Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms
Abstract
:1. Introduction
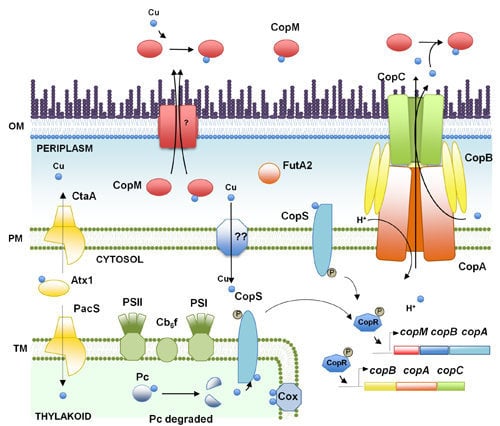
2. Copper, an Essential Element for Photosynthesis and Respiration

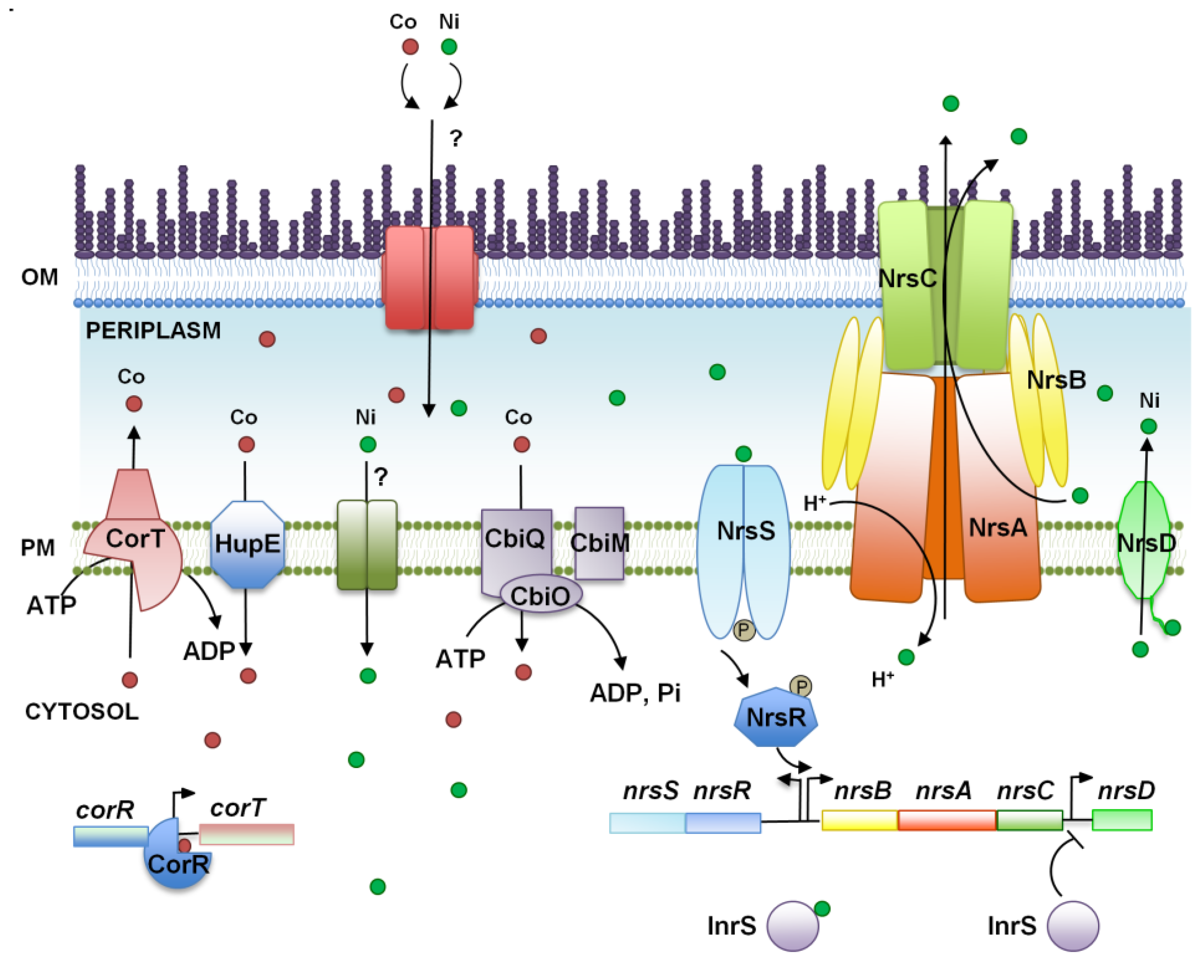
3. Nickel and Cobalt, Two Essential Metals for Cyanobacteria?
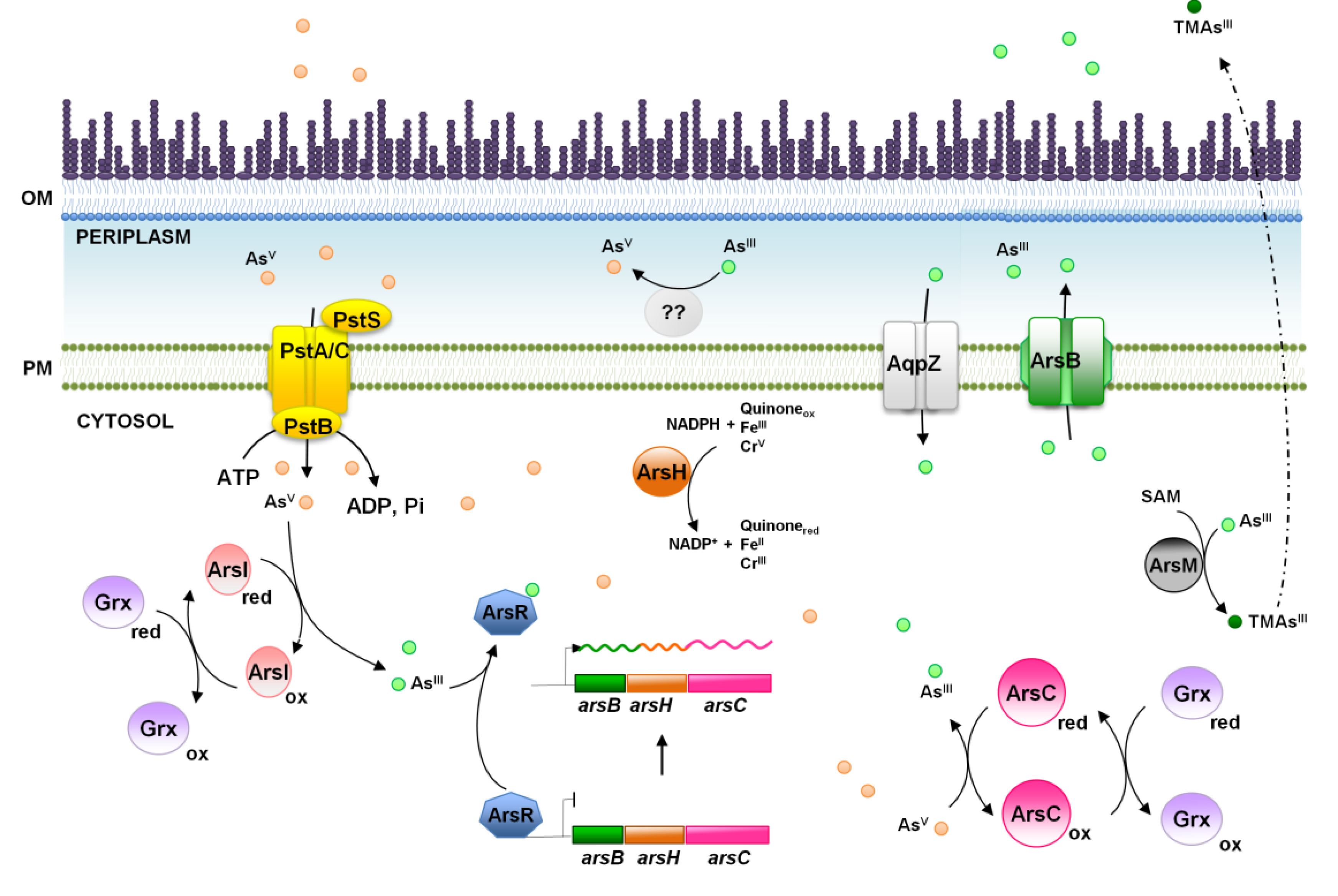
4. Arsenic, a World Wide Spread Phosphate Competitor
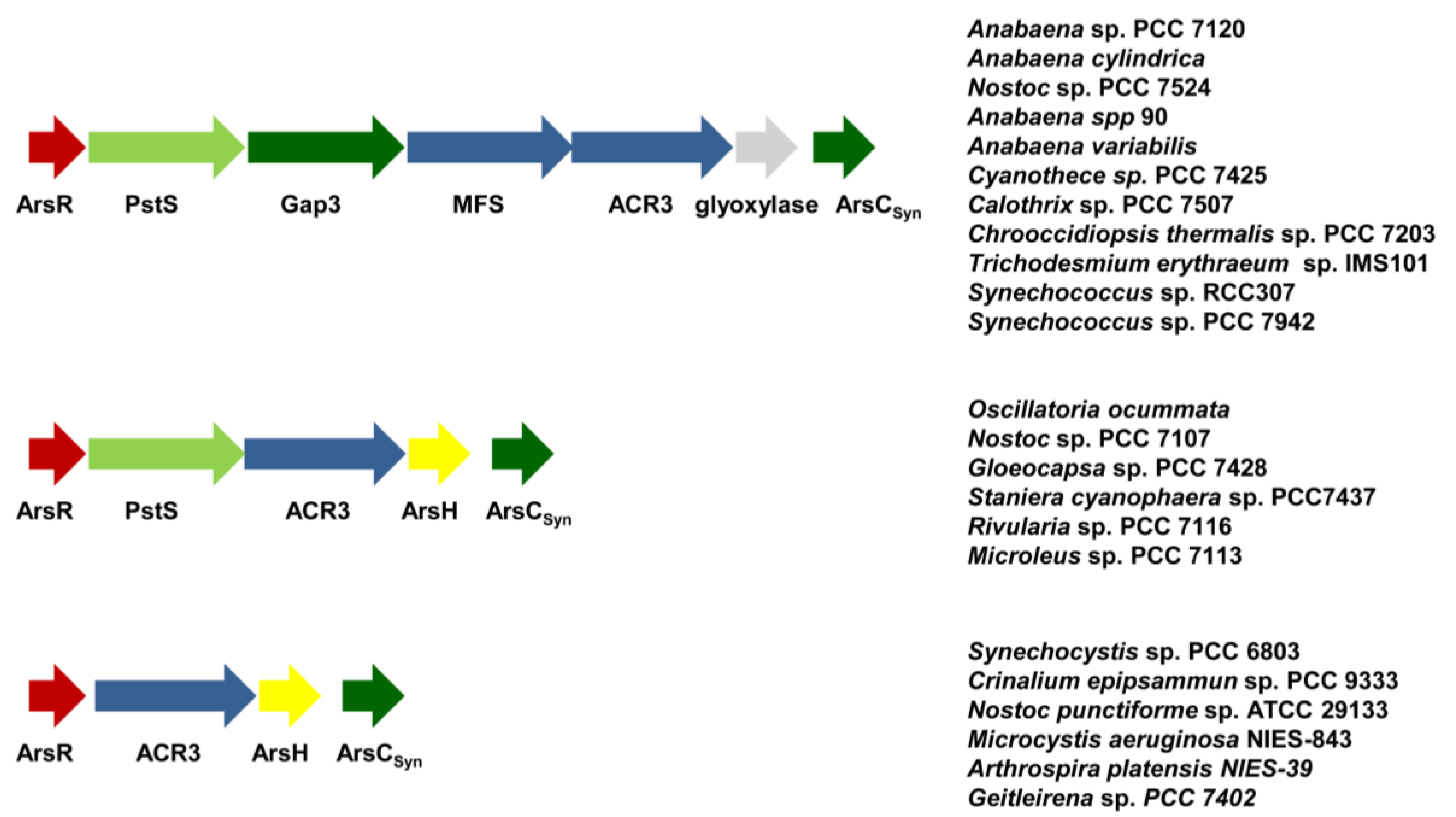
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Waldron, K.J.; Robinson, N.J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009, 7, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Menon, A.L.; Thorgersen, M.P.; Scott, J.W.; Poole, F.L., II; Jenney, F.E., Jr.; Lancaster, W.A.; Praissman, J.L.; Shanmukh, S.; Vaccaro, B.J.; et al. Microbial metalloproteomes are largely uncharacterized. Nature 2010, 466, 779–782. [Google Scholar] [CrossRef]
- Waldron, K.J.; Firbank, S.J.; Dainty, S.J.; Perez-Rama, M.; Tottey, S.; Robinson, N.J. Structure and metal loading of a soluble periplasm cuproprotein. J. Biol. Chem. 2010, 285, 32504–32511. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Guerra-García, J.M.; García-Gómez, J.C. Assessing pollution levels in sediments of a harbour with two opposing entrances. Environmental implications. J. Environ. Manag. 2005, 77, 1–11. [Google Scholar] [CrossRef]
- Peng, J.-F.; Song, Y.-H.; Yuan, P.; Cui, X.-Y.; Qiu, G.-L. The remediation of heavy metals contaminated sediment. J. Hazard. Mater. 2009, 161, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Shcolnick, S.; Keren, N. Metal Homeostasis in Cyanobacteria and Chloroplasts. Balancing Benefits and Risks to the Photosynthetic Apparatus. Plant Physiol. 2006, 141, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; López-Maury, L.; Florencio, F.J. Proteomic pattern alterations of the cyanobacterium Synechocystis sp. PCC 6803 in response to cadmium, nickel and cobalt. J. Proteomics 2014, 102, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Micheletti, E.; Zille, A.; Santos, A.; Moradas-Ferreira, P.; Tamagnini, P.; de Philippis, R. Using extracellular polymeric substances (EPS)-producing cyanobacteria for the bioremediation of heavy metals: Do cations compete for the EPS functional groups and also accumulate inside the cell? Microbiology 2011, 157, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Burnat, M.; Diestra, E.; Esteve, I.; Solé, A. In Situ Determination of the Effects of Lead and Copper on Cyanobacterial Populations in Microcosms. PLoS One 2009, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merchant, S.S.; Helmann, J.D.; Robert, K.P. Chapter 2—Elemental Economy: Microbial Strategies for Optimizing Growth in the Face of Nutrient Limitation. In Advances in Microbial Physiology; Academic Press: Waltham, MA, USA, 2012; Volume 60, pp. 91–210. [Google Scholar]
- Hillier, W.; Babcock, G.T. Photosynthetic Reaction Centers. Plant Physiol. 2001, 125, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krausz, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.P.; Millard, A.; Ksibe, A.; Scanlan, D.J.; Schmid, R.; Blindauer, C.A. Mining genomes of cyanobacteria for elements of zinc homeostasis. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Dupont, C.L.; Johnson, D.A.; Phillippy, K.; Paulsen, I.T.; Brahamsha, B.; Palenik, B. Genetic Identification of a High-Affinity Ni Transporter and the Transcriptional Response to Ni Deprivation in Synechococcus sp. Strain WH8102. Appl. Environ. Microbiol. 2012, 78, 7822–7832. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.L.; Ahlgren, N.; Moffett, J.W.; Chisholm, S.W. Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 2002, 47, 976–988. [Google Scholar] [CrossRef]
- Twining, B.S.; Baines, S.B. The trace metal composition of marine phytoplankton. Ann. Rev. Mar. Sci. 2013, 5, 191–215. [Google Scholar] [CrossRef] [PubMed]
- Trick, C.; Kerry, A. Isolation and purification of siderophores produced by cyanobacteria, Synechococcus sp. PCC 7942 and Anabaena variabilis ATCC 29413. Curr. Microbiol. 1992, 24, 241–245. [Google Scholar] [CrossRef]
- Itou, Y.; Okada, S.; Murakami, M. Two structural isomeric siderophores from the freshwater cyanobacterium Anabaena cylindrica (NIES-19). Tetrahedron 2001, 57, 9093–9099. [Google Scholar] [CrossRef]
- De Philippis, R.; Sili, C.; Paperi, R.; Vincenzini, M. Exopolysaccharide-producing cyanobacteria and their possible exploitation: A review. J. Appl. Phycol. 2001, 13, 293–299. [Google Scholar] [CrossRef]
- Jittawuttipoka, T.; Planchon, M.; Spalla, O.; Benzerara, K.; Guyot, F.; Cassier-Chauvat, C.; Chauvat, F. Multidisciplinary evidences that Synechocystis PCC 6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PLoS One 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, E.; Pereira, S.; Mannelli, F.; Moradas-Ferreira, P.; Tamagnini, P.; de Philippis, R. Sheathless mutant of Cyanobacterium Gloeothece sp. strain PCC 6909 with increased capacity to remove copper ions from aqueous solutions. Appl. Environ. Microbiol. 2008, 74, 2797–2804. [Google Scholar] [CrossRef] [PubMed]
- Huckle, J.W.; Morby, A.P.; Turner, J.S.; Robinson, N.J. Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol. Microbiol. 1993, 7, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Whitehall, S.K.; Cavet, J.S. Microbial metallothioneins. Adv. Microb. Physiol. 2001, 44, 183–213. [Google Scholar] [PubMed]
- Turner, J.S.; Morby, A.P.; Whitton, B.A.; Gupta, A.; Robinson, N.J. Construction of Zn2+/Cd2+ hypersensitive cyanobacterial mutants lacking a functional metallothionein locus. J. Biol. Chem. 1993, 268, 4494–4498. [Google Scholar] [PubMed]
- Liu, T.; Nakashima, S.; Hirose, K.; Shibasaka, M.; Katsuhara, M.; Ezaki, B.; Giedroc, D.P.; Kasamo, K. A novel cyanobacterial SmtB/ArsR family repressor regulates the expression of a CPx-ATPase and a metallothionein in response to both Cu(I)/Ag(I) and Zn(II)/Cd(II). J. Biol. Chem. 2004, 279, 17810–17818. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Nakashima, S.; Hirose, K.; Uemura, Y.; Shibasaka, M.; Katsuhara, M.; Kasamo, K. A metallothionein and CPx-ATPase handle heavy-metal tolerance in the filamentous cyanobacterium Oscillatoria brevis. FEBS Lett. 2003, 542, 159–163. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, M.; Lopez-Maury, L.; Florencio, F.J.; Reyes, J.C. A Gene Cluster Involved in Metal Homeostasis in the Cyanobacterium Synechocystis sp. Strain PCC 6803. J. Bacteriol. 2000, 182, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; López-Maury, L.; Reyes, J.C.; Florencio, F.J. The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012, 159, 1806–1818. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; Florencio, F.J.; Reyes, J.C. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003, 185, 5363–5371. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, J.C.; Cavet, J.S.; Robinson, N.J. Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J. Biol. Chem. 1999, 274, 25827–25832. [Google Scholar] [CrossRef] [PubMed]
- Thelwell, C.; Robinson, N.J.; Turner-Cavet, J.S. An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. USA 1998, 95, 10728–10733. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Golden, J.W.; Giedroc, D.P. A Zinc(II)/Lead(II)/Cadmium(II)-Inducible Operon from the Cyanobacterium Anabaena Is Regulated by AztR, an α3N ArsR/SmtB Metalloregulator. Biochemistry 2005, 44, 8673–8683. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.K.; Dupont, C.L.; Johnson, D.A.; Paulsen, I.T.; Palenik, B. Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response relative to those of open-ocean strains. Appl. Environ. Microbiol. 2009, 75, 5047–5057. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; You, L.; Liu, D.; Hollinshead, W.; Tang, Y.; Zhang, F. Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory. Mar. Drugs 2013, 11, 2894–2916. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A. Zinc-Handling in Cyanobacteria: An Update. Chem. Biodivers. 2008, 5, 1990–2013. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Cavet, J.S. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008, 65, 217–247. [Google Scholar] [PubMed]
- Rademacher, C.; Masepohl, B. Copper-responsive gene regulation in bacteria. Microbiology 2012, 158, 2451–2464. [Google Scholar] [CrossRef] [PubMed]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Chillappagari, S.; Seubert, A.; Trip, H.; Kuipers, O.P.; Marahiel, M.A.; Miethke, M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 2010, 192, 2512–2524. [Google Scholar] [CrossRef] [PubMed]
- Tottey, S.; Patterson, C.J.; Banci, L.; Bertini, I.; Felli, I.C.; Pavelkova, A.; Dainty, S.J.; Pernil, R.; Waldron, K.J.; Foster, A.W.; et al. Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc. Natl. Acad. Sci. USA 2012, 109, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; López-Maury, L.; Florencio, F.J. Global Transcriptional Profiles of the Copper Responses in the Cyanobacterium Synechocystis sp. PCC 6803. PLoS One 2014, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macomber, L.; Rensing, C.; Imlay, J.A. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 2007, 189, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Tottey, S.; Rich, P.R.; Rondet, S.A.; Robinson, N.J. Two Menkes-type atpases supply copper for photosynthesis in Synechocystis. PCC 6803. J. Biol. Chem. 2001, 276, 19999–20004. [Google Scholar] [CrossRef] [PubMed]
- Tottey, S.; Rondet, S.A.; Borrelly, G.P.; Robinson, P.J.; Rich, P.R.; Robinson, N.J. A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer, and alternative sites for copper acquisition. J. Biol. Chem. 2002, 277, 5490–5497. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, K.; Kashiwagi, S.; Mizuno, T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Mol. Microbiol. 1994, 13, 369–377. [Google Scholar] [CrossRef] [PubMed]
- González-Guerrero, M.; Raimunda, D.; Cheng, X.; Argüello, J.M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Raimunda, D.; González-Guerrero, M.; Leeber, B., III; Argüello, J. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. BioMetals 2011, 24, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Gugel, I.L.; Hilger, D.; Rengstl, B.; Jung, H.; Nickelsen, J. Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis. Plant Cell 2012, 24, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Schottkowski, M.; Gkalympoudis, S.; Tzekova, N.; Stelljes, C.; Schunemann, D.; Ankele, E.; Nickelsen, J. Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 2009, 284, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Rengstl, B.; Oster, U.; Stengel, A.; Nickelsen, J. An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for Photosystem II biogenesis. J. Biol. Chem. 2011, 286, 21944–21951. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; Tottey, S.; Yanagisawa, S.; Dennison, C.; Robinson, N.J. A periplasmic iron-binding protein contributes toward inward copper supply. J. Biol. Chem. 2007, 282, 3837–3846. [Google Scholar] [CrossRef] [PubMed]
- Lutkenhaus, J. Role of a major outer membrane protein in Escherichia coli. J. Bacteriol. 1977, 131, 631–637. [Google Scholar] [PubMed]
- Speer, A.; Rowland, J.L.; Haeili, M.; Niederweis, M.; Wolschendorf, F. Porins increase copper susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 2013, 195, 5133–5140. [Google Scholar] [CrossRef] [PubMed]
- Nicolaisen, K.; Hahn, A.; Valdebenito, M.; Moslavac, S.; Samborski, A.; Maldener, I.; Wilken, C.; Valladares, A.; Flores, E.; Hantke, K.; et al. The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta 2010, 1798, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Moffett, J.W.; Brand, L.E. Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol. Oceanogr. 1996, 41, 388–395. [Google Scholar] [CrossRef]
- Stevanovic, M.; Lehmann, C.; Schleiff, E. The response of the TonB-dependent transport network in Anabaena sp. PCC 7120 to cell density and metal availability. BioMetals 2013, 26, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.V.; Hervas, M.; de la Rosa, M.A.; Navarro, J.A. The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J. Biol. Chem. 2004, 279, 7229–7233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; McSpadden, B.; Pakrasi, H.B.; Whitmarsh, J. Copper-mediated regulation of cytochrome c553 and plastocyanin in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 1992, 267, 19054–19059. [Google Scholar] [PubMed]
- Briggs, L.M.; Pecoraro, V.L.; McIntosh, L. Copper-induced expression, cloning, and regulatory studies of the plastocyanin gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 1990, 15, 633–642. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, M.A.; Navarro, J.A.; Dı́az-Quintana, A.; de la Cerda, B.; Molina-Heredia, F.P.; Balme, A.; Murdoch, P.D.S.; Dı́az-Moreno, I.; Durán, R.V.; Hervás, M.; et al. An evolutionary analysis of the reaction mechanisms of photosystem I reduction by cytochrome c6 and plastocyanin. Bioelectrochemistry 2002, 55, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; López-Maury, L.; Florencio, F.J. CopM is a novel copper binding protein involved in copper resistance in Synechocystis sp. PCC 6803. MicrobiologyOpen 2014, in press. [Google Scholar]
- López-Maury, L.; Giner-Lamia, J.; Florencio, F.J. Redox control of copper homeostasis in cyanobacteria. Plant Signal. Behav. 2012, 7, 1712–1714. [Google Scholar] [CrossRef] [PubMed]
- Osanai, T.; Imamura, S.; Asayama, M.; Shirai, M.; Suzuki, I.; Murata, N.; Tanaka, K. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res. 2006, 13, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.K.; Brahamsha, B.; Busby, K.; Palenik, B. Genomic island genes in a coastal marine Synechococcus strain confer enhanced tolerance to copper and oxidative stress. ISME J. 2012, 7, 1139–1149. [Google Scholar] [CrossRef]
- Rodriguez, I.B.; Ho, T.-Y. Diel nitrogen fixation pattern of Trichodesmium: The interactive control of light and Ni. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Ragsdale, S.W. Nickel-based Enzyme Systems. J. Biol. Chem. 2009, 284, 18571–18575. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Shimizu, S. Cobalt proteins. Eur. J. Biochem. 1999, 261, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Collier, J.L.; Brahamsha, B.; Palenik, B. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amiohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: Molecular-Genetic and biochemical analysis of the enzyme. Microbiology 1999, 145, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Quintero, M.J.; Muro-Pastor, A.M.; Herrero, A.; Flores, E. Arginine catabolism in the cyanobacterium Synechocystis sp. Strain PCC 6803 involves the urea cycle and arginase pathway. J. Bacteriol. 2000, 182, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Mitamura, O.; Kawashima, M.; Maeda, H. Urea degradation by picophytoplankton in the euphotic zone of Lake Biwa. Limnology 2000, 1, 19–26. [Google Scholar] [CrossRef]
- Saito, M.A.; McIlvin, M.R.; Moran, D.M.; Goepfert, T.J.; DiTullio, G.R.; Post, A.F.; Lamborg, C.H. Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science 2014, 345, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Valladares, A.; Montesinos, M.L.; Herrero, A.; Flores, E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 2002, 43, 703–715. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, C.L.; Germer, F.; Schulz, R.; Appel, J.; Jones, A.K. The [NiFe]-Hydrogenase of the Cyanobacterium Synechocystis sp. PCC 6803 Works Bidirectionally with a Bias to H2 Production. J. Am. Chem. Soc. 2011, 133, 11308–11319. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, D.; Wawrousek, K.; Eckert, C.; Yu, J.; Maness, P.-C. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 2011, 102, 8368–8377. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Boehm, M.; Carrieri, D.; Yu, J.; Dubini, A.; Nixon, P.J.; Maness, P.-C. Genetic Analysis of the Hox Hydrogenase in the Cyanobacterium Synechocystis sp. PCC 6803 Reveals Subunit Roles in Association, Assembly, Maturation, and Function. J. Biol. Chem. 2012, 287, 43502–43515. [Google Scholar] [CrossRef] [PubMed]
- Gutekunst, K.; Chen, X.; Schreiber, K.; Kaspar, U.; Makam, S.; Appel, J. The bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J. Biol. Chem. 2014, 289, 1930–1937. [Google Scholar] [CrossRef] [PubMed]
- Leitão, E.; Oxelfelt, F.; Oliveira, P.; Moradas-Ferreira, P.; Tamagnini, P. Analysis of the hupSL Operon of the Nonheterocystous Cyanobacterium Lyngbya majuscula CCAP 1446/4: Regulation of Transcription and Expression under a Light-Dark Regimen. Appl. Environ. Microbiol. 2005, 71, 4567–4576. [Google Scholar] [CrossRef] [PubMed]
- Wunschiers, R.; Batur, M.; Lindblad, P. Presence and expression of hydrogenase specific C-terminal endopeptidases in cyanobacteria. BMC Microbiol. 2003, 3. [Google Scholar] [CrossRef]
- Zhang, X.; Sherman, D.M.; Sherman, L.A. The Uptake Hydrogenase in the Unicellular Diazotrophic Cyanobacterium Cyanothece sp. Strain PCC 7822 Protects Nitrogenase from Oxygen Toxicity. J. Bacteriol. 2014, 196, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Khetkorn, W.; Lindblad, P.; Incharoensakdi, A. Inactivation of uptake hydrogenase leads to enhanced and sustained hydrogen production with high nitrogenase activity under high light exposure in the cyanobacterium Anabaena siamensis TISTR 8012. J. Biol. Eng. 2012, 6. [Google Scholar] [CrossRef]
- Happe, T.; Schütz, K.; Böhme, H. Transcriptional and Mutational Analysis of the Uptake Hydrogenase of the Filamentous Cyanobacterium Anabaena Variabilis ATCC 29413. J. Bacteriol. 2000, 182, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Neupane, K.; Shearer, J.; Palenik, B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008, 10, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Suttisansanee, U.; Lau, K.; Lagishetty, S.; Rao, K.N.; Swaminathan, S.; Sauder, J.M.; Burley, S.K.; Honek, J.F. Structural Variation in Bacterial Glyoxalase I Enzymes: Investigation of the metalloenzyme glyoxalase I from clostridium acetobutylicum. J. Biol. Chem. 2011, 286, 38367–38374. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Vishnoi, A.; Ariyadasa, T.U.; Bhattacharya, A.; Singla-Pareek, S.L.; Sopory, S.K. Episodes of horizontal gene-transfer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, Y.; Yabuta, Y.; Yamaji, R.; Shigeoka, S.; Nakano, Y.; Watanabe, F.; Inui, H. Occurrence of Pseudovitamin B12 and Its Possible Function as the Cofactor of Cobalamin-Dependent Methionine Synthase in a Cyanobacterium Synechocystis sp. PCC6803. J. Nutr. Sci. Vitaminol. 2009, 55, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Buck, K.N.; Palenik, B.; Barbeau, K. Nickel utilization in phytoplankton assemblages from contrasting oceanic regimes. Deep Sea Res. Part I Oceanogr. Res. Pap. 2010, 57, 553–566. [Google Scholar] [CrossRef]
- Zhang, Y.; Rodionov, D.; Gelfand, M.; Gladyshev, V. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Vitreschak, A.G.; Mironov, A.A.; Gelfand, M.S. Comparative Genomics of the Vitamin B12 Metabolism and Regulation in Prokaryotes. J. Biol. Chem. 2003, 278, 41148–41159. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, D.; Gutekunst, K.; Klissenbauer, M.; Schulz-Friedrich, R.; Appel, J. Mutagenesis of hydrogenase accessory genes of Synechocystis sp. PCC 6803. FEBS J. 2006, 273, 4516–4527. [Google Scholar] [CrossRef] [PubMed]
- Eitinger, T.; Rodionov, D.A.; Grote, M.; Schneider, E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: Diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 2011, 35, 3–67. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Hebbeln, P.; Gelfand, M.S.; Eitinger, T. Comparative and Functional Genomic Analysis of Prokaryotic Nickel and Cobalt Uptake Transporters: Evidence for a Novel Group of ATP-Binding Cassette Transporters. J. Bacteriol. 2006, 188, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, M.; Rubio, M.A.; Santamaria-Gomez, J.; Olmedo-Verd, E.; Robinson, N.J.; Luque, I. Characterization of the response to zinc deficiency in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2012, 194, 2426–2436. [Google Scholar] [CrossRef] [PubMed]
- Mirus, O.; Strauss, S.; Nicolaisen, K.; von Haeseler, A.; Schleiff, E. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol. 2009, 7. [Google Scholar] [CrossRef] [PubMed]
- An, Y.J.; Ahn, B.E.; Han, A.R.; Kim, H.M.; Chung, K.M.; Shin, J.H.; Cho, Y.B.; Roe, J.H.; Cha, S.S. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009, 37, 3442–3451. [Google Scholar] [CrossRef] [PubMed]
- Iwig, J.S.; Chivers, P.T. Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors. Nat. Prod. Rep. 2010, 27, 658–667. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; García-Domínguez, M.; Florencio, F.J.; Reyes, J.C. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 2002, 43, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Patterson, C.J.; Pernil, R.; Dainty, S.J.; Chakrabarti, B.; Henry, C.E.; Money, V.A.; Foster, A.W.; Robinson, N.J. Co(ll)-detection does not follow Kco(ll) gradient: Channelling in Co(ll)-sensing. Metallomics 2013, 5, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.W.; Patterson, C.J.; Pernil, R.; Hess, C.R.; Robinson, N.J. Cytosolic Ni(II) Sensor in Cyanobacterium: Nickel Detection Follows Nickel Affinity Across Four Families of Metal Sensors. J. Biol. Chem. 2012, 287, 12142–12151. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ramesh, A.; Ma, Z.; Ward, S.K.; Zhang, L.; George, G.N.; Talaat, A.M.; Sacchettini, J.C.; Giedroc, D.P. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 2007, 3, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.W.; Pernil, R.; Patterson, C.J.; Robinson, N.J. Metal-specificity of cyanobacterial nickel-responsive repressor InrS: Cells maintain zinc and copper below the detection-threshold for InrS. Mol. Microbiol. 2014, 92, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, D.K. Public health. Worldwide occurrences of arsenic in ground water. Science 2002, 296, 2143–2145. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, D.S.; Viola, R.E. Arsenate replacing phosphate: Alternative life chemistries and ion promiscuity. Biochemistry 2011, 50, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Fekry, M.I.; Tipton, P.A.; Gates, K.S. Kinetic Consequences of Replacing the Internucleotide Phosphorus Atoms in DNA with Arsenic. ACS Chem. Biol. 2011, 6, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Kamerlin, S.C.; Sharma, P.K.; Prasad, R.B.; Warshel, A. Why nature really chose phosphate. Q. Rev. Biophys. 2013, 46, 1–132. [Google Scholar] [CrossRef] [PubMed]
- Westheimer, F.H. Why Nature Chose Phosphates. Science 1987, 235, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shen, J.; Carbrey, J.M.; Mukhopadhyay, R.; Agre, P.; Rosen, B.P. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc. Natl. Acad. Sci. USA 2002, 99, 6053–6058. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.L.; Liu, Z.; Rosen, B.P. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J. Biol. Chem. 2004, 279, 18334–18341. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, R.; Chery, C.C.; Wawrzycka, D.; van Hulle, M.; Cornelis, R.; Thevelein, J.M.; Tamas, M.J. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol. Microbiol. 2001, 40, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Bernstam, L.; Nriagu, J. Molecular aspects of arsenic stress. J. Toxicol. Environ. Health B Crit. Rev. 2000, 3, 293–322. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Kulp, T.R.; Blum, J.S.; Hoeft, S.E.; Baesman, S.; Miller, L.G.; Stolz, J.F. A Microbial Arsenic Cycle in a Salt-Saturated, Extreme Environment. Science 2005, 308, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Kulp, T.R.; Hoeft, S.E.; Asao, M.; Madigan, M.T.; Hollibaugh, J.T.; Fisher, J.C.; Stolz, J.F.; Culbertson, C.W.; Miller, L.G.; Oremland, R.S.; et al. Arsenic(III) Fuels Anoxygenic Photosynthesis in Hot Spring Biofilms from Mono Lake, California. Science 2008, 321, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Van Lis, R.; Nitschke, W.; Duval, S.; Schoepp-Cothenet, B. Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 2013, 1827, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Rensing, C.; Rosen, B.P.; Zhu, Y.G. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 2012, 17, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.J.; Li, J.; Waters, S.B.; Xing, W.; Adair, B.M.; Drobna, Z.; Devesa, V.; Styblo, M. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp. Biol. Med. 2007, 232, 3–13. [Google Scholar]
- Sánchez-Riego, A.M.; López-Maury, L.; Florencio, F.J. Genomic Responses to Arsenic in the Cyanobacterium Synechocystis sp. PCC 6803. PLoS One 2014, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hervas, M.; Lopez-Maury, L.; Leon, P.; Sanchez-Riego, A.M.; Florencio, F.J.; Navarro, J.A. ArsH from the cyanobacterium Synechocystis sp. PCC 6803 is an efficient NADPH-dependent quinone reductase. Biochemistry 2012, 51, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.M.; Yan, Y.; Xu, H.J.; Wang, N.; Zhang, X.; Ye, J. ArsH from Synechocystis sp. PCC 6803 reduces chromate and ferric iron. FEMS Microbiol. Lett. 2014, 356, 105–112. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; Sanchez-Riego, A.M.; Reyes, J.C.; Florencio, F.J. The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2009, 191, 3534–3543. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.X.; Chen, J.; Qin, J.; Sun, G.X.; Rosen, B.P.; Zhu, Y.G. Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 2011, 156, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Rai, R.; Rai, L.C. Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PCC7120 under arsenic stress. J. Proteomics 2012, 75, 921–937. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, P.; Pal, R. Response of cyanobacteria to arsenic toxicity. J. Appl. Phycol. 2011, 23, 293–299. [Google Scholar] [CrossRef]
- Li, R.; Haile, J.D.; Kennelly, P.J. An arsenate reductase from Synechocystis sp. strain PCC 6803 exhibits a novel combination of catalytic characteristics. J. Bacteriol. 2003, 185, 6780–6789. [Google Scholar] [CrossRef] [PubMed]
- Ordoñez, E.; van Belle, K.; Roos, G.; de Galan, S.; Letek, M.; Gil, J.A.; Wyns, L.; Mateos, L.M.; Messens, J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 2009, 284, 15107–15116. [Google Scholar] [CrossRef] [PubMed]
- Messens, J.; Silver, S. Arsenate reduction: Thiol cascade chemistry with convergent evolution. J. Mol. Biol. 2006, 362, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, D.J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W.R.; Post, A.F.; Hagemann, M.; Paulsen, I.; Partensky, F.; et al. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 249–299. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Florencio, F.J. The cyanobacterial thioredoxin gene is required for both photoautotrophic and heterotrophic growth. Plant Physiol. 1996, 111, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.G.; Buchanan, B.B. Thioredoxin is essential for photosynthetic growth. The thioredoxin m gene of Anacystis nidulans. J. Biol. Chem. 1989, 264, 4008–4014. [Google Scholar] [PubMed]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Bobrowicz, P.; Wysocki, R.; Owsianik, G.; Goffeau, A.; Ułaszewski, S. Isolation of Three Contiguous Genes, ACR1, ACR2 and ACR3, Involved in Resistance to Arsenic Compounds in the Yeast Saccharomyces cerevisiae. Yeast 1997, 13, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Indriolo, E.; Na, G.; Ellis, D.; Salt, D.E.; Banks, J.A. A Vacuolar Arsenite Transporter Necessary for Arsenic Tolerance in the Arsenic Hyperaccumulating Fern Pteris vittata is Missing in Flowering Plants. Plant Cell Online 2010, 22, 2045–2057. [Google Scholar] [CrossRef]
- Wurl, O.; Zimmer, L.; Cutter, G.A. Arsenic and phosphorus biogeochemistry in the ocean: Arsenic species as proxies for P-limitation. Limnol. Oceanogr. 2013, 58, 729–740. [Google Scholar] [CrossRef]
- Zhang, S.; Rensing, C.; Zhu, Y.-G. Cyanobacteria-Mediated Arsenic Redox Dynamics is Regulated by Phosphate in Aquatic Environments. Environ. Sci. Technol. 2014, 48, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.-X.; Wang, L.H.; Bai, R.; Huang, H.; Sun, G.-X. Accumulation and Transformation of Arsenic in the Blue-Green Alga Synechocysis sp. PCC 6803. Water Air Soil Pollut. 2012, 223, 1183–1190. [Google Scholar] [CrossRef]
- Pitt, F.D.; Mazard, S.; Humphreys, L.; Scanlan, D.J. Functional characterization of Synechocystis sp. strain PCC 6803 pst1 and pst2 gene clusters reveals a novel strategy for phosphate uptake in a freshwater cyanobacterium. J. Bacteriol. 2010, 192, 3512–3523. [Google Scholar] [CrossRef] [PubMed]
- Dyhrman, S.T.; Haley, S.T. Arsenate Resistance in the Unicellular Marine Diazotroph Crocosphaera watsonii. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Kawakami, H.; Bada, A.; Okonogi, Y.; Matsuto, S. Effects of phosphate on arsenate inhibition in a marine cyanobacterium, Phormidium sp. Appl. Organomet. Chem. 1990, 4, 269–279. [Google Scholar] [CrossRef]
- Takahashi, A.; Kawakami, H.; Iwakiri, K.; Matsuto, S. Some characteristics of arsenate transport in a marine cyanobacterium, Synechococcus sp. Appl. Organomet. Chem. 2001, 15, 291–298. [Google Scholar] [CrossRef]
- Thiel, T. Phosphate transport and arsenate resistance in the cyanobacterium Anabaena variabilis. J. Bacteriol. 1988, 170, 1143–1147. [Google Scholar] [PubMed]
- Markley, C.T. Arsenate uptake, sequestration and reduction by a freshwater cyanobacterium: A potential biologic control of arsenic in South Texas. Master’s Thesis, Texas A&M University, College Station, TX, USA, 2004. [Google Scholar]
- Elias, M.; Wellner, A.; Goldin-Azulay, K.; Chabriere, E.; Vorholt, J.A.; Erb, T.J.; Tawfik, D.S. The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 2012, 491, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, R.; Clemens, S.; Augustyniak, D.; Golik, P.; Maciaszczyk, E.; Tamas, M.J.; Dziadkowiec, D. Metalloid tolerance based on phytochelatins is not functionally equivalent to the arsenite transporter Acr3p. Biochem. Biophys. Res. Commun. 2003, 304, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Harada, E.; von Roepenack-Lahaye, E.; Clemens, S. A cyanobacterial protein with similarity to phytochelatin synthases catalyzes the conversion of glutathione to gamma-glutamylcysteine and lacks phytochelatin synthase activity. Phytochemistry 2004, 65, 3179–3185. [Google Scholar] [CrossRef] [PubMed]
- Malasarn, D.; Kropat, J.; Hsieh, S.I.; Finazzi, G.; Casero, D.; Loo, J.A.; Pellegrini, M.; Wollman, F.A.; Merchant, S.S. Zinc deficiency impacts CO2 assimilation and disrupts copper homeostasis in Chlamydomonas reinhardtii. J. Biol. Chem. 2013, 288, 10672–10683. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huertas, M.J.; López-Maury, L.; Giner-Lamia, J.; Sánchez-Riego, A.M.; Florencio, F.J. Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms. Life 2014, 4, 865-886. https://doi.org/10.3390/life4040865
Huertas MJ, López-Maury L, Giner-Lamia J, Sánchez-Riego AM, Florencio FJ. Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms. Life. 2014; 4(4):865-886. https://doi.org/10.3390/life4040865
Chicago/Turabian StyleHuertas, María José, Luis López-Maury, Joaquín Giner-Lamia, Ana María Sánchez-Riego, and Francisco Javier Florencio. 2014. "Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms" Life 4, no. 4: 865-886. https://doi.org/10.3390/life4040865
APA StyleHuertas, M. J., López-Maury, L., Giner-Lamia, J., Sánchez-Riego, A. M., & Florencio, F. J. (2014). Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms. Life, 4(4), 865-886. https://doi.org/10.3390/life4040865