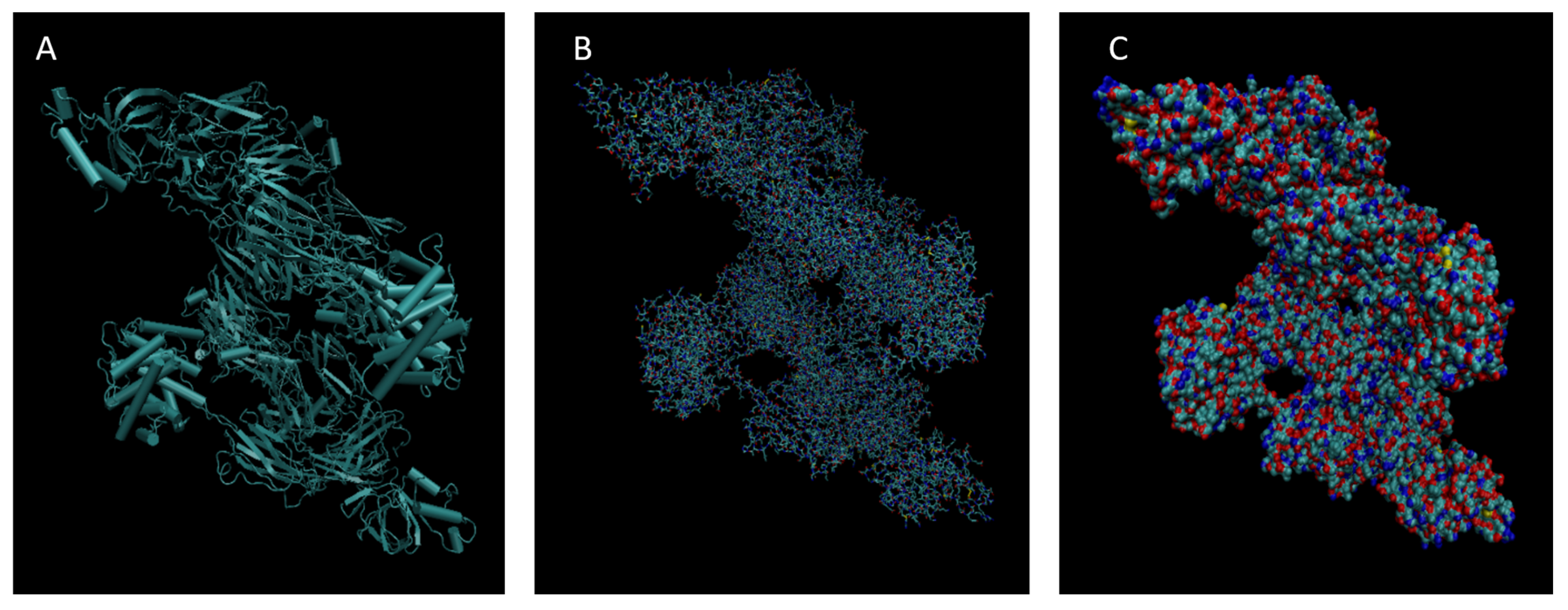
Figure 1.
Cartoon (A), Bond (B), and Surface Model (C) of Albumin.
Figure 1.
Cartoon (A), Bond (B), and Surface Model (C) of Albumin.
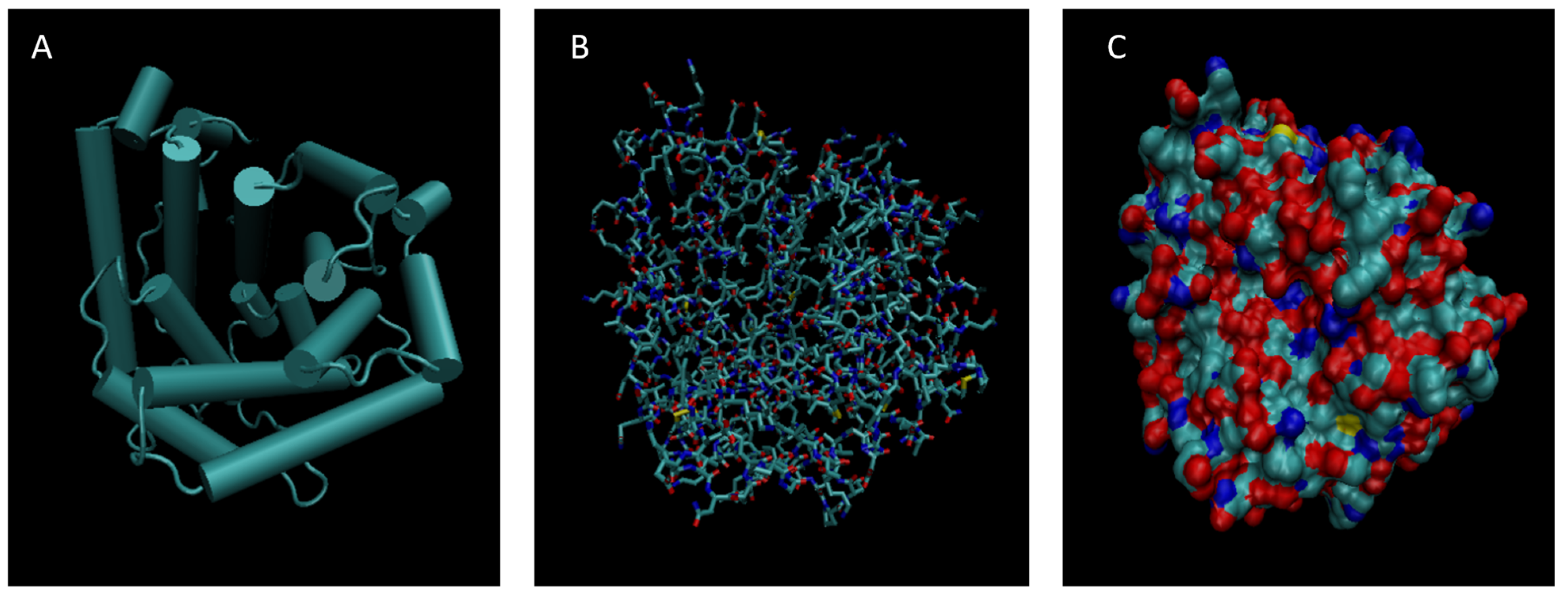
Figure 2.
Cartoon (A), Bond (B), and Surface Model (C) of Alpha-1-microglobulin.
Figure 2.
Cartoon (A), Bond (B), and Surface Model (C) of Alpha-1-microglobulin.
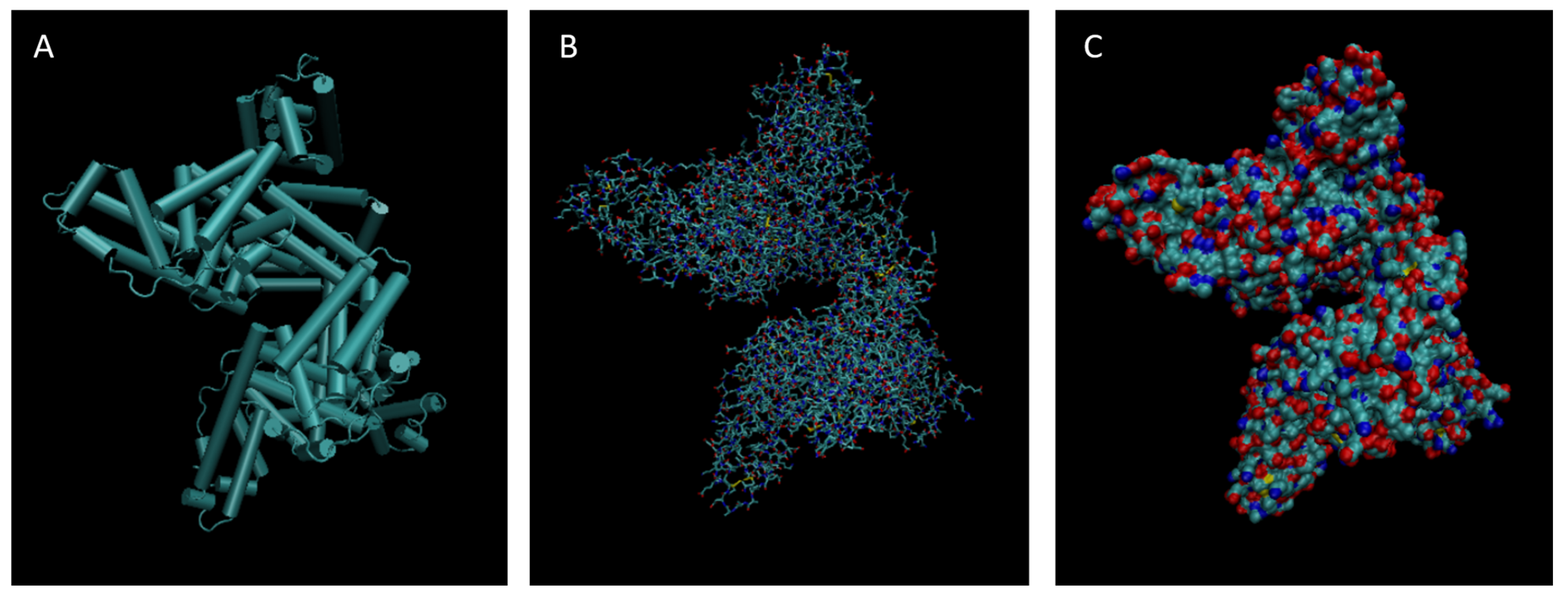
Figure 3.
Cartoon (A), Bond (B), and Surface Model (C) of Alpha-2-macroglobulin.
Figure 3.
Cartoon (A), Bond (B), and Surface Model (C) of Alpha-2-macroglobulin.
Figure 4.
Cartoon (A), Bond (B), and Surface Model (C) of Apolipoprotein E.
Figure 4.
Cartoon (A), Bond (B), and Surface Model (C) of Apolipoprotein E.
Figure 5.
Cartoon (A), Bond (B), and Surface Model (C) of Complement Component 3.
Figure 5.
Cartoon (A), Bond (B), and Surface Model (C) of Complement Component 3.
Figure 6.
Cartoon (A), Bond (B), and Surface Model (C) of Haptoglobin.
Figure 6.
Cartoon (A), Bond (B), and Surface Model (C) of Haptoglobin.
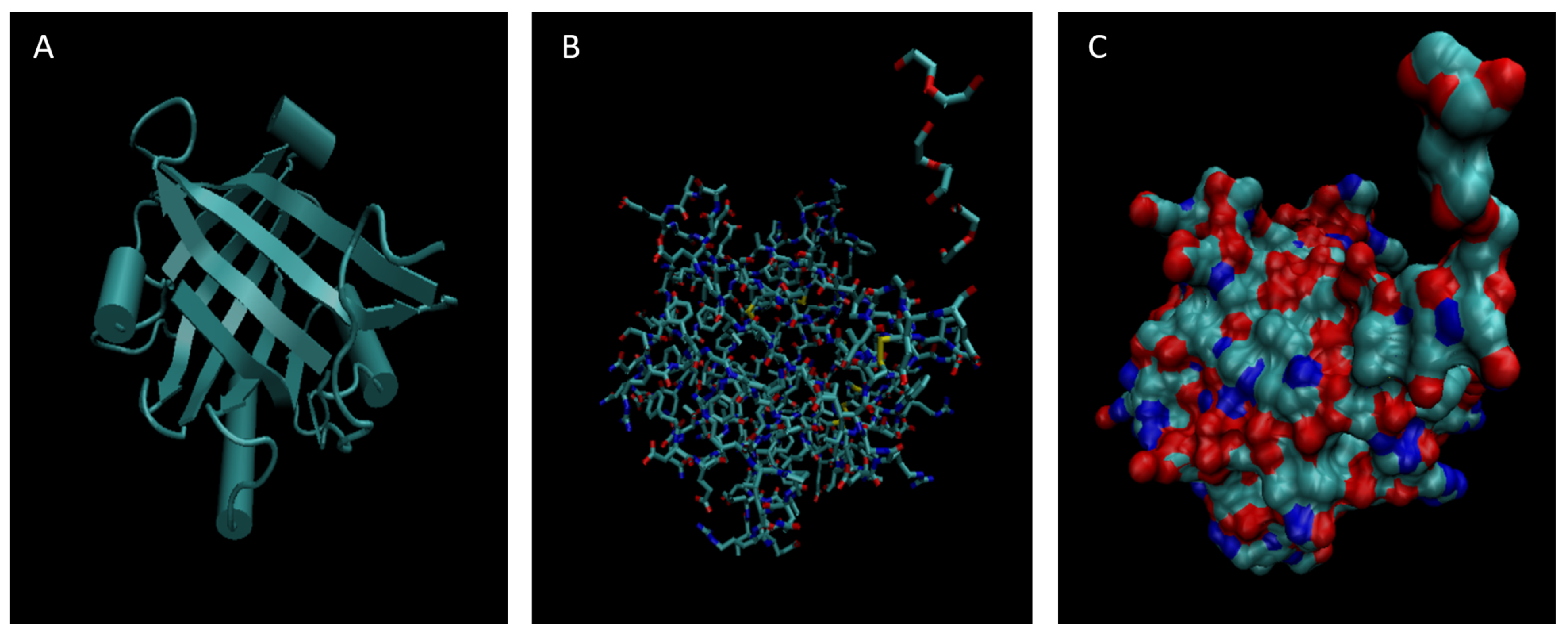
Figure 7.
Cartoon (A), Bond (B), and Surface Model (C) of Orosomucoid 1.
Figure 7.
Cartoon (A), Bond (B), and Surface Model (C) of Orosomucoid 1.
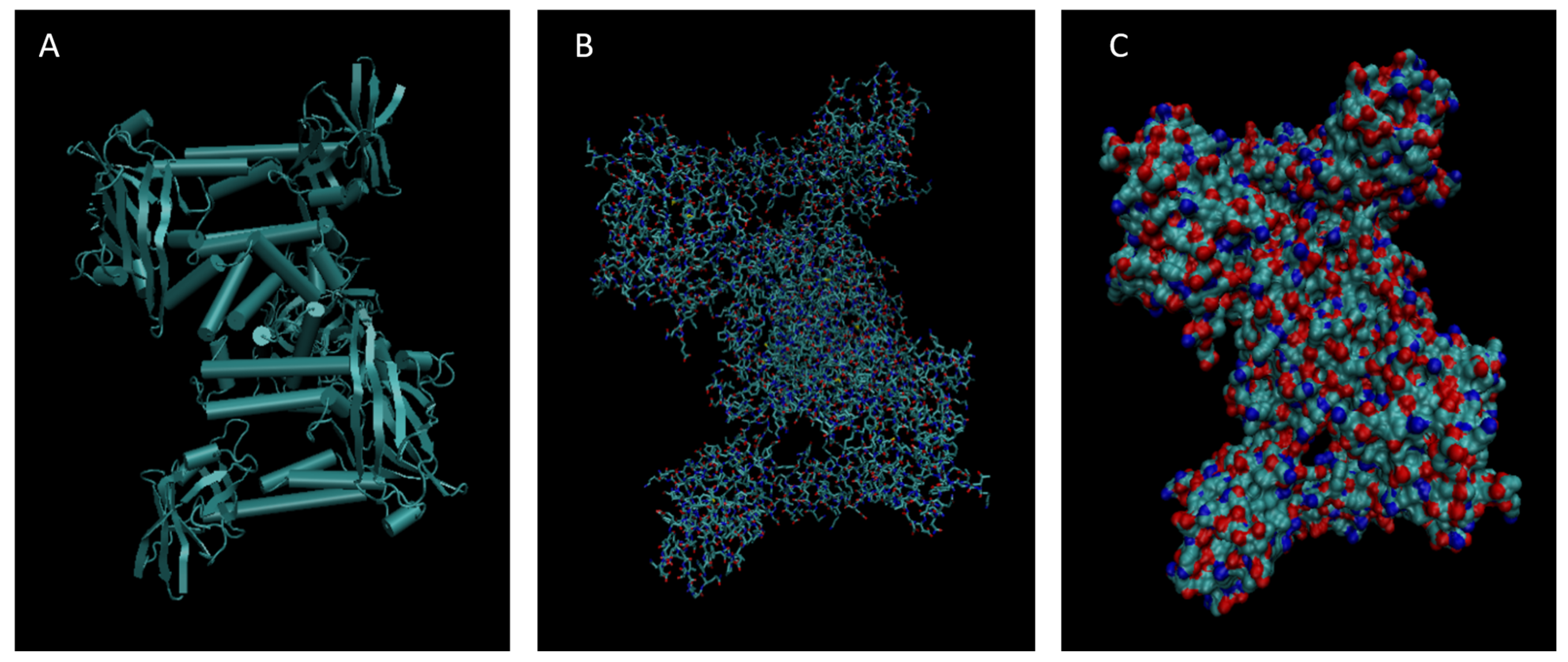
Figure 8.
Cartoon (A), Bond (B), and Surface Model (C) of Complement Component 4B.
Figure 8.
Cartoon (A), Bond (B), and Surface Model (C) of Complement Component 4B.
Figure 9.
Cartoon (A), Bond (B), and Surface Model (C) of Retinol-binding protein 4.
Figure 9.
Cartoon (A), Bond (B), and Surface Model (C) of Retinol-binding protein 4.
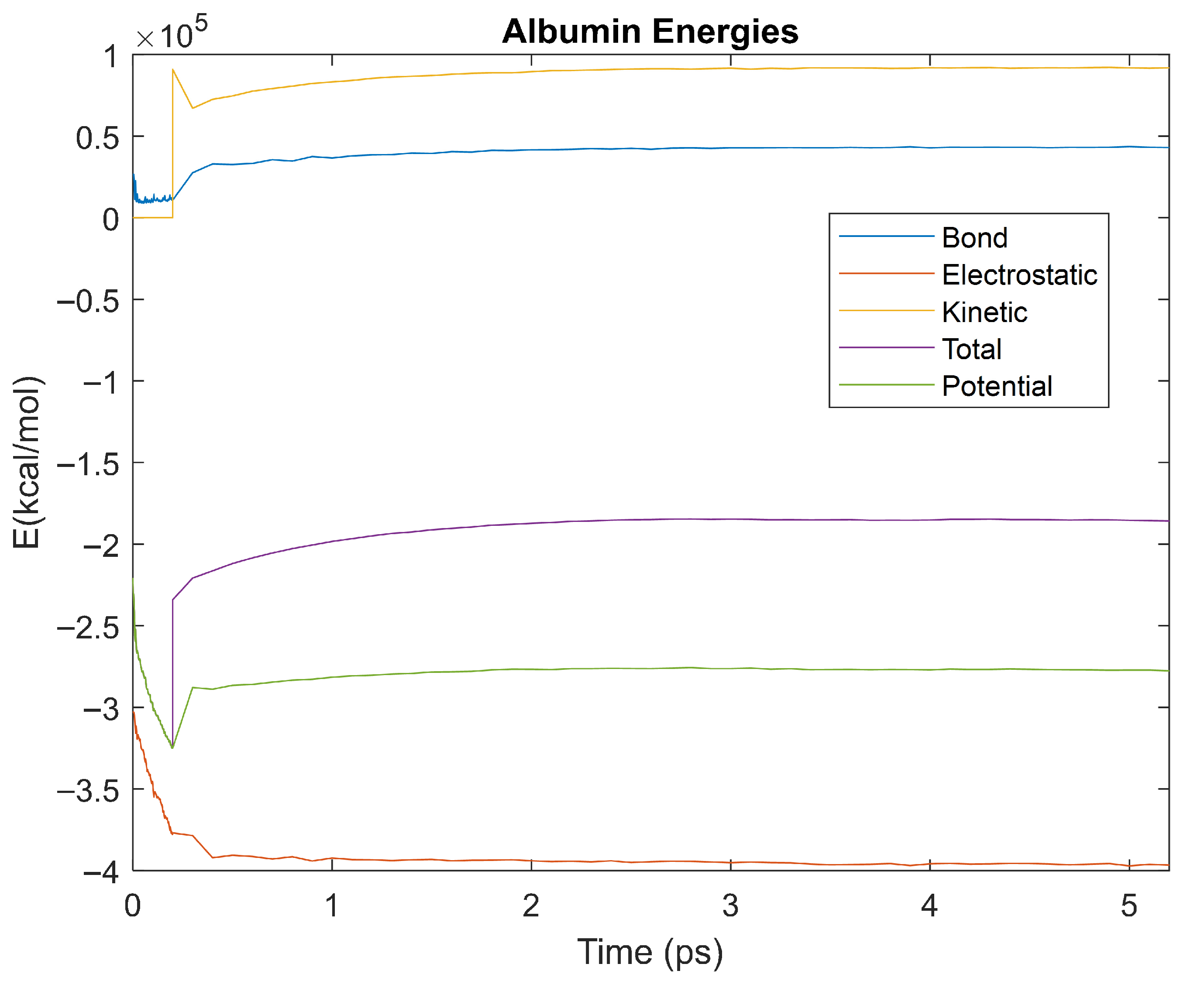
Figure 10.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 310 K 5 ps simulation.
Figure 10.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 310 K 5 ps simulation.
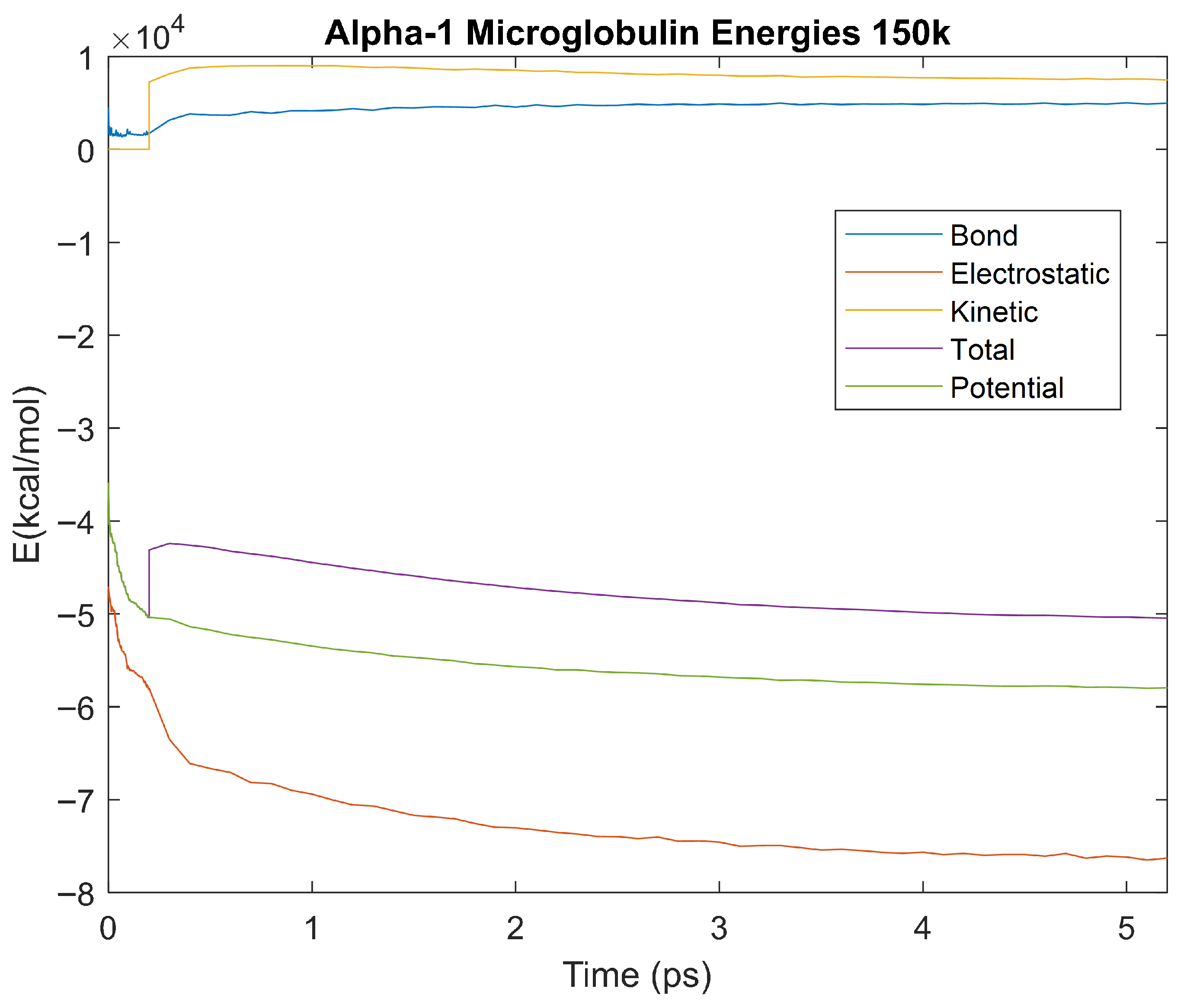
Figure 11.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 310 K 5 ps simulation.
Figure 11.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 310 K 5 ps simulation.
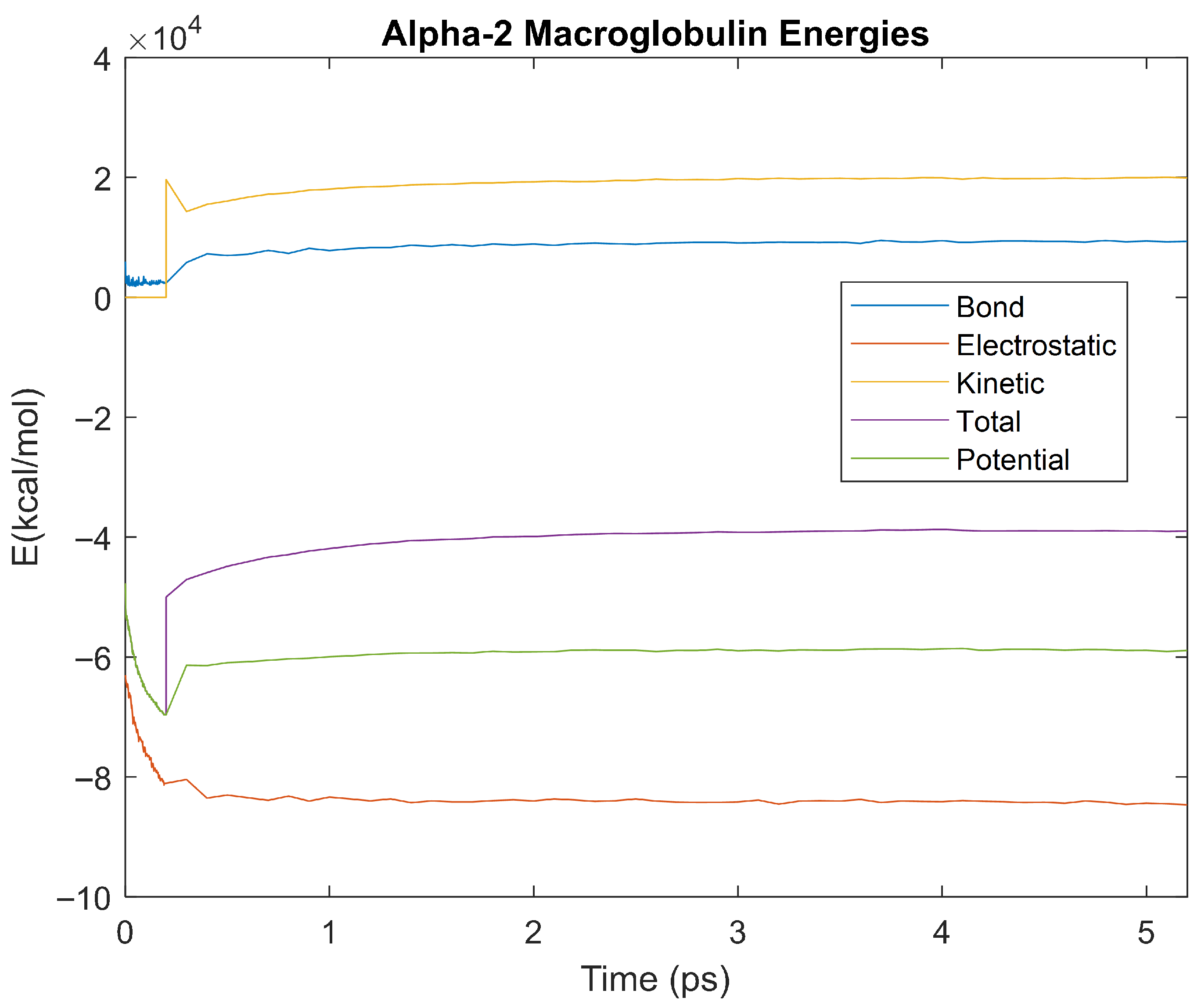
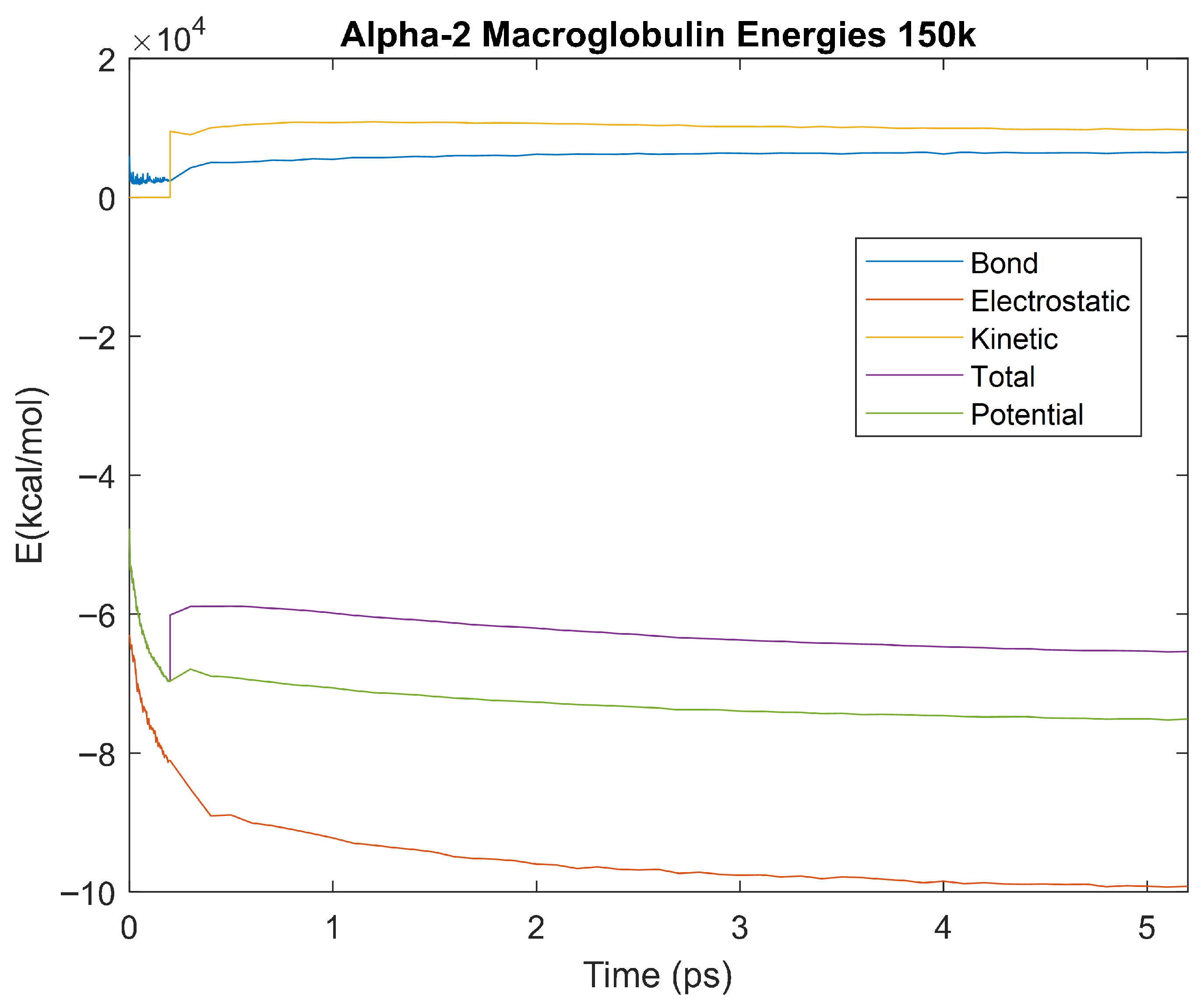
Figure 12.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 310 K 5 ps simulation.
Figure 12.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 310 K 5 ps simulation.
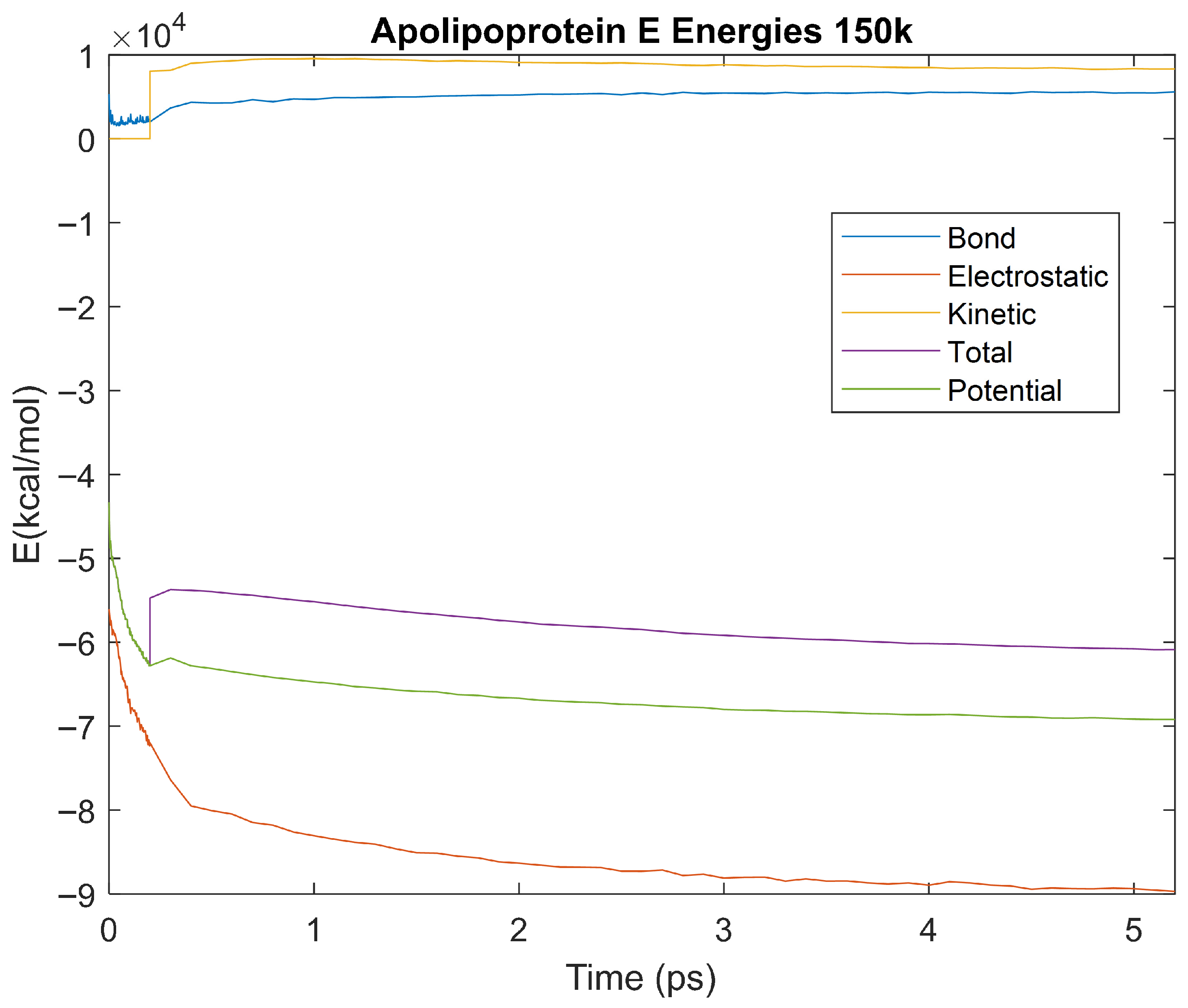
Figure 13.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 310 K 5 ps simulation.
Figure 13.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 310 K 5 ps simulation.
Figure 14.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 310 K 5 ps simulation.
Figure 14.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 310 K 5 ps simulation.
Figure 15.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 310 K 5 ps simulation.
Figure 15.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 310 K 5 ps simulation.
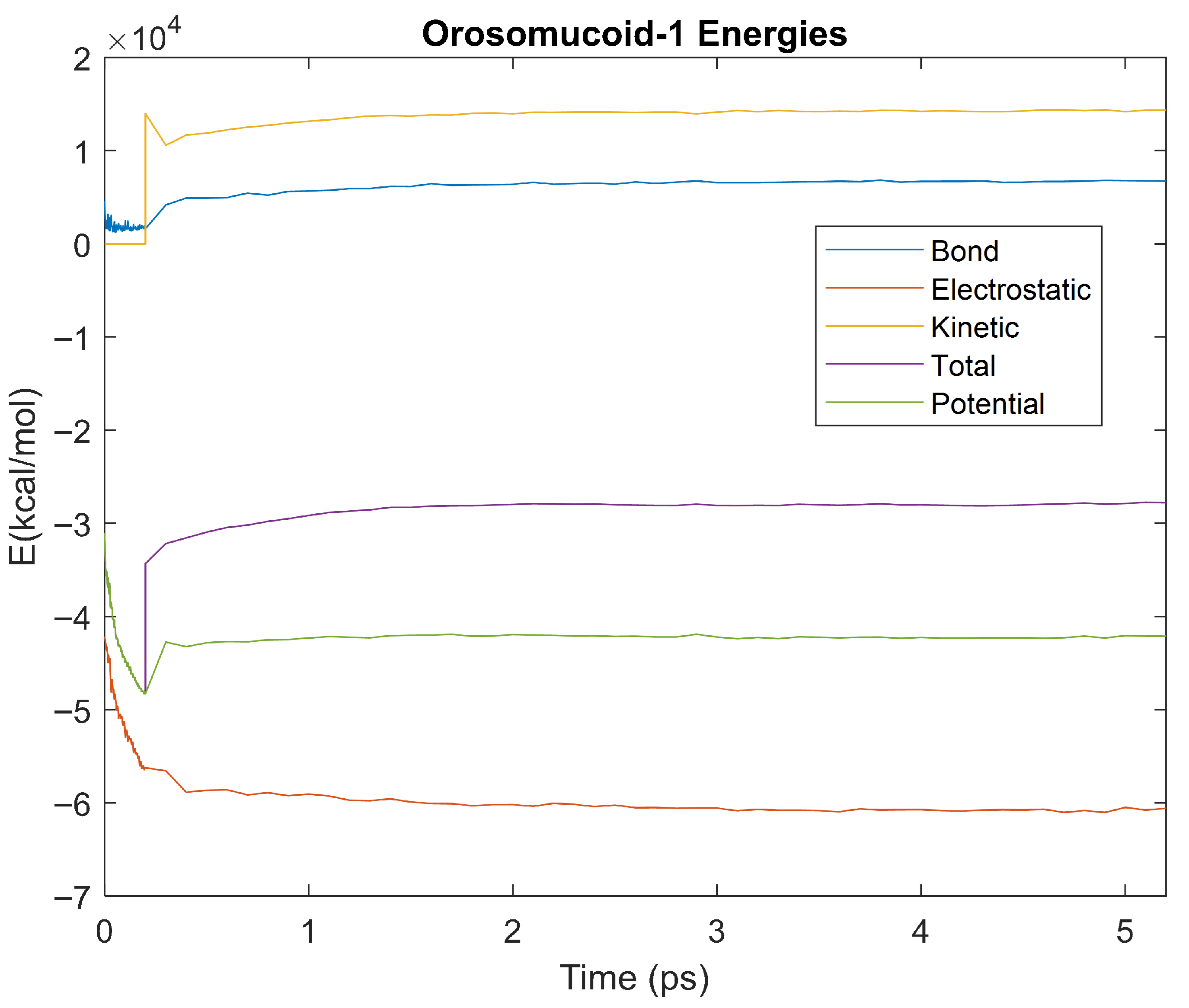
Figure 16.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid 1 over the 310 K 5 ps simulation.
Figure 16.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid 1 over the 310 K 5 ps simulation.
Figure 17.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 310 K 5 ps simulation.
Figure 17.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 310 K 5 ps simulation.
Figure 18.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-binding Protein 4 over the 310 K 5 ps simulation.
Figure 18.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-binding Protein 4 over the 310 K 5 ps simulation.
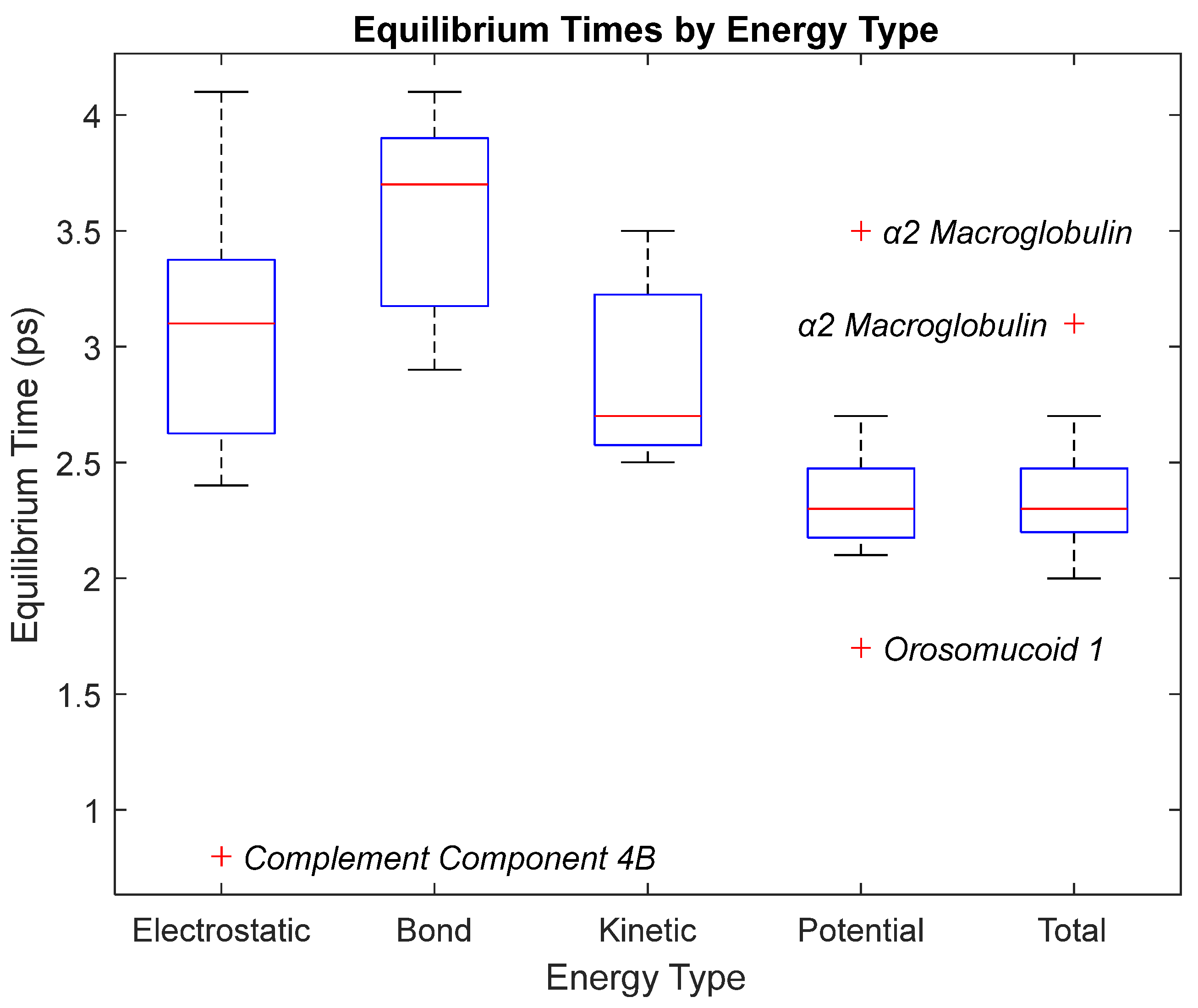
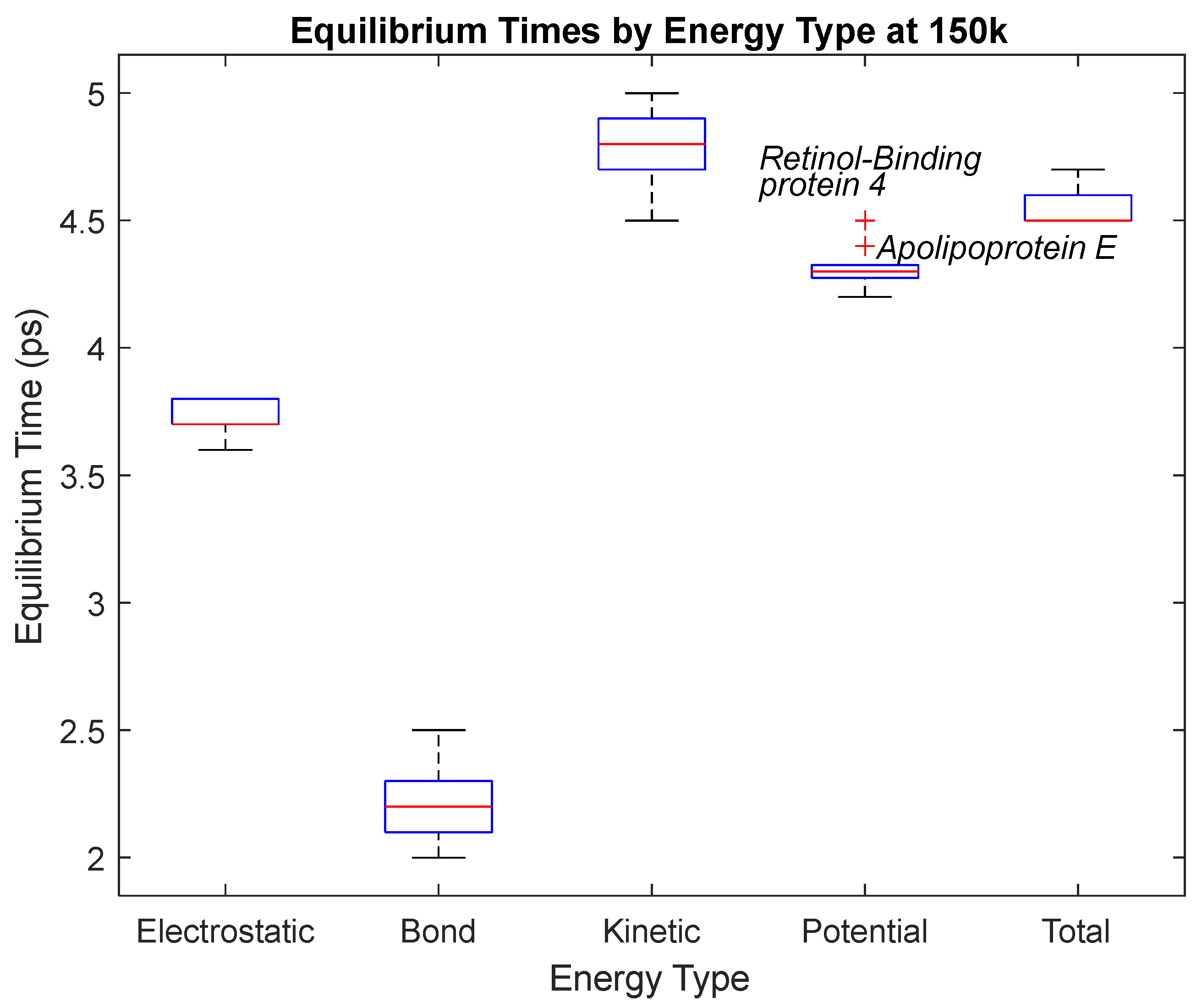
Figure 19.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 3. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarker.
Figure 19.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 3. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarker.
Figure 20.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 150 K 5 ps simulation.
Figure 20.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 150 K 5 ps simulation.
Figure 21.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 150 K 5 ps simulation.
Figure 21.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 150 K 5 ps simulation.
Figure 22.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 150 K 5 ps simulation.
Figure 22.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 150 K 5 ps simulation.
Figure 23.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 150 K 5 ps simulation.
Figure 23.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 150 K 5 ps simulation.
Figure 24.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 150 K 5 ps simulation.
Figure 24.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 150 K 5 ps simulation.
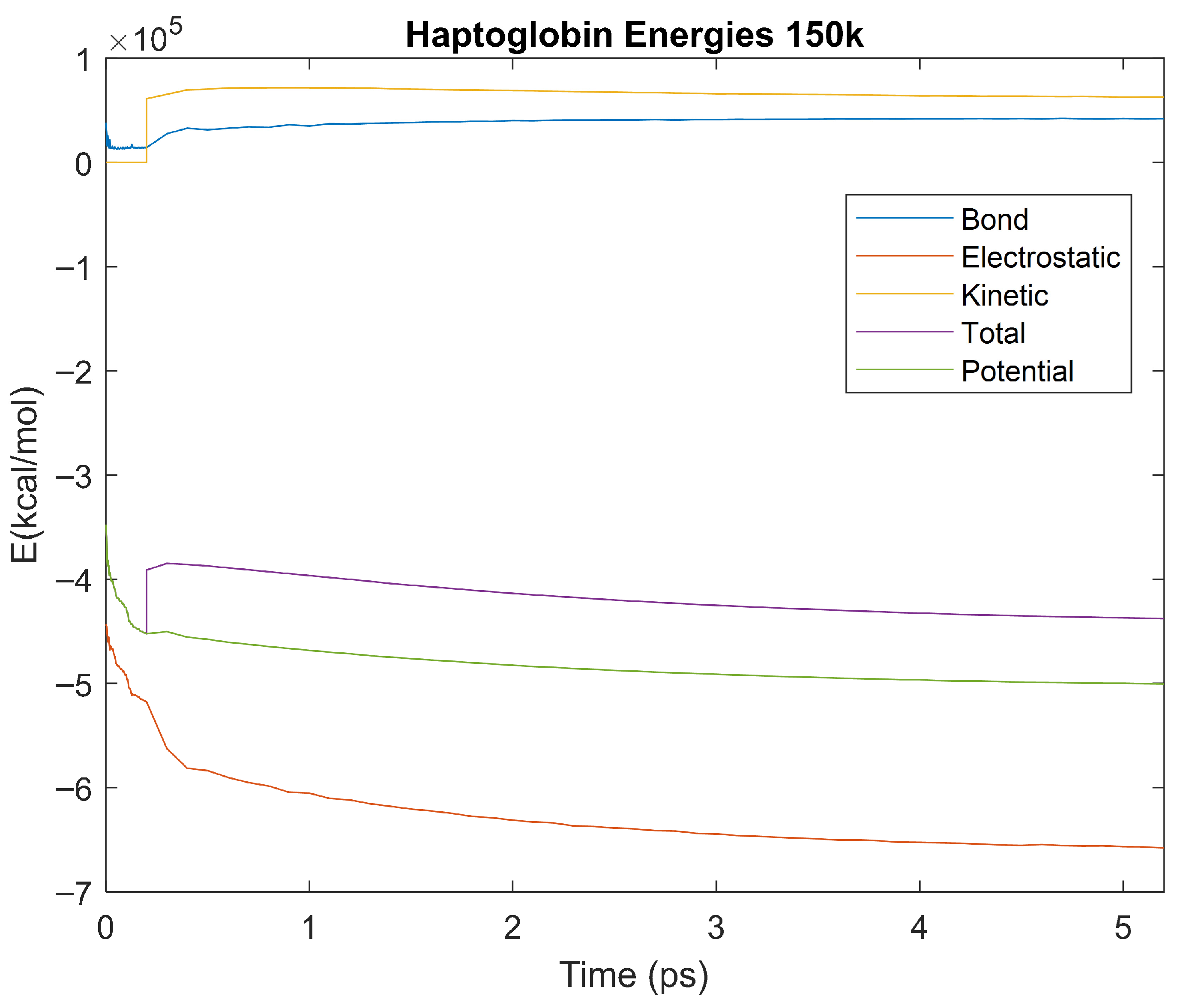
Figure 25.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 150 K 5 ps simulation.
Figure 25.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 150 K 5 ps simulation.
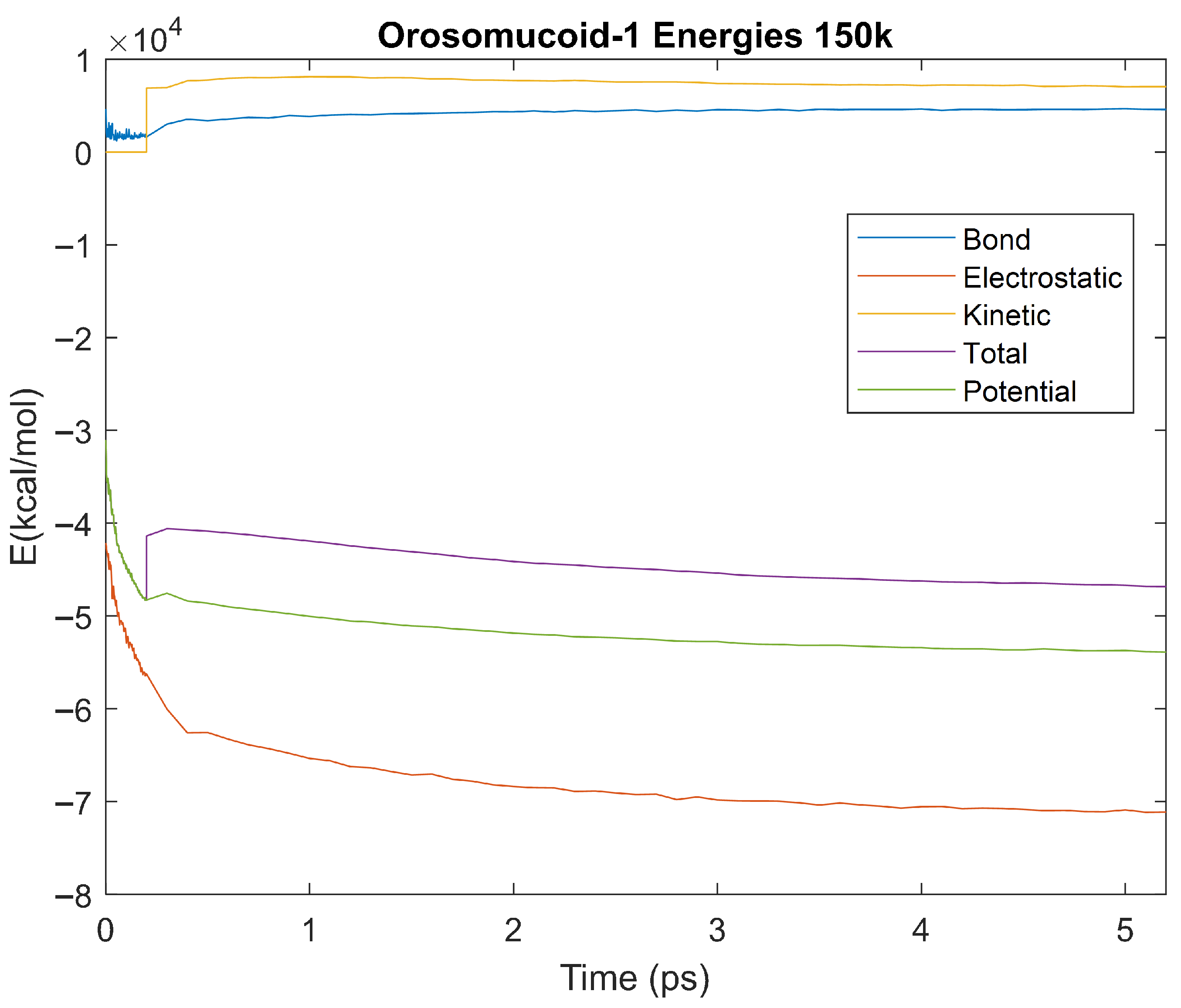
Figure 26.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 150 K 5 ps simulation.
Figure 26.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 150 K 5 ps simulation.
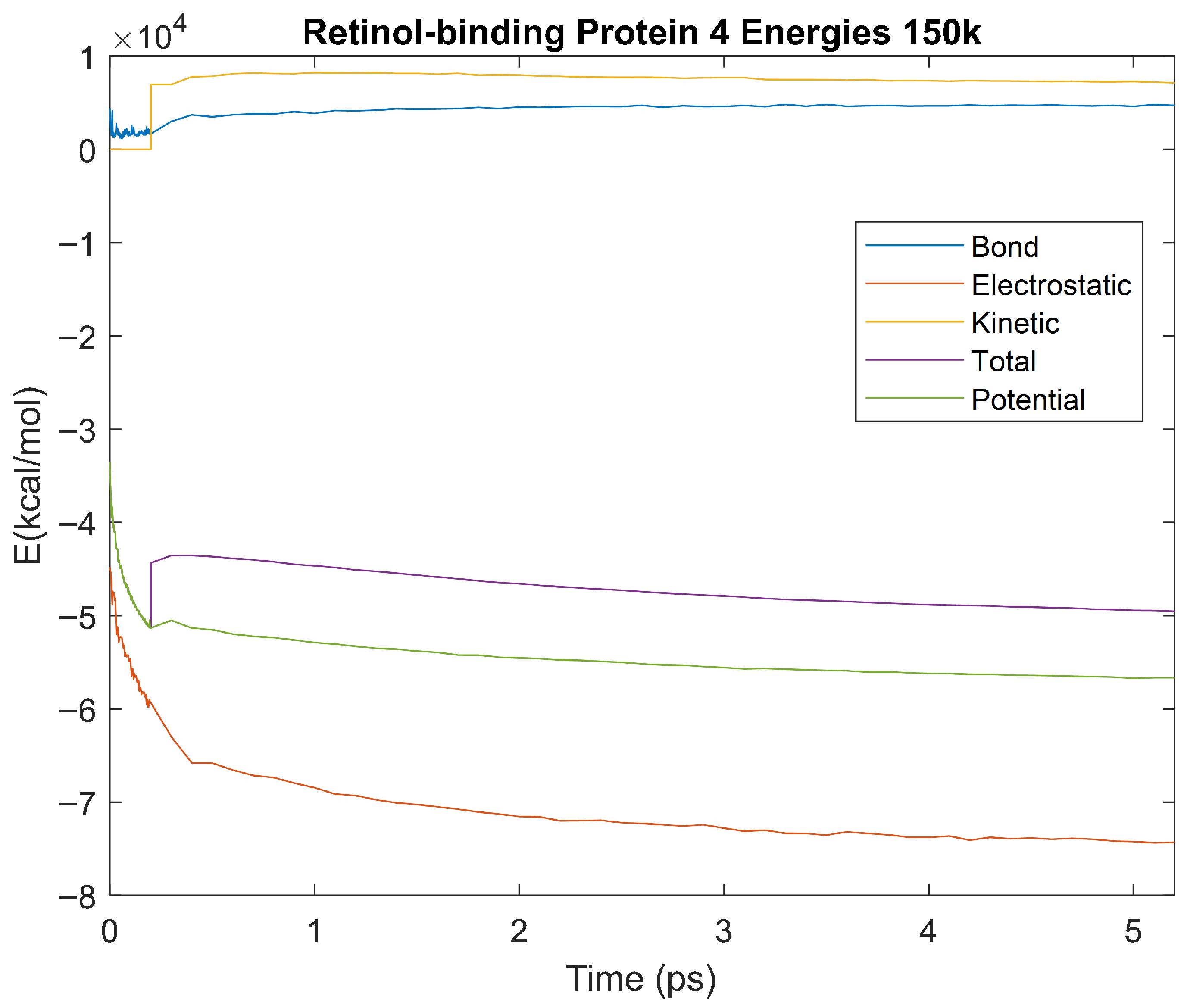
Figure 27.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-Binding Protein 4 over the 150 K 5 ps simulation.
Figure 27.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-Binding Protein 4 over the 150 K 5 ps simulation.
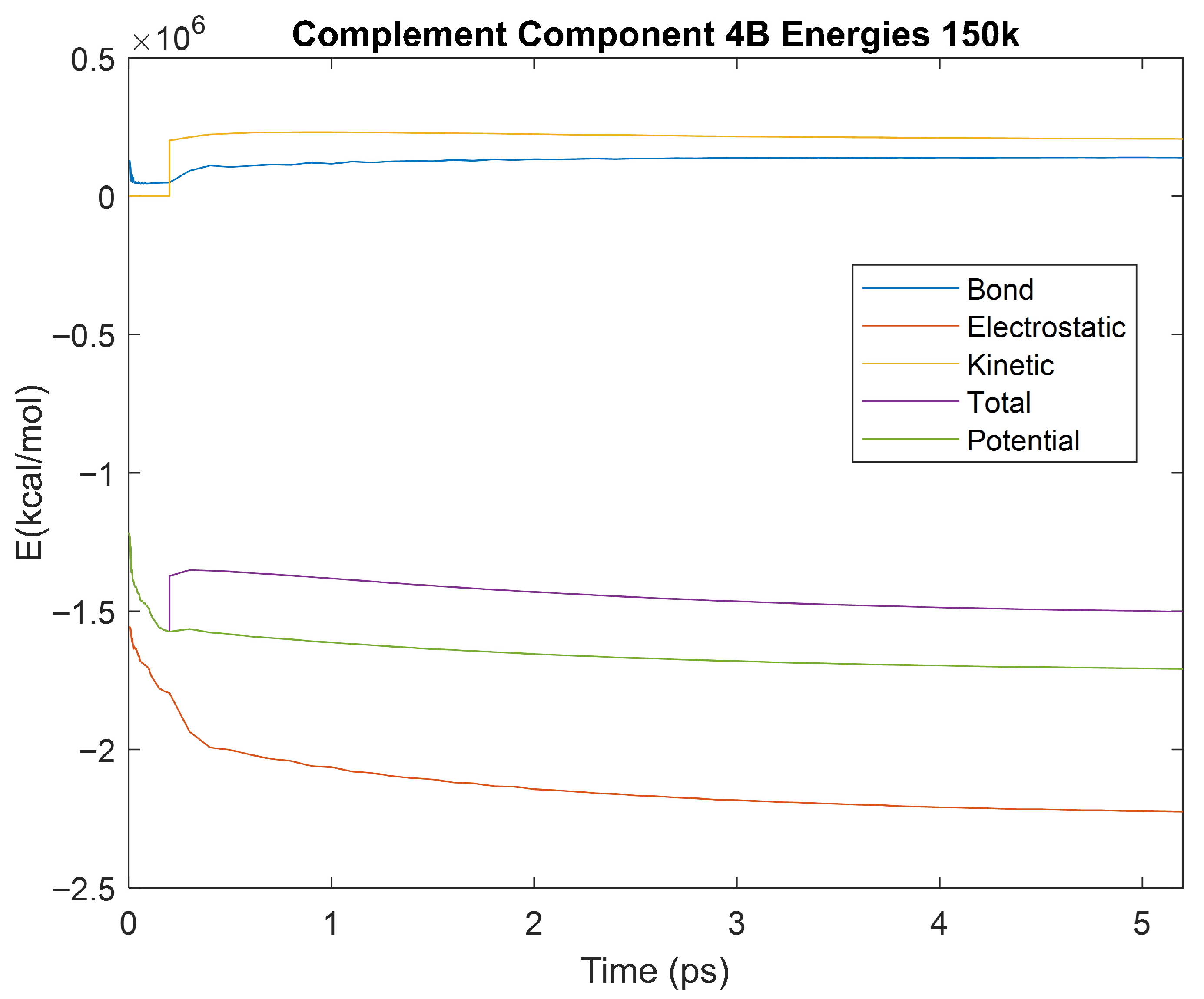
Figure 28.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 150 K 5 ps simulation.
Figure 28.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 150 K 5 ps simulation.
Figure 29.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 4. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarkers.
Figure 29.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 4. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarkers.
Figure 30.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 0 K 5 ps simulation.
Figure 30.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 0 K 5 ps simulation.
Figure 31.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 0 K 5 ps simulation.
Figure 31.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Albumin over the 0 K 5 ps simulation.
Figure 32.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 0 K 5 ps simulation.
Figure 32.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-1 Microglobulin over the 0 K 5 ps simulation.
Figure 33.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 0 K 5 ps simulation.
Figure 33.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Apolipoprotein E over the 0 K 5 ps simulation.
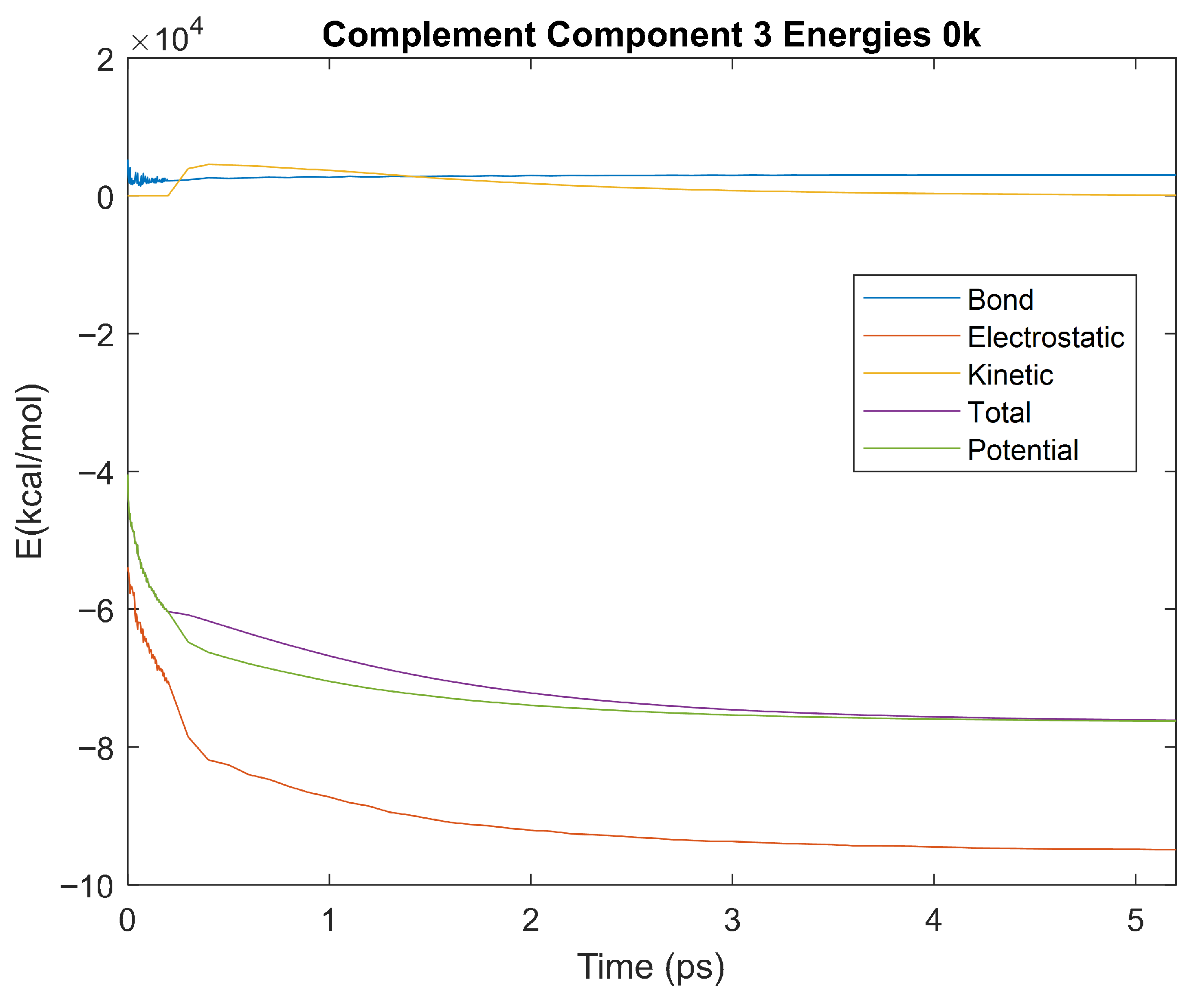
Figure 34.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 0 K 5 ps simulation.
Figure 34.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 3 over the 0 K 5 ps simulation.
Figure 35.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 0 K 5 ps simulation.
Figure 35.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Haptoglobin over the 0 K 5 ps simulation.
Figure 36.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 0 K 5 ps simulation.
Figure 36.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 0 K 5 ps simulation.
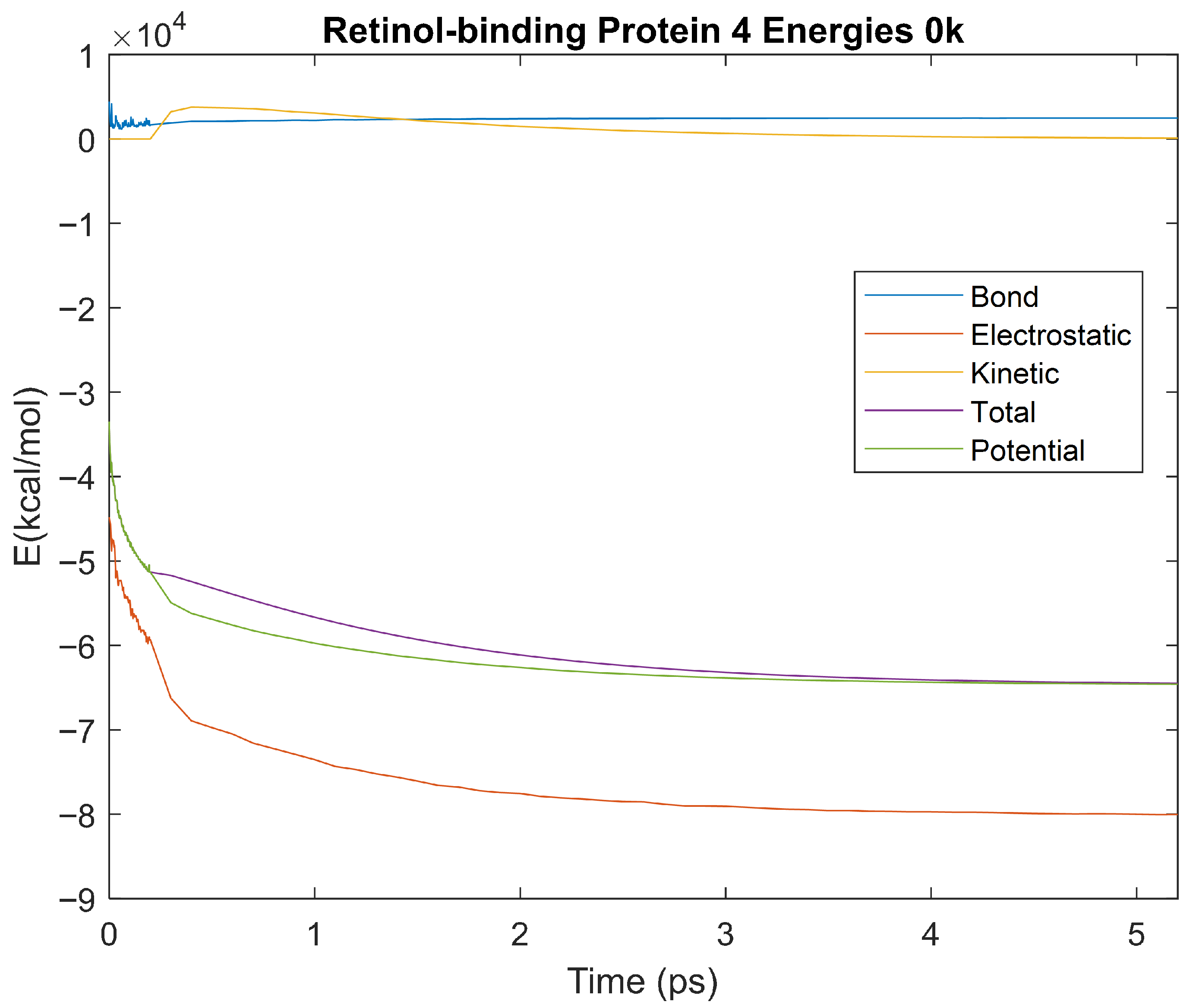
Figure 37.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-Binding Protein 4 over the 0 K 5 ps simulation.
Figure 37.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Retinol-Binding Protein 4 over the 0 K 5 ps simulation.
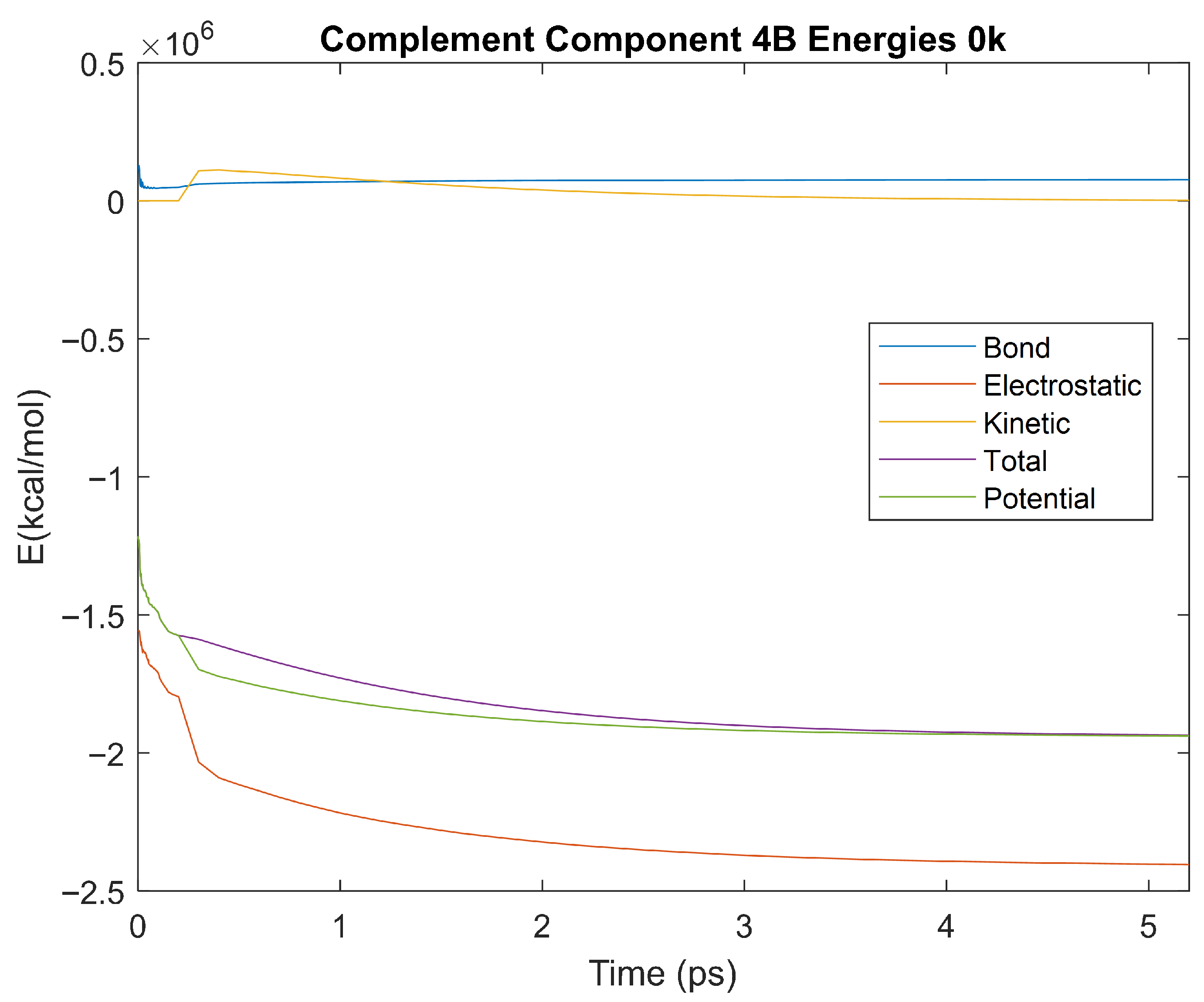
Figure 38.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 0 K 5 ps simulation.
Figure 38.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 0 K 5 ps simulation.
Figure 39.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 5. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarker.
Figure 39.
A box plot of the equilibrium times for each energy type. Data for this plot can be seen in
Table 5. The red line represents the median time, the blue box is the interquartile range (25th–75th percentile), and the ends of the whiskers represent the 0th and 100th percentile, also called the maximum and minimum. Outliers are marked with red crosses and are labeled with their respective biomarker.
Figure 40.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 310 K 1 ns simulation.
Figure 40.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Alpha-2 Macroglobulin over the 310 K 1 ns simulation.
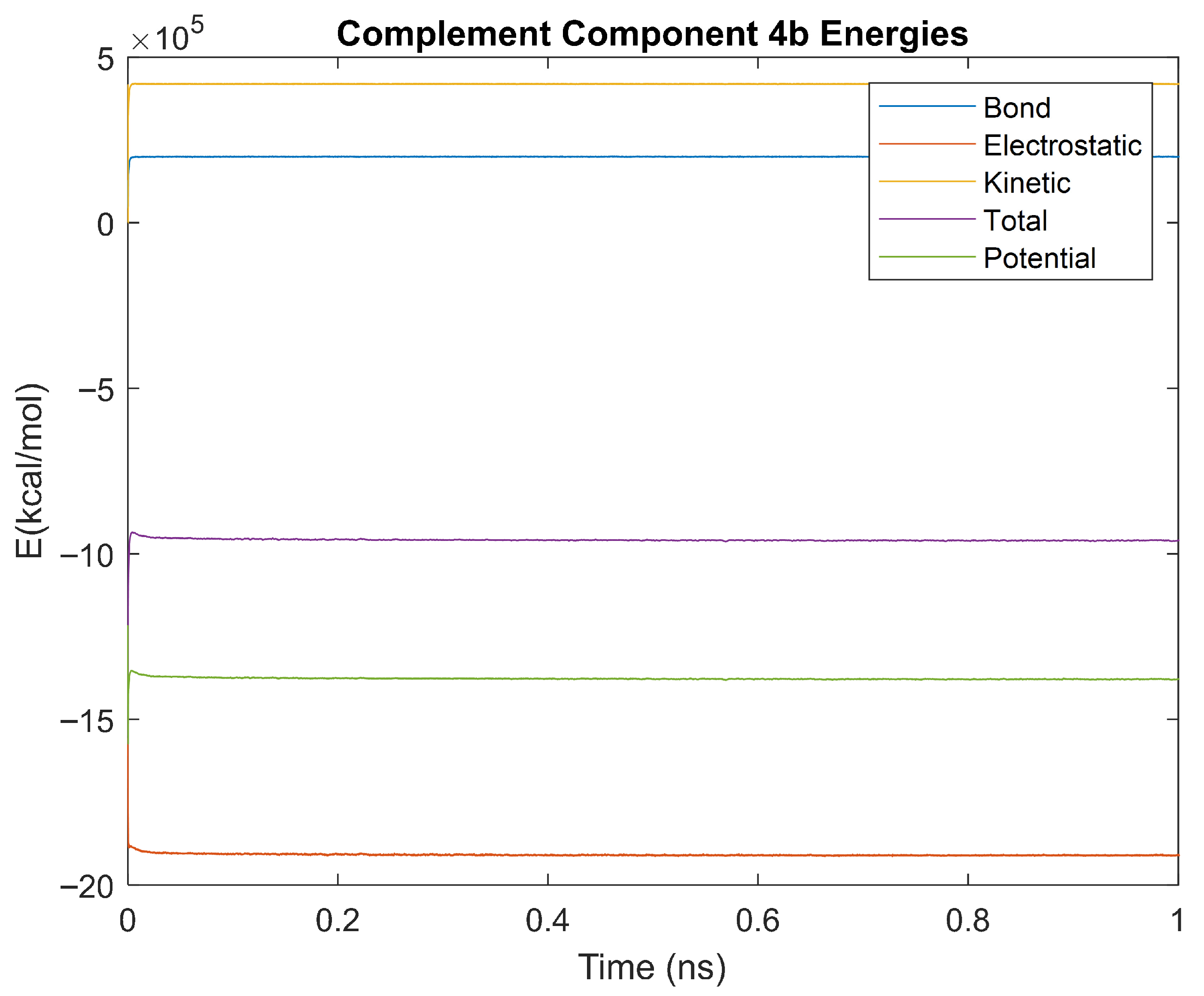
Figure 41.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 310 K 1 ns simulation.
Figure 41.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Complement Component 4B over the 310 K 1 ns simulation.
Figure 42.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 310 K 1 ns simulation.
Figure 42.
A plot of Bond, Electrostatic, Kinetic, Total, and Potential Energies for Orosomucoid-1 over the 310 K 1 ns simulation.
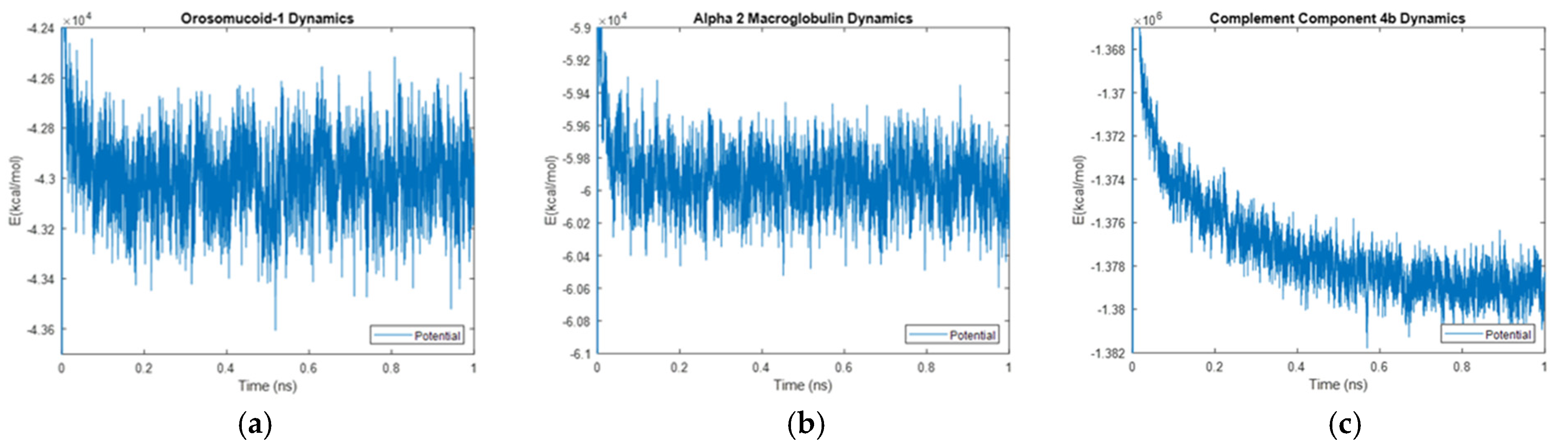
Figure 43.
Molecular interactions in (a) Orosomucoid-1, (b) Alpha-2-Macroglobulin, and (c) Complement Component 4B. The dynamic potential energy drop with time in Complement Component 4B is shown in part (c). These studies in themselves form the basis for an accompanying article that will be submitted in the future.
Figure 43.
Molecular interactions in (a) Orosomucoid-1, (b) Alpha-2-Macroglobulin, and (c) Complement Component 4B. The dynamic potential energy drop with time in Complement Component 4B is shown in part (c). These studies in themselves form the basis for an accompanying article that will be submitted in the future.
Table 1.
GI number (#), specificity, sensitivity, and
p-value for proteins found to be upregulated in OA in a complete protein analysis of synovial fluid [
1].
Table 1.
GI number (#), specificity, sensitivity, and
p-value for proteins found to be upregulated in OA in a complete protein analysis of synovial fluid [
1].
| Protein | GI # | Specificity | Sensitivity | p Value |
|---|
| Albumin | 4502027 | 0.950 | 0.718 | 7.96 × 10−7 |
| α1-Microglobulin/bikunin precursor | 4502067 | 0.950 | 0.718 | 7.96 × 10−7 |
| Fibrinogen, α chain isoform α-E preprotein | 4503689 | 0.950 | 0.718 | 7.96 × 10−7 |
| Fibrinogen, γ chain isoform γ-A precursor | 4503715 | 1.000 | 0.744 | 1.43 × 10−8 |
| α2-Macroglobulin | 4557225 | 0.950 | 0.718 | 7.96 × 10−7 |
| Apolipoprotein E | 4557325 | 1.000 | 0.744 | 1.43 × 10−8 |
| Apolipoprotein H (β2-glycoprotein I) | 4557327 | 1.000 | 0.744 | 1.43 × 10−8 |
| Complement component 3 (gel slice 3) | 4557385 | 0.950 | 0.718 | 7.96 × 10−7 |
| Complement component 3 (gel slice 5) | 4557385 | 1.000 | 0.744 | 1.43 × 10−8 |
| Ceruloplasmin (ferroxidase) | 4557485 | 0.950 | 0.718 | 7.96 × 10−7 |
| Haptoglobin | 4826762 | 0.950 | 0.718 | 7.96 × 10−7 |
| Orosomucoid 1 | 9257232 | 0.850 | 0.667 | 2.51 × 10−4 |
| Group specific component (vitamin D binding protein) | 32483410 | 1.000 | 0.744 | 1.43 × 10−8 |
| Complement component 4B preprotein | 50345296 | 1.000 | 0.744 | 1.43 × 10−8 |
| PREDICTED: similar to apolipoprotein A-1 precursor | 5147611 | 0.950 | 0.718 | 7.96 × 10−7 |
| Retinol-binding protein 4, plasma precursor | 55743122 | 0.900 | 0.692 | 1.87 × 10−5 |
Table 2.
Properties of the protein biomarkers that were simulated using the VMD and NAMD software. The gene and weight for each protein were gathered from the UniProt data bank [
35,
38,
39,
40,
42,
44,
46,
48,
50]. Amino acid length was gathered via the GI number from the NCBI databank [
51,
52,
53,
54,
55,
56,
57,
58,
59].
Table 2.
Properties of the protein biomarkers that were simulated using the VMD and NAMD software. The gene and weight for each protein were gathered from the UniProt data bank [
35,
38,
39,
40,
42,
44,
46,
48,
50]. Amino acid length was gathered via the GI number from the NCBI databank [
51,
52,
53,
54,
55,
56,
57,
58,
59].
| Protein | Gene | Weight (Da) | Amino Acid Length (From GI #) | Citation |
|---|
| Albumin | ALB | 69,367 | 609 | [35,51] |
| α1-Microglobulin | AMBP (HCP, ITIL) | 38,999 | 352 | [38,52] |
| α2-Macroglobulin | A2M (CPAMD5) | 163,291 | 1474 | [39,53] |
| Apolipoprotein E | APOE | 36,154 | 317 | [40,54] |
| Complement component 3 | C3 (CPAMD1) | 187,148 | 1663 | [42,55] |
| Haptoglobin | HP | 45,205 | 406 | [44,56] |
| Orosomucoid 1 | ORM1 (AGP1) | 23,540 | 201 | [46,57] |
| Complement component 4B preprotein | C4B (CO4, CPAMD3) | 192,751 | 1744 | [48,58] |
| Retinol-binding protein 4, plasma precursor | RBP4 | 23,010 | 201 | [50,59] |
Table 3.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 310 K.
Table 3.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 310 K.
| Amount of Time (ps) Taken to Reach Equilibrium (310 K) |
|---|
| Biomarker | Electrostatic | Bond | Kinetic | Potential | Total |
|---|
| Albumin | 2.4 | 3.1 | 2.7 | 2.2 | 2.3 |
| α1-Microglobulin | 4.1 | 2.9 | 2.5 | 2.4 | 2.4 |
| α2-Macroglobulin | 2.7 | 3.9 | 3.2 | 3.5 | 3.1 |
| Apolipoprotein E | 3.6 | 4.1 | 3.5 | 2.3 | 2.4 |
| Complement component 3 | 3.1 | 3.5 | 2.6 | 2.1 | 2.2 |
| Haptoglobin | 3.2 | 3.7 | 2.5 | 2.3 | 2.3 |
| Orosomucoid 1 | 2.9 | 3.9 | 3.3 | 1.7 | 2.0 |
| Complement component 4B | 0.8 | 3.2 | 2.9 | 2.7 | 2.7 |
| Retinol-binding protein 4 | 3.3 | 3.9 | 2.7 | 2.3 | 2.2 |
Table 4.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 150 K.
Table 4.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 150 K.
| Amount of Time (ps) Taken to Reach Equilibrium (150 K) |
|---|
| Biomarker | Electrostatic | Bond | Kinetic | Potential | Total |
|---|
| Albumin | 3.6 | 2.2 | 4.8 | 4.2 | 4.5 |
| α1-Microglobulin | 3.8 | 2.3 | 4.7 | 4.3 | 4.5 |
| α2-Macroglobulin | 3.8 | 2.1 | 4.7 | 4.3 | 4.5 |
| Apolipoprotein E | 3.7 | 2.1 | 4.9 | 4.4 | 4.6 |
| Complement component 3 | 3.8 | 2.5 | 4.5 | 4.3 | 4.5 |
| Haptoglobin | 3.7 | 2.1 | 4.8 | 4.3 | 4.6 |
| Orosomucoid 1 | 3.8 | 2.3 | 4.9 | 4.3 | 4.6 |
| Complement component 4B | 3.7 | 2.2 | 4.8 | 4.2 | 4.5 |
| Retinol-binding protein 4 | 3.7 | 2 | 5 | 4.5 | 4.7 |
Table 5.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 0 K.
Table 5.
The time taken in picoseconds for each of the protein biomarkers to reach equilibrium for each respective energy at 0 K.
| Amount of Time (ps) Taken to Reach Equilibrium (0 K) |
|---|
| Biomarker | Electrostatic | Bond | Kinetic | Potential | Total |
|---|
| Albumin | 3.2 | 1.8 | 4.1 | 3.2 | 3.7 |
| α1-Microglobulin | 3.3 | 1.9 | 4.0 | 3.3 | 3.8 |
| α2-Macroglobulin | 3.2 | 1.7 | 4.1 | 3.2 | 3.7 |
| Apolipoprotein E | 3.2 | 1.8 | 4.1 | 3.2 | 3.7 |
| Complement component 3 | 3.3 | 1.7 | 4.1 | 3.2 | 3.7 |
| Haptoglobin | 3.3 | 1.9 | 4.0 | 3.2 | 3.7 |
| Orosomucoid 1 | 3.3 | 1.8 | 4.1 | 3.2 | 3.7 |
| Complement component 4B | 3.4 | 1.9 | 4.0 | 3.1 | 3.6 |
| Retinol-binding protein 4 | 3.2 | 1.9 | 4.1 | 3.2 | 3.7 |
Table 6.
Values obtained from the analysis of the energy data noise for alpha-2-macroglobulin. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
Table 6.
Values obtained from the analysis of the energy data noise for alpha-2-macroglobulin. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
| Alpha-2 Macroglobulin Energy Noise (0.1–1 ns) |
|---|
| | Bond | Electrostatic | Kinetic | Total | Potential |
|---|
| RMS (kcal/mol): | 9320.9968 | 85,387.3186 | 19,778.495 | 40,165.0394 | 59,942.976 |
| Standard Deviation (kcal/mol): | 102.8123 | 231.4206 | 110.415 | 193.1473 | 160.236 |
| Percent Change (kcal/mol): | 0.01103 | 0.0027102 | 0.0055826 | 0.0048088 | 0.0026731 |
Table 7.
Values obtained from the analysis of the energy data noise for complement component 4B. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
Table 7.
Values obtained from the analysis of the energy data noise for complement component 4B. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
| Complement Component 4B Energy Noise (0.1–1 ns) |
|---|
| | Bond | Electrostatic | Kinetic | Total | Potential |
|---|
| RMS (kcal/mol): | 199,128.5033 | 1,909,673.714 | 418,972.4289 | 958,798.3667 | 1,377,769.945 |
| Standard Deviation (kcal/mol): | 478.31444 | 1578.9485 | 507.70323 | 1636.9161 | 1533.8466 |
| Percent Change (kcal/mol): | 0.002402 | 0.00082682 | 0.0012118 | 0.0017073 | 0.0011133 |
Table 8.
Values obtained from the analysis of the energy data noise for orosomucoid-1. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
Table 8.
Values obtained from the analysis of the energy data noise for orosomucoid-1. Energy data between 0.1 ns and 1 ns from the 1 ns simulation was considered in the analysis.
| Orosomucoid-1 Energy Noise (0.1–1 ns) |
|---|
| | Bond | Electrostatic | Kinetic | Total | Potential |
|---|
| RMS (kcal/mol): | 6711.66627 | 61,459.1381 | 14,222.678 | 28,782.0271 | 43,004.1156 |
| Standard Deviation (kcal/mol): | 87.60533 | 198.0308 | 95.3199 | 168.9017 | 139.2274 |
| Percent Change (kcal/mol): | 0.013053 | 0.0032222 | 0.006702 | 0.0058683 | 0.0032375 |