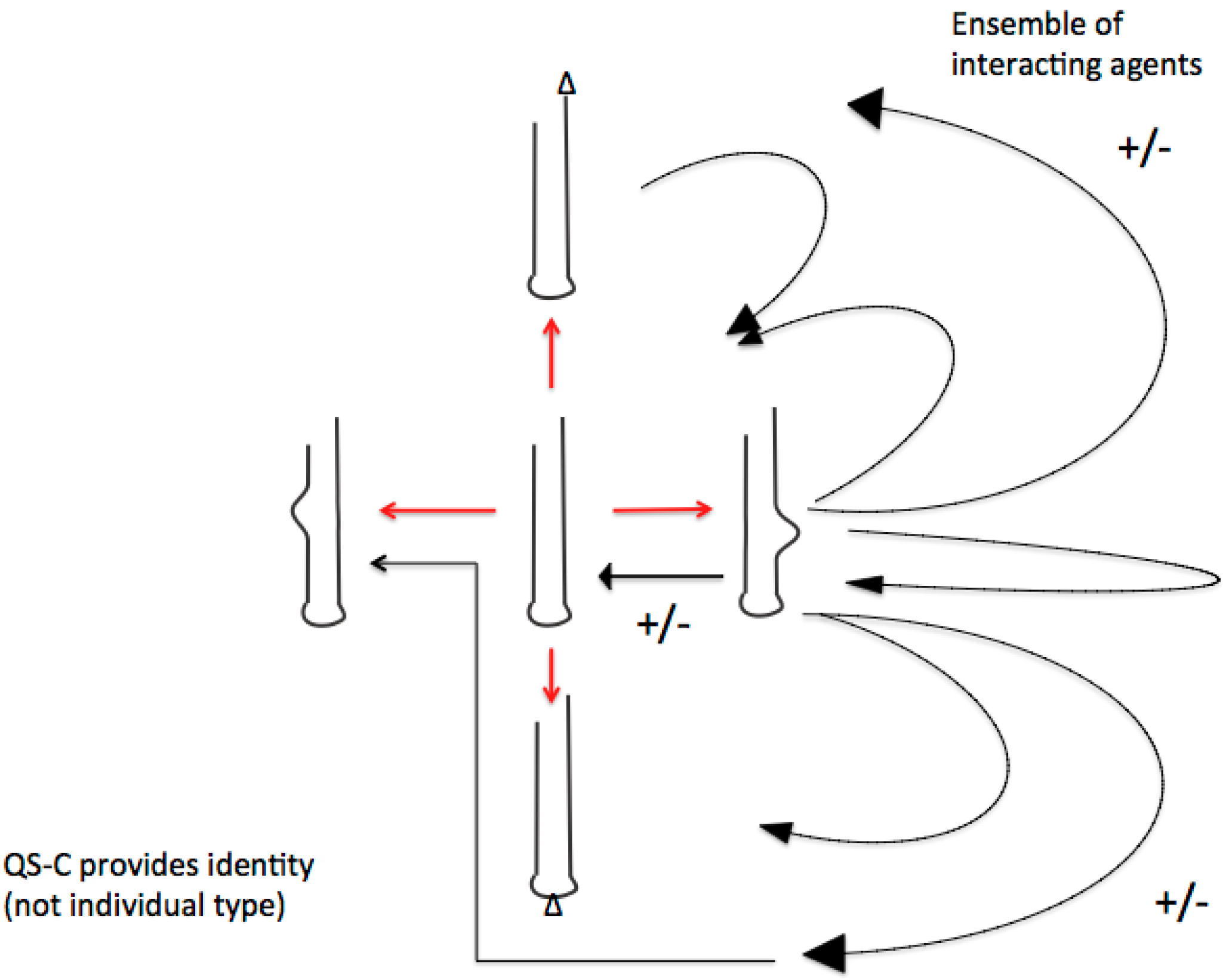
2.2. Addiction Modules: Successful Cooperative Motif for Invading DNA Habitats
2.3. Evolution of New Motifs through Kissing and Pseudoknotting
3. Basic Modules for RNA Group Building
Two of a Perfect Pair: Micro RNAs and siRNAs
4. The Biotic Split: From Molecular Random Assemblies to RNA Group Behavior
4.1. The Editosome Competence
4.2. The Spliceosome Competence
4.3. The Ribosome Competence
4.4. Bypassing Translation Leads to New Amino Acid Identities
4.5. The Secret Life of RNA Stem Loop Groups Which Build tRNAs
5. Inheritable Conservation of New Experiences
6. Endogenization Experts and Persistent Symbiotic Lifestyle
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Witzany, G. Biocommunication and Natural Genome Editing; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Witzany, G. Pragmatic turn in biology: From biological molecules to genetic content operators. World J. Biol. Chem. 2014, 5, 279–285. [Google Scholar] [PubMed]
- Witzany, G. (Ed.) Biocommunication in Soil Microorganisms; Springer: Heidelberg, Germany, 2011.
- Witzany, G.; Baluska, F. (Eds.) Biocommunication of Plants; Springer: Heidelberg, Germany, 2012.
- Witzany, G. (Ed.) Biocommunication of Fungi; Springer: Dordrecht, The Netherlands, 2012.
- Witzany, G. (Ed.) Biocommunication of Animals; Springer: Dordrecht, The Netherlands, 2014.
- Witzany, G.; Baluska, F. Life’s code script does not code itself. The machine metaphor for living organisms is outdated. EMBO Rep. 2012, 13, 1054–1056. [Google Scholar] [PubMed]
- Brookfield, J.F.Y. The ecology of the genome. Mobile DNA elements and their hosts. Nat. Rev. Genet. 2005, 6, 128–136. [Google Scholar] [PubMed]
- Mauricio, R. Can ecology help genomics: The genome as ecosystem? Genetica 2005, 123, 205–209. [Google Scholar] [PubMed]
- Le Rouzic, A.; Dupas, S.; Capy, P. Genome ecosystem and transposable elements species. Gene 2007, 390, 214–220. [Google Scholar] [PubMed]
- Vennera, S.; Feschotte, C.; Biémonta, C. Transposable elements dynamics: Toward a community ecology of the genome. Trends Genet. 2009, 25, 317–323. [Google Scholar]
- Witzany, G. Language and Communication as Universal Requirements for Life. In Astrobiology: An Evolutionary Approach; Kolb, V., Ed.; CRC Press: Oxford, UK, 2014; pp. 349–374. [Google Scholar]
- Shapiro, J.A. How life changes itself: The Read-Write (RW) genome. Phys. Life Rev. 2013, 10, 287–323. [Google Scholar] [PubMed]
- Shapiro, J.A. Constraint and opportunity in genome innovation. RNA Biol. 2014, 11, 186–196. [Google Scholar]
- Villarreal, L.P.; Witzany, G. The DNA Habitat and its RNA Inhabitants: At the Dawn of RNA Sociology. Genomics Insights 2013, 6, 1–12. [Google Scholar]
- Witzany, G. The agents of natural genome editing. J. Mol. Cell Biol. 2011, 3, 181–189. [Google Scholar] [PubMed]
- Przybilski, R.; Hammann, C. The tolerance to exchanges of the Watson-Crick base pair in the hammerhead ribozyme core is determined by surrounding elements. RNA 2007, 13, 1625–1630. [Google Scholar] [PubMed]
- Lambowitz, A.M.; Zimmerly, S. Group II Introns: Mobile Ribozymes that Invade DNA. Cold Spring Harb. Perspect. Biol. 2010, 3, a003616. [Google Scholar] [CrossRef]
- Villarreal, L.P.; Witzany, G. Rethinking quasispecies theory: From fittest type to cooperative consortia. World J. Biol. Chem. 2013, 4, 70–79. [Google Scholar]
- Smit, S.; Yarus, M.; Knight, R. Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA 2006, 12, 1–14. [Google Scholar] [CrossRef]
- Higgs, P.G.; Lehman, N. The RNA world: molecular cooperation at the origins of life. Nat. Rev. Genet. 2014. [Google Scholar] [CrossRef]
- Gwiazda, S.; Salomon, K.; Appel, B.; Müller, S. RNA self-ligation: From oligonucleotides to full length ribozymes. Biochimie 2012, 94, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Appel, B.; Krellenberg, T.; Petkovic, S. The Many Faces of the Hairpin Ribozyme: Structural and Functional Variants of a Small Catalytic RNA. IUBMB Life 2012, 64, 36–47. [Google Scholar] [PubMed]
- Villarreal, L.P. The Addiction Module as a Social Force. In Viruses: Essential Agents of Life; Witzany, G., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 107–145. [Google Scholar]
- Kobayashi, I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001, 29, 3742–3756. [Google Scholar] [CrossRef] [PubMed]
- Mruk, I.; Kobayashi, I. To be or not to be: Regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014, 42, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Viral Ancestors of Antiviral Systems. Viruses 2011, 3, 1933–1958. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Viruses and Host Evolution: Virus Mediated Self Identity. In Self and Nonself; Lopez-Larrea, C., Ed.; Springer: New York, NY, USA, 2012; pp. 185–217. [Google Scholar]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Wagner, E.G. RNA antitoxins. Curr. Opin. Microbiol. 2007, 10, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Origin of Group Identity: Viruses, Addiction and Cooperation; Springer: New York, NY, USA, 2009. [Google Scholar]
- Lincoln, T.A.; Joyce, G.F. Self-sustained replication of an RNA enzyme. Science 2009, 323, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 2011, 19, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, R.; Saito, H.; Ikawa, Y.; Fujita, Y.; Inoue, T. Self-replication reactions dependent on tertiary interaction motifs in an RNA ligase ribozyme. J. Mol. Evol. 2011, 73, 221–229. [Google Scholar] [CrossRef]
- Witzany, G. Noncoding RNAs: Persistent viral agents as modular tools for cellular needs. Ann. N. Y. Acad. Sci. 2009, 1178, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Steitz, J.; Borah, S.; Cazalla, D.; Fok, V.; Lytle, R.; Mitton-Fry, R.; Riley, K.; Samji, T. Noncoding RNPs of Viral Origin. Cold Spring Harb. Perspect. Biol. 2011, 3, a005165. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. The viral origins of telomeres, telomerases and their important role in eukaryogenesis and genome maintenance. Biosemiotics 2008, 2, 191–206. [Google Scholar] [CrossRef]
- Alliegro, M.C. The centrosome and spindle as a ribonucleoprotein complex. Chromosome Res. 2011, 19, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Alliegro, M.A.; Henry, J.J.; Alliegro, M.C. Rediscovery of the nucleolinus, a dynamic RNA-rich organelle associated with the nucleolus, spindle, and centrosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 13718–13723. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.W.; Chetnani, B.; Mondragon, A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip. Rev. RNA 2013. [Google Scholar] [CrossRef]
- Staple, D.W.; Butcher, S.E. Pseudoknots: RNA Structures with Diverse Functions. PLoS Biol. 2005, 3, e213. [Google Scholar] [CrossRef] [PubMed]
- Brierley, I.; Gilbert, R.J.C.; Pennell, S. RNA pseudoknots and the regulation of protein synthesis. Biochem. Soc. Trans. 2008, 36, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Briones, C.; Stich, M.; Manrubia, S.C. The dawn of the RNA World: Toward functional complexity through ligation of random RNA oligomers. RNA 2009, 15, 743–749. [Google Scholar] [CrossRef]
- Peselis, A.; Serganov, A. Structure and function of pseudoknots involved in gene expression control. Wiley Interdiscip. Rev. RNA 2014. [Google Scholar] [CrossRef]
- Thapar, R.; Denmon, A.P.; Nikonowicz, E.P. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2013. [Google Scholar] [CrossRef]
- Dixon, R.J.; Eperon, I.A.; Samani, N.J. Complementary intron sequence motifs associated with human exon repetition: A role for intragenic, inter-transcript interactions in gene expression. Bioinformatics 2007, 23, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Tomizawa, J. Evolution of Functional Structures of RNA. In The RNA World, 2nd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; pp. 419–445. [Google Scholar]
- Forsdyke, D.R. A Stem-Loop“Kissing” Model for the Initiation of Recombination and the Origin of Introns. Mol. Biol. Evol. 1995, 12, 949–959. [Google Scholar] [PubMed]
- Mattick, J.S. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann. N. Y. Acad. Sci. 2009, 1178, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Hannon, G.J. Small RNAs as Guardians of the Genome. Cell 2009, 136, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Stanton, L.W. Long non-coding RNAs in stem cell pluripotency. Wiley Interdiscip. Rev. RNA 2013, 4, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.P.; Dinger, M.E.; Mercer, T.R.; Mattick, J.S. The eukaryotic genome as an RNA machine. Science 2008, 319, 1787–1789. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W. Imprinted expression of small nucleolar RNAs in brain: Time for RNomics. Proc. Natl. Acad. Sci. USA 2000, 97, 14035–14037. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.J. Mammalian Small Nucleolar RNAs Are Mobile Genetic Elements. PLoS Genet. 2006, 2, 1984–1997. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 2008, 136, 215–233. [Google Scholar] [CrossRef]
- Fire, A. Nucleic acid structure and intracellular immunity: Some recent ideas from the world of RNAi. Q. Rev. Biophys. 2005, 38, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer, E.J.; Carthew, R.W. Silence from within: endogenous siRNAs and miRNAs. Cell 2005, 122, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Gordon, K.H.; Buck, A.H.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Piriyapongsa, J.; Jordan, I.K. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 2008, 14, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P. The great leap forward. Major evolutionary jumps might be caused by changes in gene regulation rather than the emergence of new genes. EMBO Rep. 2008, 9, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.C.; Jensen, R.P.; Lehman, N. The Chemical Origin of Behavior is Rooted in Abiogenesis. Life 2012, 2, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Gott, J.M. Expanding genome capacity via RNA editing. Comptes Rendus Biol. 2003, 326, 901–908. [Google Scholar] [CrossRef]
- Takenaka, M.; Verbitskiy, D.; van der Merwe, J.A.; Zehrmann, A.; Brennicke, A. The process of RNA editing in plant mitochondria. Mitochondrion 2008, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.C. Editing Informational Content of Expressed DNA Sequences and Their Transcripts. In RNA Editing; Goeringer, H.U., Ed.; Springer: Berlin, Germany, 2008; pp. 249–265. [Google Scholar]
- Grosjean, H.; Bjork, G.R. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial isoleucyl-tRNA—an alternative way of RNA editing. Trends Biochem. Sci. 2004, 29, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D. Editing of tRNA for Structure and Function. In RNA Editing; Goeringer, H.U., Ed.; Springer: Berlin, Germany, 2008; pp. 33–50. [Google Scholar]
- Homann, M. Editing Reactions from the Perspective of RNA Structure. In RNA Editing; Goeringer, H.U., Ed.; Springer: Berlin, Germany, 2008; pp. 1–32. [Google Scholar]
- Jantsch, M.F.; Oehmann, M. RNA Editing by Adenosine Deaminases that Act on RNA (ADARs). In RNA Editing; Goeringer, H.U., Ed.; Springer: Berlin, Germany, 2008; pp. 51–84. [Google Scholar]
- Hesselberth, J.R. Lives that introns lead after splicing. Wiley Interdiscip. Rev. RNA 2013. [Google Scholar] [CrossRef]
- Carnes, J.; Stuart, K. Working Together: The RNA Editing Machinery in Trypanosoma brucei. In RNA Editing; Goeringer, H.U., Ed.; Springer: Berlin, Germany, 2008; pp. 143–164. [Google Scholar]
- House, A.E.; Lynch, K.W. Regulation of alternative splicing: More than just the ABCs. J. Biol. Chem. 2008, 283, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Matlin, A.J.; Moore, M.J. Spliceosome assembly and composition. Adv. Exp. Med. Biol. 2007, 623, 14–35. [Google Scholar] [PubMed]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Weiner, A. Another Bridge between Kingdoms: tRNA Splicing in Archaea and Eukaryotes. Cell 1997, 89, 1003–1006. [Google Scholar] [PubMed]
- Pyle, A.M.; Lambowitz, A.M. Group II Introns: Ribozymes that Splice RNA and Invade DNA. In The RNA World, 3rd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2006; pp. 469–506. [Google Scholar]
- Forterre, P.; Prangishvili, D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N. Y. Acad. Sci. 2009, 1178, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Turowksi, T.W.; Tollervey, D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip. Rev. RNA 2014. [Google Scholar] [CrossRef]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Belousoff, M.J.; Davidovich, C.; Zimmerman, E.; Caspi, Y.; Wekselman, I.; Rozenszajn, L.; Shapira, T.; Sade-Falk, O.; Taha, L.; Bashan, A.; et al. Ancient machinery embedded in the contemporary ribosome. Biochem. Soc. Trans. 2010, 38, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.B.; Steitz, T.A. The Roles of RNA in the Synthesis of Protein. Cold Spring Harb. Perspect. Biol. 2011, 3, a003780. [Google Scholar] [CrossRef] [PubMed]
- Hamann, C.; Westhof, E. Searching genomes for ribozymes and riboswitches. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Harish, A.; Caetano-Anolles, G. Ribosomal History Reveals Origins of Modern Protein Synthesis. PLoS One 2012, 7, e32776. [Google Scholar] [PubMed]
- Lang, B.F.; Jakubkova, M.; Hegedusova, E.; Daoud, R.; Forget, L.; Brejova, B.; Vinar, T.; Kosa, P.; Fricova, D.; Nebohacova, M.; et al. Massive programmed translational jumping in mitochondria. Proc. Natl. Acad. Sci. USA 2014, 111, 5926–5931. [Google Scholar] [CrossRef] [PubMed]
- Todd, G.C.; Walter, N.G. Secondary structure of bacteriophage T4 gene 60 mRNA: Implications for translational bypassing. RNA 2013, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Daoud, R.; Forget, L.; Lang, B.F. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012, 40, 1728–1736. [Google Scholar] [CrossRef] [PubMed]
- Maizels, A.; Weiner, A.M. The Genomic Tag Hypothesis: Modern Viruses as Molecular Fossils of Ancient Strategies for Genomic Replication. In The RNA World, 2nd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; pp. 577–602. [Google Scholar]
- Maizels, N.; Weiner, A.M.; Yue, D.; Shi, P.Y. New evidence for the genomic tag hypothesis: Archaeal CCA-adding enzymes and tRNA substrates. Biol. Bull. 1999, 196, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Calvin, K.; Hall, M.; Yuan, J.; Podar, M.; Li, H.; Söll, D. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl. Acad. Sci. USA 2005, 102, 17934–17939. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Steitz, T.A. Mechanism of transfer RNA maturation by CCA—adding enzyme without using an oligonucleotide template. Nature 2004, 430, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Koonin, E.V. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol. Direct 2007, 2. [Google Scholar] [CrossRef]
- Sun, F.J.; Caetano-Anollés, G. Evolutionary patterns in the sequence and structure of transfer RNA: Early origins of Archaea and viruses. PLoS Comput. Biol. 2008, 4, e1000018. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M. Have tRNA, will travel. Proc. Natl. Acad. Sci. USA 2005, 102, 11127–11128. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, G.; Wegrzyn, A. Is tRNA only a translation factor or also a regulator of other processes? J. Appl. Genet. 2008, 49, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Söll, D. Transfer RNA genes in pieces. EMBO Rep. 2008, 9, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Dalla-Favera, R.; Basso, K. RNAs with multiple personalities. Wiley Interdiscip. Rev. RNA 2013. [Google Scholar] [CrossRef]
- Huda, A.; Mariño-Ramírez, L.; Jordan, I.K. Epigenetic histone modifications of human transposable elements: Genome defense versus exaptation. Mob. DNA 2010, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The central role of RNA in the genetic programming of complex organisms. Anais da Academia Brasileira de Ciências 2010, 82, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Taft, R.J.; Faulkner, G.J. A global view of genomic information—moving beyond the gene and the master regulator. Trends Genet. 2010, 26, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 2013, 23, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Mattick, J. The role of regulatory RNA in cognitive evolution. Trends Cogn. Sci. 2012, 16, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Blaze, J.; Roth, T.L. Epigenetic mechanisms in learning and memory. Wiley Interdiscip. Rev. Cogn. Sci. 2012. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.S.; Beitzel, B.F.; Schroeder, A.R.W.; Shinn, P.; Chen, H.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Retroviral DNA Integration: ASLV, HIV, and MLV Show Distinct Target Site Preferences. PLoS Biol. 2004, 2, e234. [Google Scholar] [CrossRef] [PubMed]
- Diemer, G.S.; Stedman, K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct 2012, 7. [Google Scholar] [CrossRef]
- Villarreal, L.P. Viruses and the Evolution of Life; American Society for Microbiology Press: Washington, DC, USA, 2005. [Google Scholar]
- Ryan, F. Virolution; Harper Collins: London, UK, 2009. [Google Scholar]
- Witzany, G. (Ed.) Viruses: Essential Agents of Life; Springer: Dordrecht, The Netherlands, 2012.
- Villarreal, L.P. The Source of Self. Genetic Parasites and the Origin of Adaptive Immunity. Ann. N. Y. Acad. Sci. 2009, 1178, 194–232. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. From Molecular Entities to Competent Agents: Viral Infection-Derived Consortia Act as Natural Genetic Engineers. In Viruses: Essential Agents of Life; Witzany, G., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 407–419. [Google Scholar]
- Geuking, M.B.; Weber, J.; Dewannieux, M.; Gorelik, E.; Heidmann, T.; Hengartner, H.; Zinkernagel, R.M.; Hangartner, L. Recombination of Retrotransposon and Exogenous RNA Virus Results in Nonretroviral cDNA Integration. Science 2009, 323, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Havecker, E.R.; Baranov, P.V.; Atkins, J.F.; Voytas, D.F. Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA 2003, 9, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.B.; Nagy, P.D. Defective Interfering RNAs: Foes of Viruses and Friends of Virologists. Viruses 2009, 1, 895–919. [Google Scholar] [CrossRef] [PubMed]
- Klenerman, P.; Hengartner, H.; Zinkernagel, R.M. A non-retroviral RNA virus persists in DNA form. Nature 1997, 390, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.; Belfort, M. Social networking between mobile introns and their host genes. Mol. Microbiol. 2010, 78, 1–4. [Google Scholar] [PubMed]
- Villarreal, L.P.; Witzany, G. Viruses are essential agents within the roots and stem of the tree of life. J. Theor. Biol. 2010, 262, 698–710. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witzany, G. RNA Sociology: Group Behavioral Motifs of RNA Consortia. Life 2014, 4, 800-818. https://doi.org/10.3390/life4040800
Witzany G. RNA Sociology: Group Behavioral Motifs of RNA Consortia. Life. 2014; 4(4):800-818. https://doi.org/10.3390/life4040800
Chicago/Turabian StyleWitzany, Guenther. 2014. "RNA Sociology: Group Behavioral Motifs of RNA Consortia" Life 4, no. 4: 800-818. https://doi.org/10.3390/life4040800
APA StyleWitzany, G. (2014). RNA Sociology: Group Behavioral Motifs of RNA Consortia. Life, 4(4), 800-818. https://doi.org/10.3390/life4040800



