DNA Methylation: A Key Regulator in Male and Female Reproductive Outcomes
Abstract
1. Introduction
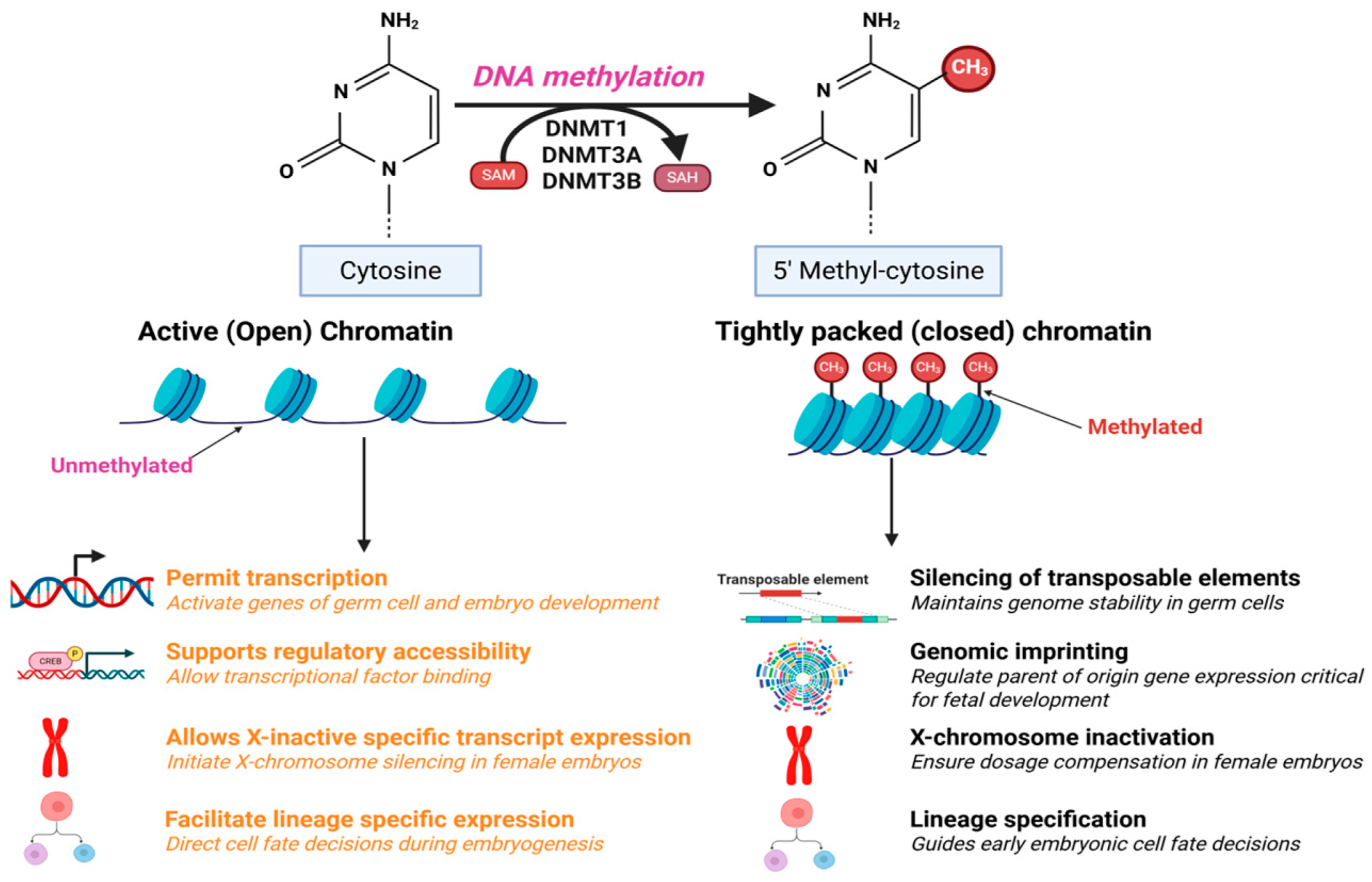
2. Role of DNA Methylation in Transcription
3. Role of DNA Methylation in Male and Female Reproductive Gamates
4. Implications of Alteration in DNA Methylation
4.1. Factors Responsible for Alteration in DNA Methylation
4.1.1. Age and DNA Methylation
4.1.2. Infection and DNA Methylation
4.1.3. Drugs and Steroids and DNA Methylation
4.1.4. Impact of Stress and Lifestyle Factors
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef] [PubMed]
- Ben Maamar, M.; Nilsson, E.E.; Skinner, M.K. Epigenetic transgenerational inheritance, gametogenesis and germline development. Biol. Reprod. 2021, 105, 570–592. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Surani, M.A. Epigenetic reprogramming in mouse and human primordial germ cells. Exp. Mol. Med. 2024, 56, 2578–2587. [Google Scholar] [CrossRef] [PubMed]
- Gunes, S.; Esteves, S.C. Role of genetics and epigenetics in male infertility. Andrologia 2021, 53, e13586. [Google Scholar] [CrossRef] [PubMed]
- Shacfe, G.; Turko, R.; Syed, H.H.; Masoud, I.; Tahmaz, Y.; Samhan, L.M.; Alkattan, K.; Shafqat, A.; Yaqinuddin, A. A DNA Methylation Perspective on Infertility. Genes 2023, 14, 2132. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Cheng, X.; Gu, C.; Xu, Y.; Long, X.; Zhai, J.; Sun, F.; Qian, J.; Du, Y.; Wang, H.; et al. Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development. Nat. Genet. 2023, 55, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Ichiyanagi, T.; Ichiyanagi, K.; Miyake, M.; Sasaki, H. Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Res. 2013, 41, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.L.; Zorzan, I.; Rugg-Gunn, P.J. Epigenetic regulation of early human embryo development. Cell Stem Cell 2023, 30, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bhandari, R.K. The dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes). Epigenetics 2020, 15, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Epigenetic transgenerational toxicology and germ cell disease. Int. J. Androl. 2007, 30, 393–396; discussion 396–397, discussion 396–397. [Google Scholar] [CrossRef] [PubMed]
- Tibben, B.M.; Rothbart, S.B. Mechanisms of DNA Methylation Regulatory Function and Crosstalk with Histone Lysine Methylation. J. Mol. Biol. 2024, 436, 168394. [Google Scholar] [CrossRef] [PubMed]
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.C.; Huang, H.H.; Jiang, B.C.; Yang, W.Z.; Chen, Y.N.; Yuan, H.S. Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation. Sci. Adv. 2025, 11, eadu8116. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Zhang, Z.; Zou, Z.; Lv, C.; Dong, Q.; He, Q.; Yi, Y.; Li, Y.; Wang, H.; Zhu, B. Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res. 2020, 30, 980–996. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, S.; Wu, Q.; Zhang, L.; Guo, F. Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res. 2019, 29, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Liu, H.; Xian, Q.; Gui, C.; Xu, M.; Zhou, Y. Mechanistic insights into UHRF1-mediated DNA methylation by structure-based functional clarification of UHRF1 domains. Oncol. Lett. 2023, 26, 542. [Google Scholar] [CrossRef] [PubMed]
- Rothbart, S.B.; Krajewski, K.; Nady, N.; Tempel, W.; Xue, S.; Badeaux, A.I.; Barsyte-Lovejoy, D.; Martinez, J.Y.; Bedford, M.T.; Fuchs, S.M.; et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012, 19, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Fallet, M.; Luquet, E.; David, P.; Cosseau, C. Epigenetic inheritance and intergenerational effects in mollusks. Gene 2020, 729, 144166. [Google Scholar] [CrossRef] [PubMed]
- Nava-Rivera, L.E.; Betancourt-Martínez, N.D.; Lozoya-Martínez, R.; Carranza-Rosales, P.; Guzmán-Delgado, N.E.; Carranza-Torres, I.E.; Delgado-Aguirre, H.; Zambrano-Ortíz, J.O.; Morán-Martínez, J. Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure. Sci. Rep. 2021, 11, 8276. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Riaz, F.; Wang, W.; Pu, J.; Liang, Y.; Wu, Z.; Pan, S.; Song, J.; Yang, L.; Zhang, Y.; et al. Functional significance of DNA methylation: Epigenetic insights into Sjögren’s syndrome. Front. Immunol. 2024, 15, 1289492. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yan, Y.; Youping, L.; Xiaohua, Z.; Zeqin, L. Epigenetic regulation of gene expression in response to environmental exposures: From bench to model. Sci. Total Environ. 2021, 776, 145998. [Google Scholar] [CrossRef]
- Marasca, F.; Bodega, B.; Orlando, V. How Polycomb-Mediated Cell Memory Deals with a Changing Environment: Variations in PcG complexes and proteins assortment convey plasticity to epigenetic regulation as a response to environment. Bioessays 2018, 40, e1700137. [Google Scholar] [CrossRef] [PubMed]
- Myant, K.; Termanis, A.; Sundaram, A.Y.; Boe, T.; Li, C.; Merusi, C.; Burrage, J.; de Las Heras, J.I.; Stancheva, I. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011, 21, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Nyikó, T.; Gyula, P.; Ráth, S.; Sós-Hegedűs, A.; Csorba, T.; Abbas, S.H.; Bóka, K.; Pettkó-Szandtner, A.; Móricz, Á.M.; Molnár, B.P.; et al. INCREASED DNA METHYLATION 3 forms a potential chromatin remodelling complex with HAIRPLUS to regulate DNA methylation and trichome development in tomato. Plant J. 2025, 121, e70085. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Xi, S.; Shan, J.; Maunakea, A.; Che, A.; Briones, V.; Lee, E.Y.; Geiman, T.; Huang, J.; Stephens, R.; et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc. Natl. Acad. Sci. USA 2011, 108, 5626–5631. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Amjadian, T.; Yaghmaei, P.; Nasim, H.R.; Yari, K. Impact of DNA methylation of the human mesoderm-specific transcript (MEST) on male infertility. Heliyon 2023, 9, e21099. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Sun, Υ.; Zhang, S.; Feng, X.; Xu, M.; Dai, J.; Ni, X.; Wang, X.; Wu, Q. Profiling the DNA methylation patterns of imprinted genes in abnormal semen samples by next-generation bisulfite sequencing. J. Assist. Reprod. Genet. 2020, 37, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Hu, B.; Yan, L.; Yong, J.; Wu, Y.; Gao, Y.; Guo, F.; Hou, Y.; Fan, X.; Dong, J.; et al. DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells. Cell Res. 2017, 27, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Georgios, V.; Chad, E.N.; Salih, T.; Athanasia, S.; Keijo, V.; de Dirk, G.R.; Richard, G.J.; Robert, J.S.; Steen, K.T.O. The Dnmt3L ADD Domain Controls Cytosine Methylation Establishment during Spermatogenesis. Cell Rep. 2015, 10, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E.; Hesselson, D.; Bogdanovic, O. Developmental Accumulation of Gene Body and Transposon Non-CpG Methylation in the Zebrafish Brain. Front. Cell Dev. Biol. 2021, 9, 643603. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Shirane, K.; Kurimoto, K.; Yabuta, Y.; Yamaji, M.; Satoh, J.; Sato, F.; Yamamoto, T.; Tada, T.; Hayashi, K.; Saitou, M. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. Cell Rep. 2013, 3, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Tomizawa, S.; Kobayashi, H.; Watanabe, T.; Andrews, S.; Hata, K.; Kelsey, G.; Sasaki, H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 2011, 138, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Sendžikaitė, G.; Kelsey, G. The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 2019, 63, 691–705. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ecker, J.R. Non-CG Methylation in the Human Genome. Annu. Rev. Genomics Hum. Genet. 2015, 16, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Shi, Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 2012, 139, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Rao, A.; Ko, M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 2017, 49, e323. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Yan, R.; Long, X.; Ji, D.; Song, B.; Wang, M.; Zhang, F.; Cheng, X.; Sun, F.; Zhu, R.; et al. Distinct dynamics of parental 5-hydroxymethylcytosine during human preimplantation development regulate early lineage gene expression. Nat. Cell Biol. 2024, 26, 1458–1469. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Duan, J.; Gao, X.; Zhu, W.; Lu, X.; Yang, L.; Zhang, J.; Li, G.; Ci, W.; et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 2014, 157, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Szabó, P.E. The link between 5-hydroxymethylcytosine and DNA demethylation in early embryos. Epigenomics 2023, 15, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Carrell, D.T. Epigenetics of the male gamete. Fertil. Steril. 2012, 97, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.W.S.; Leitch, H.G.; Requena, C.E.; Sun, Z.; Amouroux, R.; Roman-Trufero, M.; Borkowska, M.; Terragni, J.; Vaisvila, R.; Linnett, S.; et al. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 2018, 555, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Hammoud, A.O.; Gibson, M.; Cairns, B.R.; Carrell, D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011, 26, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Tomizawa, S.; Krueger, F.; Ruf, N.; Carli, N.; Segonds-Pichon, A.; Sato, S.; Hata, K.; Andrews, S.R.; Kelsey, G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011, 43, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.R.; Veselovska, L.; Kelsey, G. Establishment and functions of DNA methylation in the germline. Epigenomics 2016, 8, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.W.; Castillo-Fernandez, J.; Chan, A.C.; la Cour Freiesleben, N.; Zedeler, A.; Bungum, M.; Cardona, A.; Perry, J.R.B.; Skouby, S.O.; Hoffmann, E.R.; et al. Identification of a unique epigenetic profile in women with diminished ovarian reserve. Fertil. Steril. 2021, 115, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Sachs, M.; Onodera, C.; Blaschke, K.; Ebata, K.T.; Song, J.S.; Ramalho-Santos, M. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013, 3, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Seisenberger, S.; Peat, J.R.; Hore, T.A.; Santos, F.; Dean, W.; Reik, W. Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20110330. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Proudhon, C.; Duffié, R.; Ajjan, S.; Cowley, M.; Iranzo, J.; Carbajosa, G.; Saadeh, H.; Holland, M.L.; Oakey, R.J.; Rakyan, V.K.; et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell 2012, 47, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Fanourgakis, G.; Gaspa-Toneu, L.; Komarov, P.A.; Papasaikas, P.; Ozonov, E.A.; Smallwood, S.A.; Peters, A.H.F.M. DNA methylation modulates nucleosome retention in sperm and H3K4 methylation deposition in early mouse embryos. Nat. Commun. 2025, 16, 465. [Google Scholar] [CrossRef] [PubMed]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef] [PubMed]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Lanzillotti, C.; Mazziotta, C.; Tognon, M.; Martini, F. Epigenetics of Male Infertility: The Role of DNA Methylation. Front. Cell Dev. Biol. 2021, 9, 689624. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, W.; Badescu, D.; Tan, S.L.; Ragoussis, J.; Taketo, T. Effects of the Sex Chromosome Complement, XX, XO, or XY, on the Transcriptome and Development of Mouse Oocytes During Follicular Growth. Front. Genet. 2021, 12, 792604. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Srivastava, V.; Shandilya, C.; Kaushik, A.; Singh, K.K. Mitochondria: The epigenetic regulators of ovarian aging and longevity. Front. Endocrinol. 2024, 15, 1424826. [Google Scholar] [CrossRef] [PubMed]
- Saftić Martinović, L.; Mladenić, T.; Lovrić, D.; Ostojić, S.; Dević Pavlić, S. Decoding the Epigenetics of Infertility: Mechanisms, Environmental Influences, and Therapeutic Strategies. Epigenomes 2024, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Kulac, T.; Hekim, N.; Kocamanoglu, F.; Beyaz, C.; Gunes, S.; Asci, R. Methylation patterns of methylenetetrahydrofolate reductase gene promoter in infertile males. Andrologia 2021, 53, e13942. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gao, Y.; Liu, W.; Gao, S. Epigenetic regulation of cell fate transition: Learning from early embryo development and somatic cell reprogramming. Biol. Reprod. 2022, 107, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Knight, A.K.; Spencer, J.B.; Smith, A.K. DNA methylation as a window into female reproductive aging. Epigenomics 2024, 16, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Dhar, G.A.; Saha, S.; Mitra, P.; Nag Chaudhuri, R. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus 2021, 64, 259–270. [Google Scholar] [CrossRef] [PubMed]
- McSwiggin, H.M.; O’Doherty, A.M. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction 2018, 156, R9–R21. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, H.; Du, H.; Zhang, W.; Kang, X.; Luo, Y.; Zhou, X.; Li, L. Developmental features of DNA methylation in CpG islands of human gametes and preimplantation embryos. Exp. Ther. Med. 2019, 17, 4447–4456. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gan, H.; Wang, B.; Wang, X.; Li, M.; Yang, Q.; Geng, M.; Zhu, P.; Shao, S.; Tao, F. Mediating effects of DNA methylation in the association between sleep quality and infertility among women of childbearing age. BMC Public Health 2023, 23, 1802. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Martínez, E.R.; Gómez-Viais, Y.I.; García-Gómez, E.; Reyes-Mayoral, C.; Reyes-Muñoz, E.; Camacho-Arroyo, I.; Cerbón, M. DNA methylation in the pathogenesis of polycystic ovary syndrome. Reproduction 2019, 158, R27–R40. [Google Scholar] [CrossRef] [PubMed]
- Abur, U.; Gunes, S.; Ascı, R.; Altundag, E.; Akar, O.S.; Ayas, B.; Karadag Alpaslan, M.; Ogur, G. Chromosomal and Y-chromosome microdeletion analysis in 1300 infertile males and the fertility outcome of patients with AZFc microdeletions. Andrologia 2019, 51, e13402. [Google Scholar] [CrossRef] [PubMed]
- Sujit, K.M.; Singh, V.; Trivedi, S.; Singh, K.; Gupta, G.; Rajender, S. Increased DNA methylation in the spermatogenesis-associated (SPATA) genes correlates with infertility. Andrology 2020, 8, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Calicchio, R.; Doridot, L.; Miralles, F.; Méhats, C.; Vaiman, D. DNA methylation, an epigenetic mode of gene expression regulation in reproductive science. Curr. Pharm. Des. 2014, 20, 1726–1750. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.H.; Wang, Z.B.; Li, Q.N.; Meng, T.G.; Dong, M.Z.; Hou, Y.; Ouyang, Y.C.; Schatten, H.; Sun, Q.Y. Absence of mitochondrial DNA methylation in mouse oocyte maturation, aging and early embryo development. Biochem. Biophys. Res. Commun. 2019, 513, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.G.; Aston, K.I.; Cairns, B.; Smith, A.; Carrell, D.T. Paternal germ line aging: DNA methylation age prediction from human sperm. BMC Genom. 2018, 19, 763. [Google Scholar] [CrossRef] [PubMed]
- Kordowitzki, P.; Haghani, A.; Zoller, J.A.; Li, C.Z.; Raj, K.; Spangler, M.L.; Horvath, S. Epigenetic clock and methylation study of oocytes from a bovine model of reproductive aging. Aging Cell 2021, 20, e13349. [Google Scholar] [CrossRef] [PubMed]
- Kresovich, J.K.; Harmon, Q.E.; Xu, Z.; Nichols, H.B.; Sandler, D.P.; Taylor, J.A. Reproduction, DNA methylation and biological age. Hum. Reprod. 2019, 34, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
- Li Piani, L.; Vigano, P.; Somigliana, E. Epigenetic clocks and female fertility timeline: A new approach to an old issue? Front. Cell Dev. Biol. 2023, 11, 1121231. [Google Scholar] [CrossRef] [PubMed]
- Ashapkin, V.; Suvorov, A.; Pilsner, J.R.; Krawetz, S.A.; Sergeyev, O. Age-associated epigenetic changes in mammalian sperm: Implications for offspring health and development. Hum. Reprod. Update 2023, 29, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Åsenius, F.; Danson, A.F.; Marzi, S.J. DNA methylation in human sperm: A systematic review. Hum. Reprod. Update 2020, 26, 841–873. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Shao, X.; Chan, P.; Cheung, W.; Kwan, T.; Pastinen, T.; Robaire, B. High-resolution analyses of human sperm dynamic methylome reveal thousands of novel age-related epigenetic alterations. Clin. Epigenet. 2020, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Pilsner, J.R.; Saddiki, H.; Whitcomb, B.W.; Suvorov, A.; Buck Louis, G.M.; Mumford, S.L.; Schisterman, E.F.; Oluwayiose, O.A.; Balzer, L.B. Sperm epigenetic clock associates with pregnancy outcomes in the general population. Hum. Reprod. 2022, 37, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Aoki, V.W.; Liu, L.; Jones, K.P.; Hatasaka, H.H.; Gibson, M.; Peterson, C.M.; Carrell, D.T. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil. Steril. 2006, 86, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Khara, K.K.; Vlad, M.; Griffiths, M.; Kennedy, C.R. Human protamines and male infertility. J. Assist. Reprod. Genet. 1997, 14, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Agarwal, A.; Rohra, V.K.; Assidi, M.; Abu-Elmagd, M.; Turki, R.F. Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod. Biol. Endocrinol. 2015, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Elizabeth, A.K.; Carolina, G.; Anthony, J.H. Transgenerational epigenetic impacts of parental infection on offspring health and disease susceptibility. Trends Genet. 2022, 38, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Smith, M. Molecular Association of Sexually Transmitted Infections and Methylation Pattern of SNRPN Gene Promoter among Males with Abnormal Semen. Indian J. Forensic Med. Toxicol. 2021, 15, 1830–1836. [Google Scholar] [CrossRef]
- Jasmin, P.; Swarupa, M.; Neha, M.; Aman, T.; Negi, V.D. Pregnancy, infection, and epigenetic regulation: A complex scenario. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2023, 1869, 166768. [Google Scholar] [CrossRef]
- Pradhan, J.; Mallick, S.; Mishra, N.; Tiwari, A.; Negi, V.D. Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics 2016, 11, 750–760. [Google Scholar] [CrossRef]
- Fang, J.-Y.; Mikovits, J.A.; Bagni, R.; Petrow-Sadowski, C.L.; Ruscetti, F.W. Infection of Lymphoid Cells by Integration-Defective Human Immunodeficiency Virus Type 1 Increases De Novo Methylation. J. Virol. 2001, 75, 9753–9761. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Ramphal, U.; Adimulam, T.; Chinniah, R.; Ramsuran, V. Deciphering DNA Methylation in HIV Infection. Front. Immunol. 2021, 12, 795121. [Google Scholar] [CrossRef] [PubMed]
- Horner, P.J.; Flanagan, H.; Horne, A.W. Is There a Hidden Burden of Disease as a Result of Epigenetic Epithelial-to-Mesenchymal Transition Following Chlamydia trachomatis Genital Tract Infection? J. Infect. Dis. 2021, 224 (Suppl. 2), S128–S136. [Google Scholar] [CrossRef] [PubMed]
- Igietseme, J.U.; Omosun, Y.; Nagy, T.; Stuchlik, O.; Reed, M.S.; He, Q.; Partin, J.; Joseph, K.; Ellerson, D.; George, Z.; et al. Molecular Pathogenesis of Chlamydia Disease Complications: Epithelial-Mesenchymal Transition and Fibrosis. Infect. Immun. 2018, 86, e00585-17. [Google Scholar] [CrossRef] [PubMed]
- Kessler, M.; Hoffmann, K.; Fritsche, K.; Brinkmann, V.; Mollenkopf, H.-J.; Thieck, O.; Teixeira da Costa, A.R.; Braicu, E.I.; Sehouli, J.; Mangler, M.; et al. Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun. 2019, 10, 1194. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Raddatz, G.; Gutekunst, J.; Ridnik, M.; Cohen, D.; Abu-Remaileh, M.; Tuganbaev, T.; Shapiro, H.; Pikarsky, E.; Elinav, E.; et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 2020, 5, 610–619. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, L.R.; Kaul, R. Quality and quantity: Mucosal CD4+ T cells and HIV susceptibility. Curr. Opin. HIV AIDS 2012, 7, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Mwatelah, R.; McKinnon, L.R.; Baxter, C.; Abdool Karim, Q.; Abdool Karim, S.S. Mechanisms of sexually transmitted infection-induced inflammation in women: Implications for HIV risk. J. Int. AIDS Soc. 2019, 22, e25346. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Scicluna, B.P.; van der Poll, T. The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front. Immunol. 2021, 12, 696280. [Google Scholar] [CrossRef] [PubMed]
- Singer, B.D.; Mock, J.R.; Aggarwal, N.R.; Garibaldi, B.T.; Sidhaye, V.K.; Florez, M.A.; Chau, E.; Gibbs, K.W.; Mandke, P.; Tripathi, A.; et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am. J. Respir. Cell Mol. Biol. 2015, 52, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Samah, A.; Bruce, R.S.; Alvaro, G.H.; Lauretta, A.R.; Adrienne, M.A.; Romana, A.N.; Rodney, W.J.; Sandra, L.R.-Z. Epigenetic disruptions in the offspring hypothalamus in response to maternal infection. Gene 2024, 910, 148329. [Google Scholar] [CrossRef] [PubMed]
- Bryan, E.R.; Barrero, R.A.; Cheung, E.; Tickner, J.A.D.; Trim, L.K.; Richard, D.; McLaughlin, E.A.; Beagley, K.W.; Carey, A.J. DNA damage contributes to transcriptional and immunological dysregulation of testicular cells during Chlamydia infection. Am. J. Reprod. Immunol. 2021, 86, e13400. [Google Scholar] [CrossRef] [PubMed]
- Saki, J.; Sabaghan, M.; Arjmand, R.; Teimoori, A.; Rashno, M.; Saki, G.; Shojaee, S. Curcumin as an indirect methylation inhibitor modulates the effects of Toxoplasma gondii on genes involved in male fertility. EXCLI J. 2020, 19, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.; Olson, M.; Wang, K.; Fazleabas, A.; De La Fuente, R. Arginine methyltransferases mediate an epigenetic ovarian response to endometriosis. Reproduction 2015, 150, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Adedeji, O.A.; Tomoko, K.; Masayuki, S. Impact of lipopolysaccharide administration on luteinizing hormone/choriogonadotropin receptor (Lhcgr) expression in mouse ovaries. J. Reprod. Immunol. 2020, 142, 103193. [Google Scholar] [CrossRef] [PubMed]
- Estill, M.S.; Bolnick, J.M.; Waterland, R.A.; Bolnick, A.D.; Diamond, M.P.; Krawetz, S.A. Assisted reproductive technology alters deoxyribonucleic acid methylation profiles in bloodspots of newborn infants. Fertil. Steril. 2016, 106, 629–639.e10. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, Z.; Liu, Y.; Tang, B.; Wang, Y.; Yang, L.; Du, J.; Zhang, Y. Oct4 and the small molecule inhibitor, SC1, regulates Tet2 expression in mouse embryonic stem cells. Mol. Biol. Rep. 2013, 40, 2897–2906. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N. Infection-induced epigenetic changes and their impact on the pathogenesis of diseases. Semin. Immunopathol. 2020, 42, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Leisegang, K.; Dutta, S. Do lifestyle practices impede male fertility? Andrologia 2021, 53, e13595. [Google Scholar] [CrossRef] [PubMed]
- Salerno, M.; Cascio, O.; Bertozzi, G.; Sessa, F.; Messina, A.; Monda, V.; Cipolloni, L.; Biondi, A.; Daniele, A.; Pomara, C. Anabolic androgenic steroids and carcinogenicity focusing on Leydig cell: A literature review. Oncotarget 2018, 9, 19415–19426. [Google Scholar] [CrossRef] [PubMed]
- Mesbah, S.F.; Shokri, S.; Karbalay-Doust, S.; Mirkhani, H. The effect of nandrolone decanoate on the body, testis and epididymis weight and semen parameters in adult male rats. Iran. J. Med. Sci. 2007, 32, 93–99. [Google Scholar]
- Noorafshan, A.; Karbalay-Doust, S.; Ardekani, F.M. High doses of nandrolone decanoate reduce volume of testis and length of seminiferous tubules in rats. Apmis 2005, 113, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Tahtamouni, L.H.; Mustafa, N.H.; Hassan, I.M.; Ahmad, I.M.; Yasina, S.R.; Abdallaa, M.Y. Nandrolone decanoate administration to male rats induces oxidative stress, seminiferous tubules abnormalities, and sperm DNA fragmentation. Jordan J. Biol. Sci. 2010, 3, 165–174. [Google Scholar]
- Perez-Garcia, L.F.; Dolhain, R.J.E.M.; Vorstenbosch, S.; Bramer, W.; van Puijenbroek, E.; Hazes, J.M.W.; te Winkel, B. The effect of paternal exposure to immunosuppressive drugs on sexual function, reproductive hormones, fertility, pregnancy and offspring outcomes: A systematic review. Hum. Reprod. Update 2020, 26, 961–1001. [Google Scholar] [CrossRef] [PubMed]
- Gorczyca, G.; Wartalski, K.; Wiater, J.; Samiec, M.; Tabarowski, Z.; Duda, M. Anabolic Steroids-Driven Regulation of Porcine Ovarian Putative Stem Cells Favors the Onset of Their Neoplastic Transformation. Int. J. Mol. Sci. 2021, 22, 11800. [Google Scholar] [CrossRef] [PubMed]
- Songping, W.; Pamela, A.G.C.; Roderick, D.; Salma, M.; Bernard, K.-A. Sex steroid-induced DNA methylation changes and inflammation response in prostate cancer. Cytokine 2016, 86, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Arnold, J.T.; McFann, K.K.; Blackman, M.R. DHT and testosterone, but not DHEA or E2, differentially modulate IGF-I, IGFBP-2, and IGFBP-3 in human prostatic stromal cells. Am. J. Physiol.-Endocrinol. Metab. 2006, 290, E952–E960. [Google Scholar] [CrossRef] [PubMed]
- Pirompol, P.; Teekabut, V.; Weerachatyanukul, W.; Bupha-Intr, T.; Wattanapermpool, J. Supra-physiological dose of testosterone induces pathological cardiac hypertrophy. J. Endocrinol. 2016, 229, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Koninckx, P.R.; Ussia, A.; Adamyan, L.; Wattiez, A.; Gomel, V.; Martin, D.C. Pathogenesis of endometriosis: The genetic/epigenetic theory. Fertil. Steril. 2019, 111, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Bunkar, N.; Pathak, N.; Lohiya, N.K.; Mishra, P.K. Epigenetics: A key paradigm in reproductive health. Clin. Exp. Reprod. Med. 2016, 43, 59–81. [Google Scholar] [CrossRef] [PubMed]
- Doshi, T.; Mehta, S.S.; Dighe, V.; Balasinor, N.; Vanage, G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology 2011, 289, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Miaomiao, T.; Xiangguang, C.; Chen, W.; Min, S.; Jie, Z.; Sheng, B.; Chengju, W. Life cycle exposure to propiconazole reduces fecundity by disrupting the steroidogenic pathway and altering DNA methylation in zebrafish (Danio rerio). Environ. Int. 2020, 135, 105384. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gu, B.; Brichant, G.; Singh, J.P.; Yang, H.; Chang, H.; Zhao, Y.; Cheng, C.; Liu, Z.-W.; Alderman, M.H.; et al. The steroid hormone estriol (E3) regulates epigenetic programming of fetal mouse brain and reproductive tract. BMC Biol. 2022, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Akbas, G.E.; Song, J.; Taylor, H.S. A HOXA10 estrogen response element (ERE) is differentially regulated by 17 beta-estradiol and diethylstilbestrol (DES). J. Mol. Biol. 2004, 340, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Bromer, J.G.; Wu, J.; Zhou, Y.; Taylor, H.S. Hypermethylation of homeobox A10 by in utero diethylstilbestrol exposure: An epigenetic mechanism for altered developmental programming. Endocrinology 2009, 150, 3376–3382. [Google Scholar] [CrossRef] [PubMed]
- Bromer, J.G.; Zhou, Y.; Taylor, M.B.; Doherty, L.; Taylor, H.S. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010, 24, 2273–2280. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, S.S.; Agarwal, A.; Syriac, A. Marijuana, phytocannabinoids, the endocannabinoid system, and male fertility. J. Assist. Reprod. Genet. 2015, 32, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, S.S.; Cabler, S.; McAlister, D.A.; Sabanegh, E.; Agarwal, A. The effect of obesity on sperm disorders and male infertility. Nat. Rev. Urol. 2010, 7, 153–161. [Google Scholar] [CrossRef] [PubMed]
- du Plessis, S.S.; Kashou, A.; Vaamonde, D.; Agarwal, A. Is there a link between exercise and male factor infertility. Open Reprod. Sci. J. 2011, 3, 105. [Google Scholar] [CrossRef]
- Skinner, M.K.; Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; McBirney, M.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenet. Chromatin 2018, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility. Transl. Res. 2015, 165, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Durairajanayagam, D. Lifestyle causes of male infertility. Arab J. Urol. 2018, 16, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Nargund, V.H. Effects of psychological stress on male fertility. Nat. Rev. Urol. 2015, 12, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, Z.; Wang, G.; Wang, H.; Zhou, Y.; Zhao, X.; Cheng, C.Y.; Qiao, Y.; Sun, F. Sperm epigenetic alterations contribute to inter- and transgenerational effects of paternal exposure to long-term psychological stress via evading offspring embryonic reprogramming. Cell Discov. 2021, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Litzky, J.F.; Marsit, C.J. Epigenetically regulated imprinted gene expression associated with IVF and infertility: Possible influence of prenatal stress and depression. J. Assist. Reprod. Genet. 2019, 36, 1299–1313. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.L.; Gladish, N.; Gatev, E.; Jones, M.J.; Chen, Y.; MacIsaac, J.L.; Tworoger, S.S.; Austin, S.B.; Tanrikut, C.; Chavarro, J.E.; et al. Exposure to childhood abuse is associated with human sperm DNA methylation. Transl. Psychiatry 2018, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Hudson, P.S.; Benjamin, C.N.; Arjun, B.; Xianming, T.; Laura, S.; Reema Abdulrahman, S.A.; Elizabeth, M.M.; Krista, P.; Rebecca, C.F.; Christopher, M. Discrimination exposure and DNA methylation of stress-related genes in Latina mothers. Psychoneuroendocrinology 2018, 98, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.-J.; Luo, S.-M.; Lin, F.; Liang, Q.-X.; Huang, L.; Wei, Y.-C.; Hou, Y.; Han, Z.-M.; Schatten, H.; Sun, Q.-Y. DNA Methylation in Oocytes and Liver of Female Mice and Their Offspring: Effects of High-Fat-Diet–Induced Obesity. Environ. Health Perspect. 2014, 122, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Lambrot, R.; Xu, C.; Saint-Phar, S.; Chountalos, G.; Cohen, T.; Paquet, M.; Suderman, M.; Hallett, M.; Kimmins, S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 2013, 4, 2889. [Google Scholar] [CrossRef] [PubMed]
- Toschi, P.; Capra, E.; Anzalone, D.A.; Lazzari, B.; Turri, F.; Pizzi, F.; Scapolo, P.A.; Stella, A.; Williams, J.L.; Ajmone Marsan, P.; et al. Maternal peri-conceptional undernourishment perturbs offspring sperm methylome. Reproduction 2020, 159, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Donkin, I.; Versteyhe, S.; Ingerslev, L.R.; Qian, K.; Mechta, M.; Nordkap, L.; Mortensen, B.; Appel, E.V.; Jørgensen, N.; Kristiansen, V.B.; et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab. 2016, 23, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-J.; Zhu, C.-C.; Duan, X.; Liu, H.-L.; Wang, Q.; Sun, S.-C. Both diet and gene mutation induced obesity affect oocyte quality in mice. Sci. Rep. 2016, 6, 18858. [Google Scholar] [CrossRef] [PubMed]
- Alex, C.V.; Kim, D.L.; Cresandra, C.; Jaime, M.; Ashok, A. Oocyte developmental competence and embryo development: Impact of lifestyle and environmental risk factors. Reprod. Biomed. Online 2011, 22, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Sciorio, R.; Tramontano, L.; Adel, M.; Fleming, S. Decrease in Sperm Parameters in the 21st Century: Obesity, Lifestyle, or Environmental Factors? An Updated Narrative Review. J. Pers. Med. 2024, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Brieño-Enríquez, M.A.; García-López, J.; Cárdenas, D.B.; Guibert, S.; Cleroux, E.; Děd, L.; Hourcade Jde, D.; Pěknicová, J.; Weber, M.; Del Mazo, J. Exposure to endocrine disruptor induces transgenerational epigenetic deregulation of microRNAs in primordial germ cells. PLoS ONE 2015, 10, e0124296. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Tran, D.A.; Li, A.X.; Warden, C.; Bai, A.Y.; Singh, P.; Wu, X.; Pfeifer, G.P.; Szabó, P.E. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 2015, 16, 59. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar] [CrossRef] [PubMed]
- Roberto, S.; Serena, T.; Valentina, C.; Carlos, G.C.; Consuelo, M.; Alessia, A.; Maria Giulia, T.; Paolo, G. Role of oxidative stress, genome damage and DNA methylation as determinants of pathological conditions in the newborn: An overview from conception to early neonatal stage. Mutat. Res./Rev. Mutat. Res. 2020, 783, 108295. [Google Scholar] [CrossRef]
- Chamorro-García, R.; Blumberg, B. Transgenerational effects of obesogens and the obesity epidemic. Curr. Opin. Pharmacol. 2014, 19, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm-Benartzi, C.S.; Houseman, E.A.; Maccani, M.A.; Poage, G.M.; Koestler, D.C.; Langevin, S.M.; Gagne, L.A.; Banister, C.E.; Padbury, J.F.; Marsit, C.J. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ. Health Perspect. 2012, 120, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; Goldman, L.R.; Brebi-Mieville, P.; Ili-Gangas, C.; Lebron, C.; Witter, F.R.; Apelberg, B.J.; Hernández-Roystacher, M.; Jaffe, A.; Halden, R.U.; et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 2010, 5, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, D.; Sharma, S.; Kho, A.T.; Gaedigk, R.; Vyhlidal, C.A.; Leeder, J.S.; Morrow, J.; Carey, V.J.; Weiss, S.T.; Tantisira, K.G.; et al. Fetal lung and placental methylation is associated with in utero nicotine exposure. Epigenetics 2014, 9, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Akhatova, A.; Jones, C.; Coward, K.; Yeste, M. How do lifestyle and environmental factors influence the sperm epigenome? Effects on sperm fertilising ability, embryo development, and offspring health. Clin. Epigenet. 2025, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Chen, Y.; Wang, C.; Li, G.; Wei, Z.; He, X.; Cao, Y. Poor semen parameters are associated with abnormal methylation of imprinted genes in sperm DNA. Reprod. Biol. Endocrinol. 2022, 20, 155. [Google Scholar] [CrossRef] [PubMed]
- Emokpae, M.A.; Brown, S.I. Effects of lifestyle factors on fertility: Practical recommendations for modification. Reprod. Fertil. 2021, 2, R13–R26. [Google Scholar] [CrossRef] [PubMed]

| Factor | Genes/Enzymes/Pathways Affected | References |
|---|---|---|
| Age | DENND1A, TCF20, HOXD8 | [78,80] |
| Infection | NLRC5, E-cadherin, Lhcgr, Cyp19a1, IL-6 and IL-1β, like CARM1, PRMT2, and PRMT8 | [88,91,94,105,106] |
| Drugs/Steroids | FSH and LH | [114] |
| Stress | 11-β-HSD2, NR3C1IGF2, PEG3 | [138] |
| Obesity | BDNF, FTO, SH2B1, CHST8 | [142] |
| Alcohol intake and Smoking | lipid peroxidation (MDA) and EAO which included catalase (CAT), superoxide dismutase (SOD) and glutathione reductase (GR) | [110] |
| Chemical Exposure/Toxicants | HOXA10, MTHFR, imprinted genes (IGF2, PEG3) | [146,147] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adetunji, A.O.; Owusu, H.; Adewale, E.F.; Adesina, P.A.; Xedzro, C.; Saliu, T.P.; Islam, S.; Zhu, Z.; Morenikeji, O.B. DNA Methylation: A Key Regulator in Male and Female Reproductive Outcomes. Life 2025, 15, 1109. https://doi.org/10.3390/life15071109
Adetunji AO, Owusu H, Adewale EF, Adesina PA, Xedzro C, Saliu TP, Islam S, Zhu Z, Morenikeji OB. DNA Methylation: A Key Regulator in Male and Female Reproductive Outcomes. Life. 2025; 15(7):1109. https://doi.org/10.3390/life15071109
Chicago/Turabian StyleAdetunji, Adedeji O., Henrietta Owusu, Esiosa F. Adewale, Precious Adedayo Adesina, Christian Xedzro, Tolulope Peter Saliu, Shahidul Islam, Zhendong Zhu, and Olanrewaju B. Morenikeji. 2025. "DNA Methylation: A Key Regulator in Male and Female Reproductive Outcomes" Life 15, no. 7: 1109. https://doi.org/10.3390/life15071109
APA StyleAdetunji, A. O., Owusu, H., Adewale, E. F., Adesina, P. A., Xedzro, C., Saliu, T. P., Islam, S., Zhu, Z., & Morenikeji, O. B. (2025). DNA Methylation: A Key Regulator in Male and Female Reproductive Outcomes. Life, 15(7), 1109. https://doi.org/10.3390/life15071109







