Unveiling Genetic Loci for Root Morphology and Salt Response at Rice Seedling Stage via Genome-Wide Association Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Procedure
2.3. Phenotypic Data Collection
2.3.1. Salt Tolerance Level Evaluation
2.3.2. Root Morphological Trait Acquisition
2.4. Data Analysis
3. Results
3.1. Descriptive Statistical Results
3.2. Correlation Analysis Results
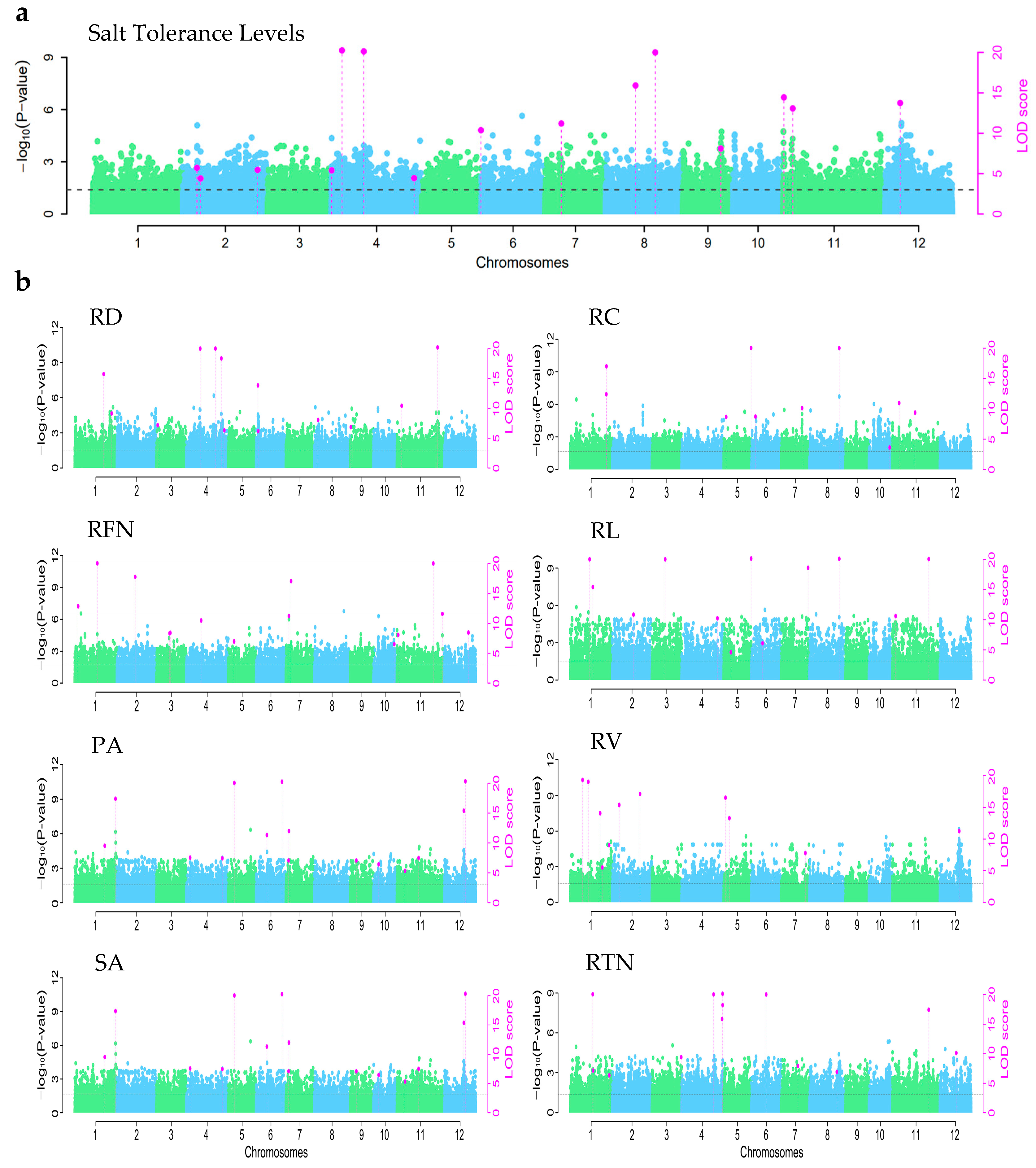
3.3. Results of Association Analysis for Rice Salt Tolerance
3.3.1. GWAS Results for the Trait of Salt Tolerance Levels in Rice
3.3.2. GWAS Results for Root Morphological Traits Under Salt Stress in Rice
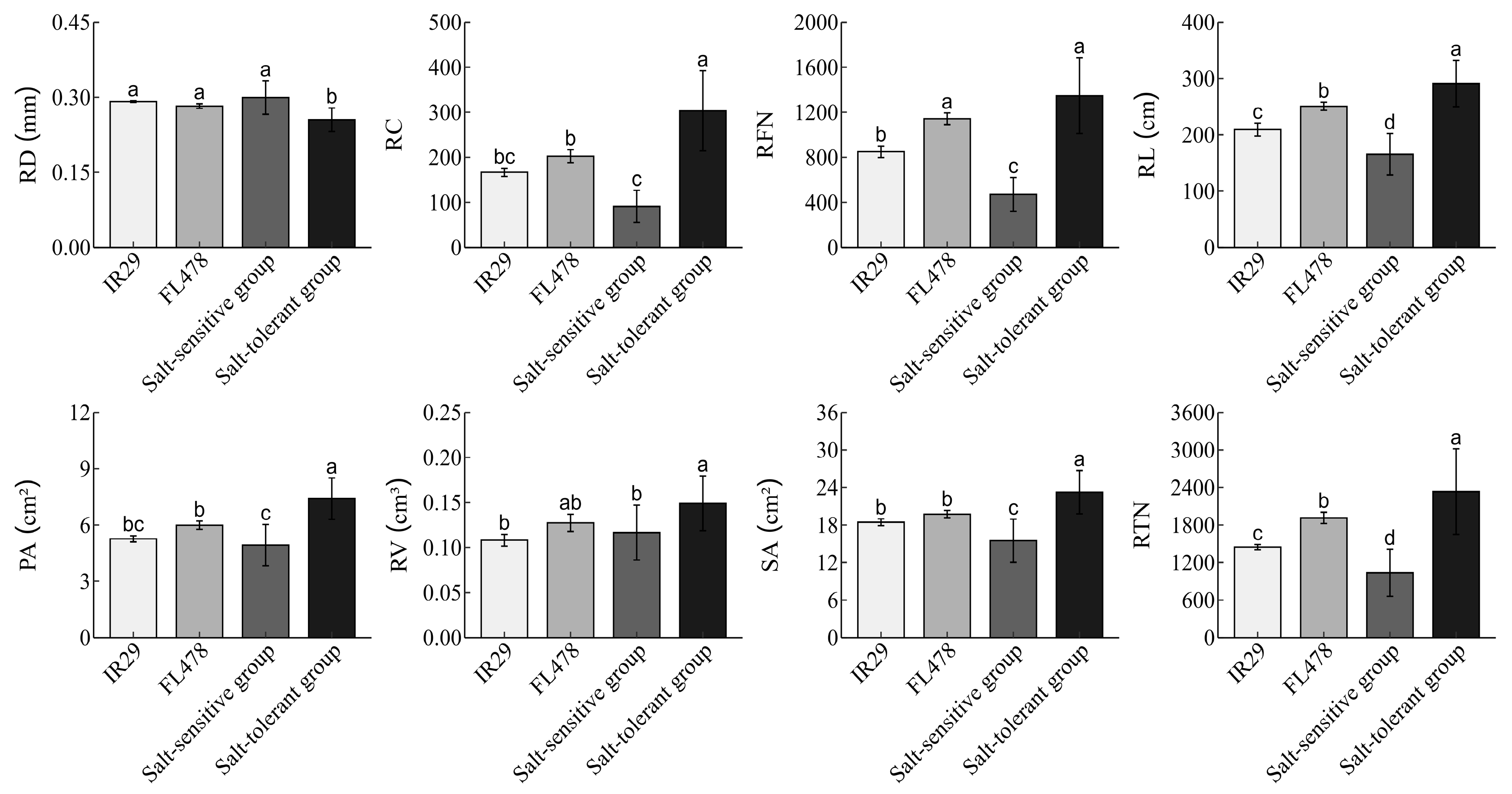
3.4. The Performance of the Selected Salt-Tolerance Varieties Compared to the Control Lines
3.5. Analysis of Favorable Alleles for the Selected Salt-Tolerance Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RD | Root diameter |
| RC | Root crossings |
| RFN | Root forks number |
| RL | Root length |
| PA | Project area |
| RV | Root volume |
| SA | Surface area |
| RTN | Root tips number |
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- Kodape, A.; Kodape, A.; Desai, R. Rice bran: Nutritional value, health benefits, and global implications for aflatoxin mitigation, cancer, diabetes, and diarrhea prevention. Food Chem. 2025, 464, 141749. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M. Genomics and transcriptomics to protect rice (Oryza sativa L.) from abiotic stressors: Pathways to achieving zero hunger. Front. Plant Sci. 2022, 13, 1002596. [Google Scholar] [CrossRef] [PubMed]
- Ahmadizadeh, M.; Babaeian-Jelodar, N.; Mohammadi-Nejad, G.; Bagheri, N.; Singh, R.K. High-density linkage mapping for agronomic and physiological traits of rice (Oryza sativa L.) under reproductive-stage salt stress. J. Genet. 2021, 100, 51. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Xu, Y.; Iqbal, A.; Liu, Y.; Zhang, Y.; Lin, Y.; Tang, L.; Wang, X.; Wang, J.; Huang, M.; et al. Comparative Transcriptome and Hormonal Analysis Reveals the Mechanisms of Salt Tolerance in Rice. Int. J. Mol. Sci. 2025, 26, 6660. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, T.; Zhu, K.; Wang, W.; Zhang, W.; Zhang, H.; Liu, L.; Zhang, Z.; Wang, Z.; Wang, B.; et al. Effects of Salt Stress on Grain Yield and Quality Parameters in Rice Cultivars with Differing Salt Tolerance. Plants 2023, 12, 3243. [Google Scholar] [CrossRef]
- Arminjon, L.; Lefort, F. Quick In Vitro Screening of PGPMs for Salt Tolerance and Evaluation of Induced Tolerance to Saline Stress in Tomato Culture. Microorganisms 2025, 13, 246. [Google Scholar] [CrossRef]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2024, 66, 303–329. [Google Scholar] [CrossRef]
- Lekklar, C.; Suriya-Arunroj, D.; Pongpanich, M.; Comai, L.; Kositsup, B.; Chadchawan, S.; Buaboocha, T. Comparative Genomic Analysis of Rice with Contrasting Photosynthesis and Grain Production under Salt Stress. Genes 2019, 10, 562. [Google Scholar] [CrossRef]
- Qin, H.; Li, Y.; Huang, R. Advances and Challenges in the Breeding of Salt-Tolerant Rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3 Biotech 2020, 10, 361. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, J.; Li, Y.; Quan, R.; Wang, J.; Huang, R.; Qin, H. Salt Stress Promotes Abscisic Acid Accumulation to Affect Cell Proliferation and Expansion of Primary Roots in Rice. Int. J. Mol. Sci. 2021, 22, 10892. [Google Scholar] [CrossRef]
- Fan, X.; Jiang, H.; Meng, L.; Chen, J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674. [Google Scholar] [CrossRef] [PubMed]
- Kojonna, T.; Suttiyut, T.; Khunpolwattana, N.; Pongpanich, M.; Suriya-Arunroj, D.; Comai, L.; Buaboocha, T.; Chadchawan, S. Identification of a Negative Regulator for Salt Tolerance at Seedling Stage via a Genome-Wide Association Study of Thai Rice Populations. Int. J. Mol. Sci. 2022, 23, 1842. [Google Scholar] [CrossRef] [PubMed]
- Susmitha, P.; Kumar, P.; Yadav, P.; Sahoo, S.; Kaur, G.; Pandey, M.K.; Singh, V.; Tseng, T.M.; Gangurde, S.S. Genome-wide association study as a powerful tool for dissecting competitive traits in legumes. Front. Plant Sci. 2023, 14, 1123631. [Google Scholar] [CrossRef]
- Ashfaq, M.; Rasheed, A.; Zhu, R.; Ali, M.; Javed, M.A.; Anwar, A.; Tabassum, J.; Shaheen, S.; Wu, X. Genome-Wide Association Mapping for Yield and Yield-Related Traits in Rice (Oryza sativa L.) Using SNPs Markers. Genes 2023, 14, 1089. [Google Scholar] [CrossRef]
- Yu, J.; Zhu, C.; Xuan, W.; An, H.; Tian, Y.; Wang, B.; Chi, W.; Chen, G.; Ge, Y.; Li, J.; et al. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef]
- Imran, S.; Tsuchiya, Y.; Tran, S.T.H.; Katsuhara, M. Identification and Characterization of Rice OsHKT1;3 Variants. Plants 2021, 10, 2006. [Google Scholar] [CrossRef]
- Le, T.D.; Gathignol, F.; Vu, H.T.; Nguyen, K.L.; Tran, L.H.; Vu, H.T.T.; Dinh, T.X.; Lazennec, F.; Pham, X.H.; Véry, A.A.; et al. Genome-Wide Association Mapping of Salinity Tolerance at the Seedling Stage in a Panel of Vietnamese Landraces Reveals New Valuable QTLs for Salinity Stress Tolerance Breeding in Rice. Plants 2021, 10, 1088. [Google Scholar] [CrossRef]
- Nayyeripasand, L.; Garoosi, G.A.; Ahmadikhah, A. Genome-Wide Association Study (GWAS) to Identify Salt-Tolerance QTLs Carrying Novel Candidate Genes in Rice During Early Vegetative Stage. Rice 2021, 14, 9. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, Y.; Testerink, C. Root dynamic growth strategies in response to salinity. Plant Cell Environ. 2022, 45, 695–704. [Google Scholar] [CrossRef]
- Zhou, Y.; Feng, C.; Wang, Y.; Yun, C.; Zou, X.; Cheng, N.; Zhang, W.; Jing, Y.; Li, H. Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding. Int. J. Mol. Sci. 2024, 25, 10940. [Google Scholar] [CrossRef]
- Kim, D.W.; Rakwal, R.; Agrawal, G.K.; Jung, Y.H.; Shibato, J.; Jwa, N.S.; Iwahashi, Y.; Iwahashi, H.; Kim, D.H.; Shim, I.S.; et al. A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 2005, 26, 4521–4539. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Ma, J.; Wei, H.; Xiao, F.; Wang, Y.; Jahan, N.; Hazman, M.; Qian, Q.; Shang, L.; Guo, L. Combining GWAS, Genome-Wide Domestication and a Transcriptomic Analysis Reveals the Loci and Natural Alleles of Salt Tolerance in Rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 912637. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Y.W.; Xiang, Y.; Liu, M.H.; Zhang, Y.M. IIIVmrMLM: The R and C++ tools associated with 3VmrMLM, a comprehensive GWAS method for dissecting quantitative traits. Mol. Plant 2022, 15, 1251–1253. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.T.; Bandillo, N.; Al Shiblawi, F.R.A.; Sharma, S.; Liu, K.; Du, Q.; Schmitz, A.J.; Zhang, C.; Véry, A.A.; Lorenz, A.J.; et al. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genet. 2017, 13, e1006823. [Google Scholar] [CrossRef]
- Raghuvanshi, R.; Srivastava, A.K.; Verulkar, S.; Suprasanna, P. Unlocking Allelic Diversity for Sustainable Development of Salinity Stress Tolerance in Rice. Curr. Genom. 2021, 22, 393–403. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A Magnesium Transporter OsMGT1 Plays a Critical Role in Salt Tolerance in Rice. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, K.; Gong, Z.; Zhang, Y.; Meng, Y.; Liu, Q. Molecular manipulations of miR398 increase rice grain yield under different conditions. Front. Plant Sci. 2022, 13, 1037604. [Google Scholar] [CrossRef]
- Rossatto, T.; do Amaral, M.N.; Benitez, L.C.; Vighi, I.L.; Braga, E.J.B.; de Magalhães Júnior, A.M.; Maia, M.A.C.; da Silva Pinto, L. Gene expression and activity of antioxidant enzymes in rice plants, cv. BRS AG, under saline stress. Physiol. Mol. Biol. Plants 2017, 23, 865–875. [Google Scholar] [CrossRef]
- Song, S.Y.; Chen, Y.; Chen, J.; Dai, X.Y.; Zhang, W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Oda, Y.; Kobayashi, N.I.; Tanoi, K.; Ma, J.F.; Itou, Y.; Katsuhara, M.; Itou, T.; Horie, T. T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress. Int. J. Mol. Sci. 2018, 19, 235. [Google Scholar] [CrossRef]
- Khan, I.; Mohamed, S.; Regnault, T.; Mieulet, D.; Guiderdoni, E.; Sentenac, H.; Véry, A.A. Constitutive Contribution by the Rice OsHKT1;4 Na+ Transporter to Xylem Sap Desalinization and Low Na+ Accumulation in Young Leaves Under Low as High External Na+ Conditions. Front. Plant Sci. 2020, 11, 1130. [Google Scholar] [CrossRef]
- Horie, T.; Sugawara, M.; Okada, T.; Taira, K.; Kaothien-Nakayama, P.; Katsuhara, M.; Shinmyo, A.; Nakayama, H. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 2011, 111, 346–356. [Google Scholar] [CrossRef]
- Ye, S.; Huang, Z.; Zhao, G.; Zhai, R.; Ye, J.; Wu, M.; Yu, F.; Zhu, G.; Zhang, X. Differential Physiological Responses to Salt Stress between Salt-Sensitive and Salt-Tolerant japonica Rice Cultivars at the Post-Germination and Seedling Stages. Plants 2021, 10, 2433. [Google Scholar] [CrossRef]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef]
- Guo, Y.; Song, Y. Differential proteomic analysis of apoplastic proteins during initial phase of salt stress in rice. Plant Signal. Behav. 2009, 4, 121–122. [Google Scholar] [CrossRef]
- Zang, J.; Sun, Y.; Wang, Y.; Yang, J.; Li, F.; Zhou, Y.; Zhu, L.; Jessica, R.; Mohammadhosein, F.; Xu, J.; et al. Dissection of genetic overlap of salt tolerance QTLs at the seedling and tillering stages using backcross introgression lines in rice. Science in China. Ser. C Life Sci. 2008, 51, 583–591. [Google Scholar] [CrossRef]
- Li, X.; Chen, R.; Chu, Y.; Huang, J.; Jin, L.; Wang, G.; Huang, J. Overexpression of RCc3 improves root system architecture and enhances salt tolerance in rice. Plant Physiol. Biochem. 2018, 130, 566–576. [Google Scholar] [CrossRef]
- Xu, S.; Zheng, J.; Du, H.; Du, X.; Li, C.; Duan, Y.; Cai, Y.; Wang, J.; Liu, H.; Yang, L.; et al. GWAS combined with linkage analysis reveals major QTLs and candidate genes of salt tolerance in Japonica rice seedlings. Front. Plant Sci. 2024, 15, 1462856. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, X.; Liu, J.; Gao, X.; Bai, J.; Hao, Y.; Cui, H. A mechanism coordinating root elongation, endodermal differentiation, redox homeostasis and stress response. Plant J. Cell Mol. Biol. 2021, 107, 1029–1039. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, H.; Wang, S.; Wang, Y.; Wang, Z.; Zheng, X.; Chen, J.; Yang, B.; Shan, H. Integrative multi-omics analysis reveals the potential mechanism by which Streptomyces pactum Act12 enhances wheat root drought tolerance by coordinating phytohormones and metabolic pathways. BMC Plant Biol. 2025, 25, 707. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, P.; Hamani, A.K.M.; Wan, S.; Gao, Y.; Wang, X. Effects of Single and Combined Drought and Salinity Stress on the Root Morphological Characteristics and Root Hydraulic Conductivity of Different Winter Wheat Varieties. Plants 2023, 12, 2694. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ren, M.; Wu, Y.; Wang, L.; Zhao, K.; Gao, H.; Li, M.; Liu, Y.; Zhu, J.; Xu, J.; et al. A root system architecture regulator modulates OsPIN2 polar localization in rice. Nat. Commun. 2025, 16, 15. [Google Scholar] [CrossRef] [PubMed]


| Root Diameter (RD) (mm) | Root Crossings (RC) | Root Forks Number (RFN) | Root Length (RL) (cm) | Project Area (PA) (cm2) | Root Volume (RV) (cm3) | Surface Area (SA) (cm2) | Root Tips Number (RTN) | |
|---|---|---|---|---|---|---|---|---|
| Mean | 0.29 | 154.65 | 795.59 | 211.80 | 6.09 | 0.14 | 19.14 | 1440.17 |
| Standard deviation | 0.03 | 72.22 | 279.92 | 52.21 | 1.42 | 0.04 | 4.45 | 469.82 |
| Skewness | 0.67 | 0.98 | 0.93 | 0.80 | 0.63 | 0.78 | 0.63 | 0.49 |
| Kurtosis | 0.67 | 0.75 | 1.07 | 1.51 | 0.96 | 0.61 | 0.96 | 0.06 |
| CV (%) | 10.34 | 46.70 | 35.18 | 24.65 | 23.32 | 28.57 | 23.25 | 32.62 |
| Minimum | 0.21 | 30.50 | 238.50 | 109.54 | 3.34 | 0.07 | 10.50 | 346.50 |
| Maximum | 0.41 | 389.50 | 1819.50 | 439.94 | 11.64 | 0.29 | 36.56 | 2792.00 |
| Marker | Chromosome | Position (bp) | LOD | PVE (%) | Favorable Allele |
|---|---|---|---|---|---|
| SNP2-49524748 | 2 | 6,253,825 | 5.71 | 3.00 | C |
| SNP2-51399642 | 2 | 8,128,719 | 4.40 | 2.38 | T |
| SNP2-74166363 | 2 | 30,895,440 | 5.46 | 1.42 | G |
| SNP4-115698850 | 4 | 76,858 | 5.41 | 3.19 | G |
| SNP4-117616752 | 4 | 1,994,760 | 45.08 | 5.59 | A |
| SNP4-123092399 | 4 | 7,470,407 | 32.88 | 6.69 | T |
| SNP4-147472833 | 4 | 31,850,841 | 4.44 | 1.82 | C |
| SNP5-180922039 | 5 | 297,97,353 | 10.37 | 5.63 | A |
| SNP7-221576010 | 7 | 9,244,103 | 11.20 | 3.51 | T |
| SNP8-252176489 | 8 | 10,146,961 | 15.91 | 8.40 | A |
| SNP8-260086690 | 8 | 18,057,162 | 20.55 | 7.18 | G |
| SNP9-286257626 | 9 | 15,785,076 | 8.11 | 4.82 | C |
| SNP11-317386697 | 11 | 694,140 | 14.44 | 3.35 | G |
| SNP11-322275422 | 11 | 5,582,865 | 13.07 | 2.10 | G |
| SNP12-354603209 | 12 | 8,889,546 | 13.74 | 2.58 | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Z.; Hao, D.; Lu, Z.; Yang, J.; Geng, Z.; Meng, C.; Cui, Y. Unveiling Genetic Loci for Root Morphology and Salt Response at Rice Seedling Stage via Genome-Wide Association Studies. Life 2025, 15, 1595. https://doi.org/10.3390/life15101595
Xue Z, Hao D, Lu Z, Yang J, Geng Z, Meng C, Cui Y. Unveiling Genetic Loci for Root Morphology and Salt Response at Rice Seedling Stage via Genome-Wide Association Studies. Life. 2025; 15(10):1595. https://doi.org/10.3390/life15101595
Chicago/Turabian StyleXue, Zifan, De Hao, Zheyu Lu, Jie Yang, Ziteng Geng, Chengsheng Meng, and Yanru Cui. 2025. "Unveiling Genetic Loci for Root Morphology and Salt Response at Rice Seedling Stage via Genome-Wide Association Studies" Life 15, no. 10: 1595. https://doi.org/10.3390/life15101595
APA StyleXue, Z., Hao, D., Lu, Z., Yang, J., Geng, Z., Meng, C., & Cui, Y. (2025). Unveiling Genetic Loci for Root Morphology and Salt Response at Rice Seedling Stage via Genome-Wide Association Studies. Life, 15(10), 1595. https://doi.org/10.3390/life15101595






