The Fusion of Wide Field Optical Coherence Tomography and AI: Advancing Breast Cancer Surgical Margin Visualization
Abstract
:1. Introduction
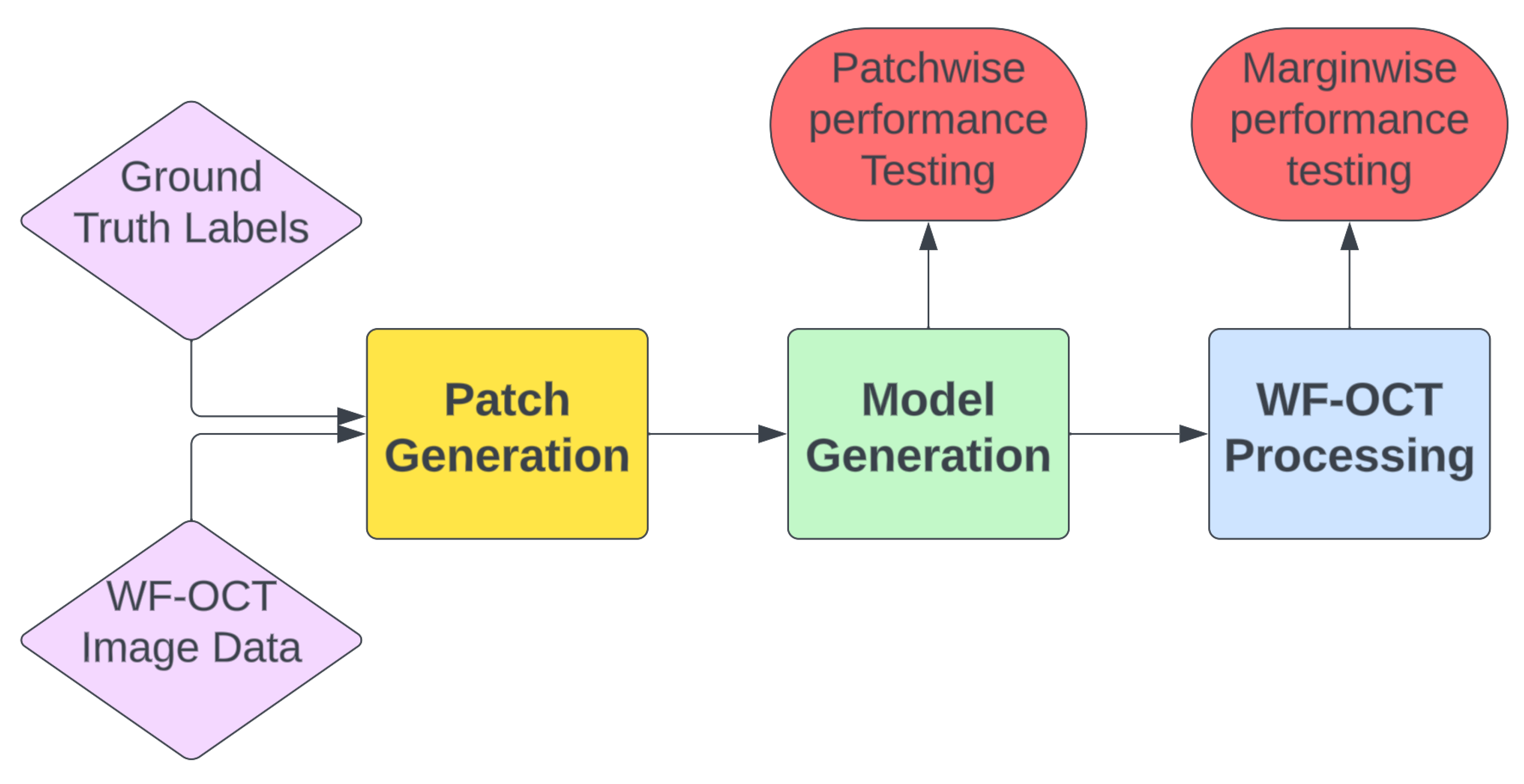
2. Materials and Methods
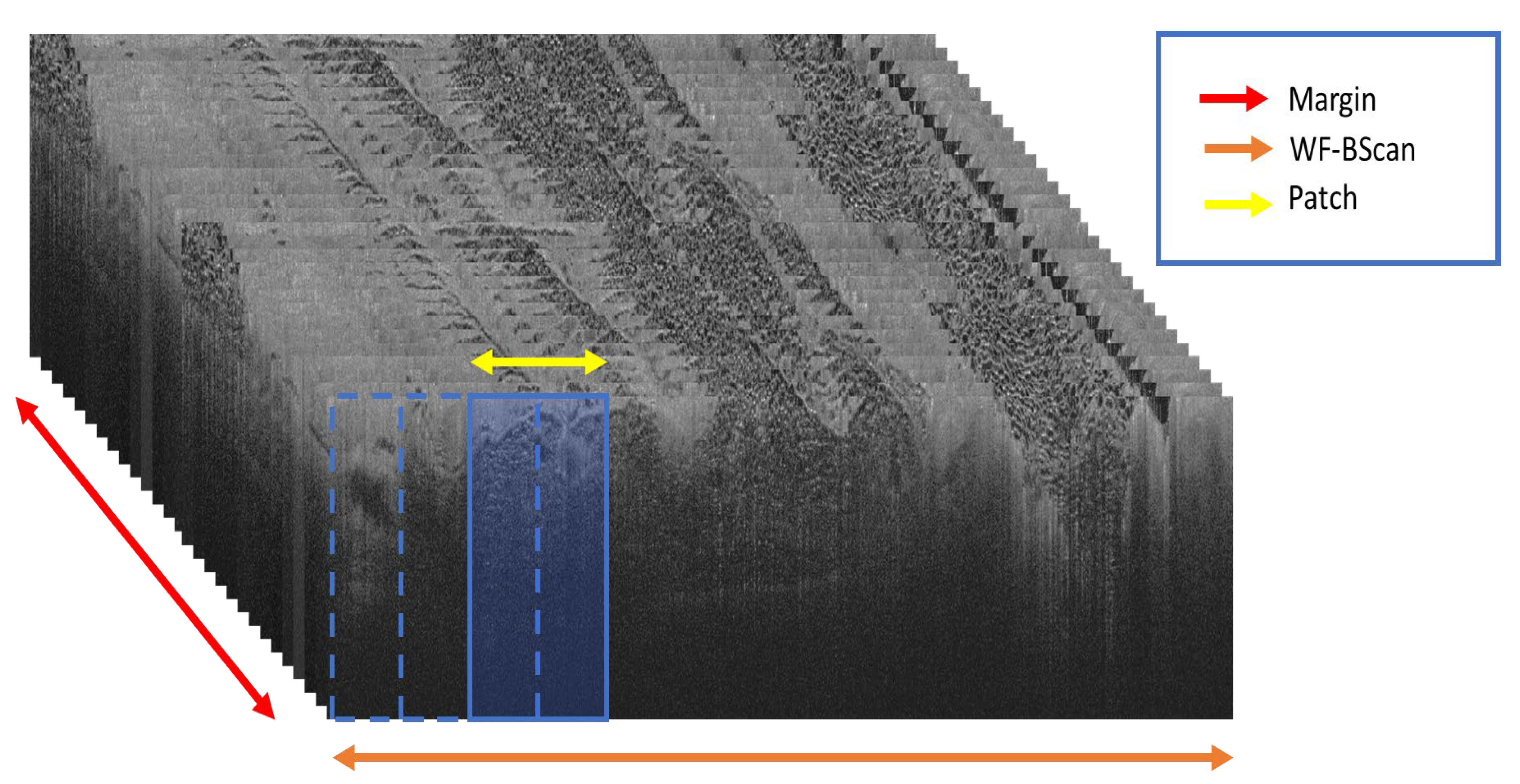
- Patch Generation: In this preliminary step, the ground truth labels are input to extract coordinates from the WF-OCT imaging data. The resulting output consists of labeled image patches, each distinctively named and characterized according to their morphological feature types. These patches are further sorted based on specific margins and unique subject directories. Concentrated data augmentation is implemented to enhance the representation of suspicious features, ensuring a balanced training dataset to the possible extent.
- Model Generation: This crucial step encompasses both the model training, with specified hyperparameters, and the evaluation of its performance. The model selection emphasizes the epoch exhibiting the lowest validation loss and peak accuracy. Following this, the chosen model undergoes testing using the distinct “test” patches to ascertain key performance metrics and the model’s overall efficacy on a blinded test set.
- Margin Processing: When a model fulfills the pre-defined performance criteria, it is tested in a simulated real-world environment using the WF-OCT Processing tool. This stage involves the simultaneous processing of designated and complete subject scans as well as the application of a clustering algorithm. The foremost aim is to identify correctly classified key suspicious features and ensure that the model presents the most accurate “Key Thumbnail Images” of the relevant patches to the clinical user. This method boosts user accessibility and efficiency in identifying suspicious features during surgical procedures.
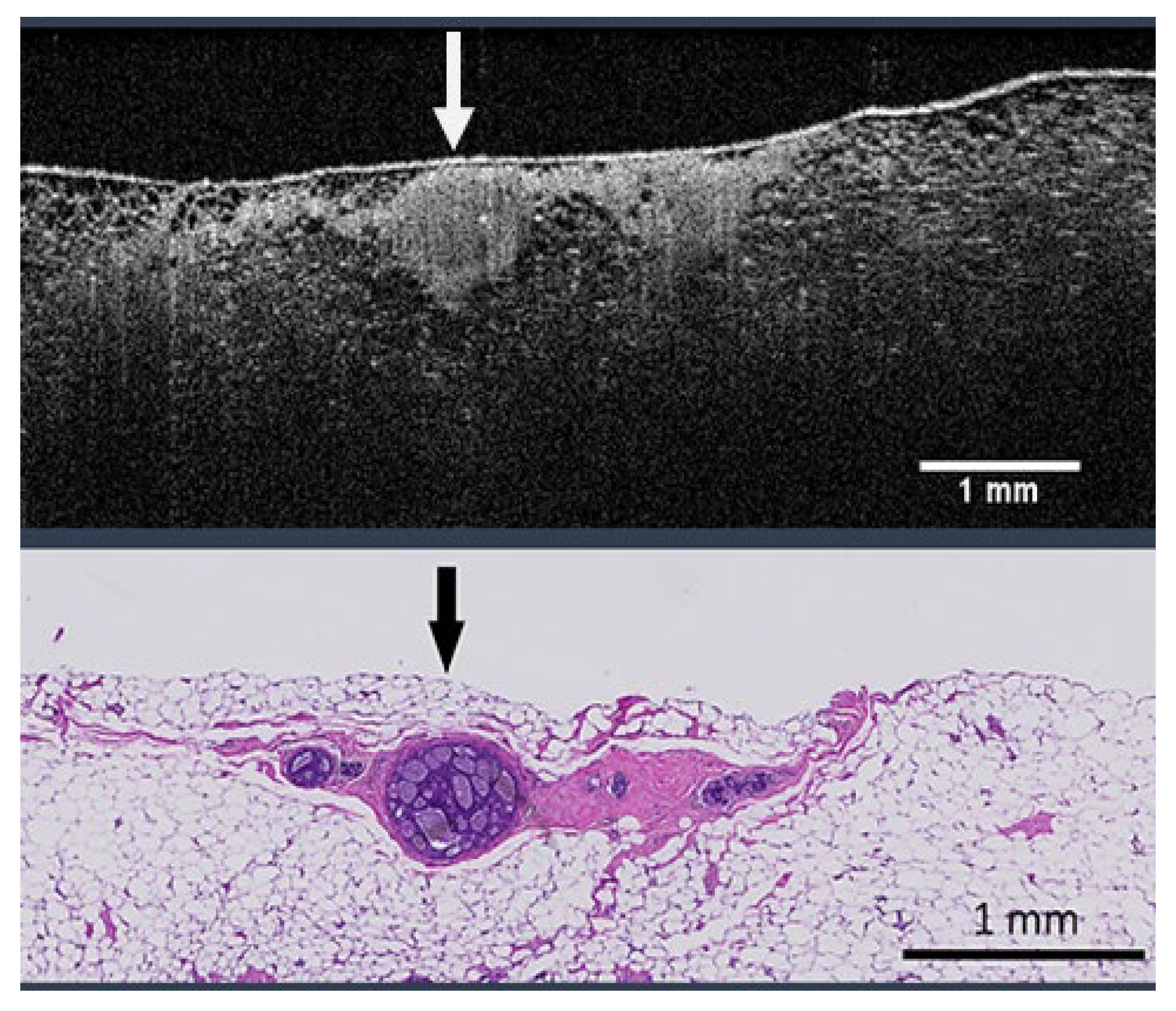
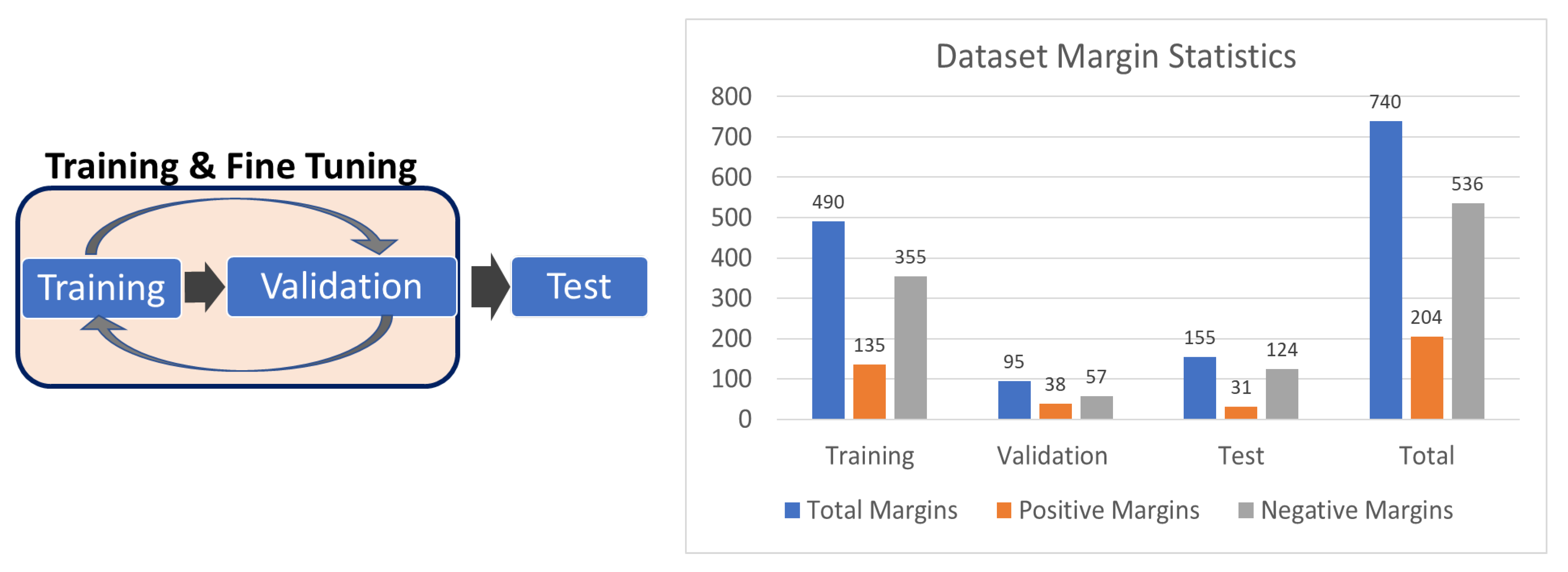
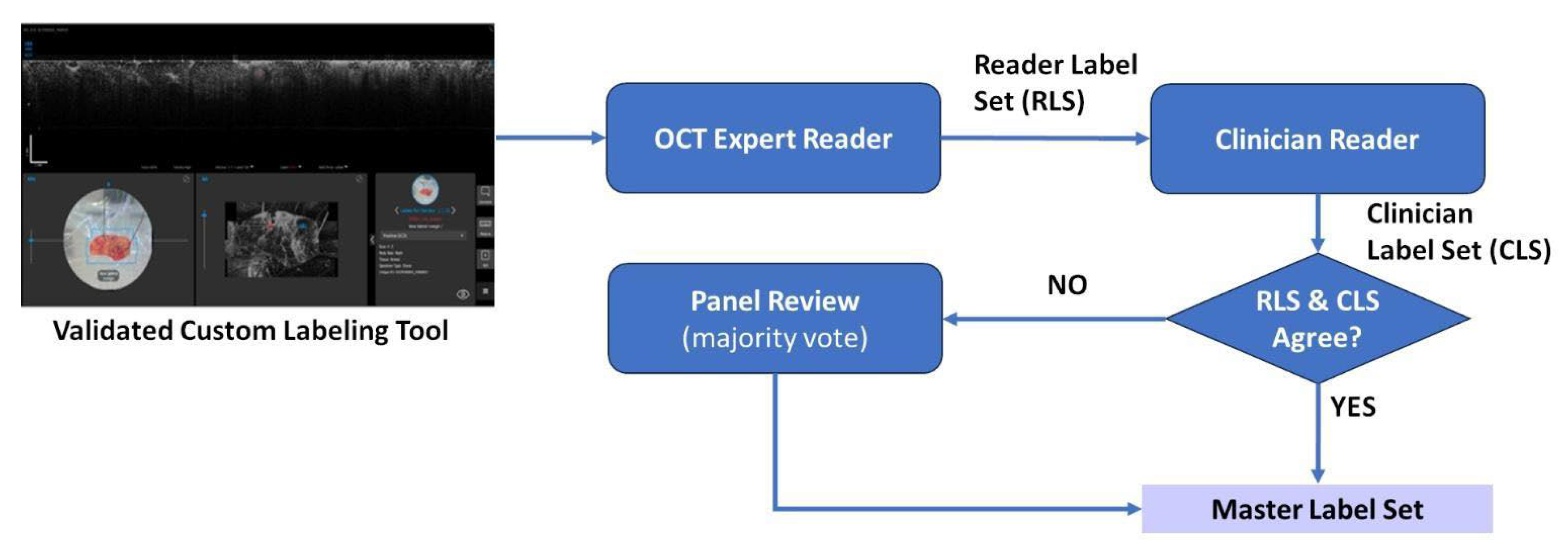
2.1. Data Collection and Curation
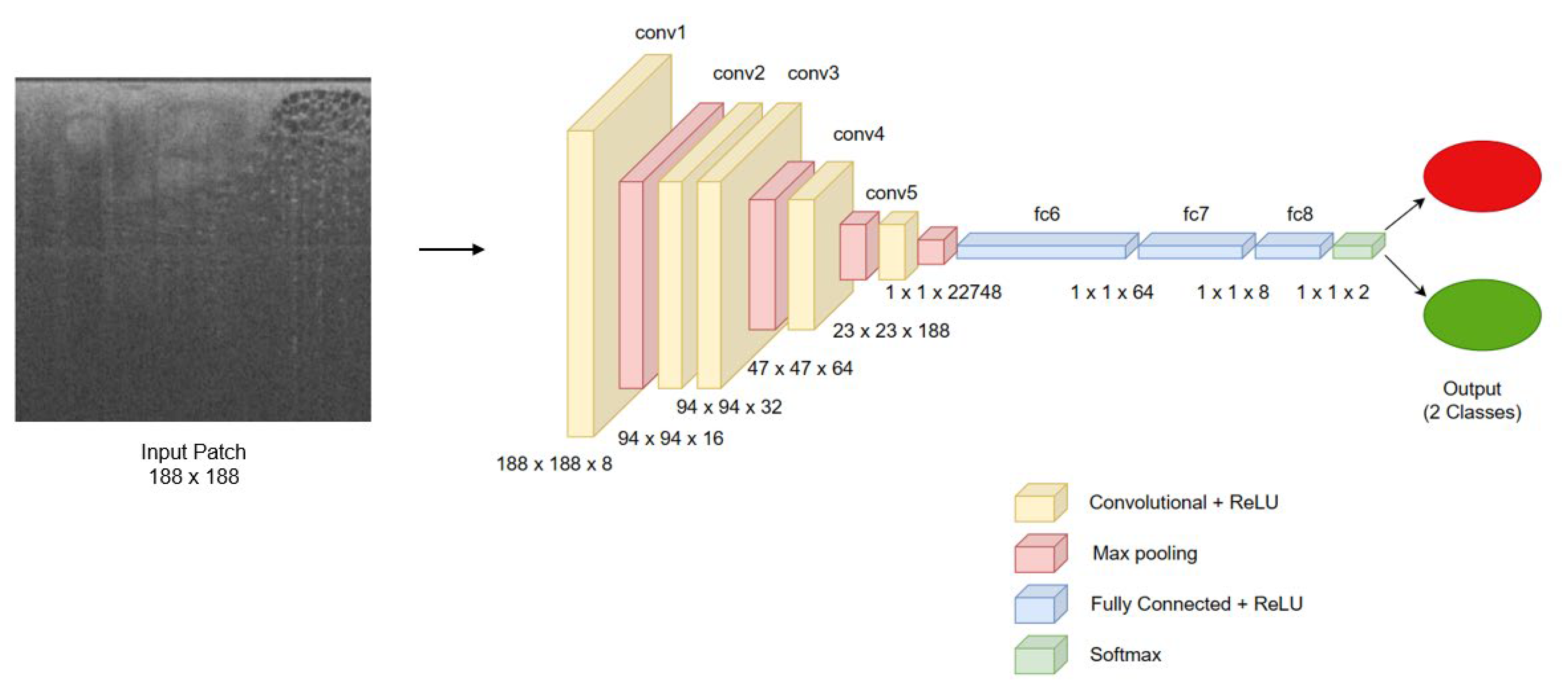
2.2. Model Development
2.3. Model Performance Assessment in a Clinical Simulation
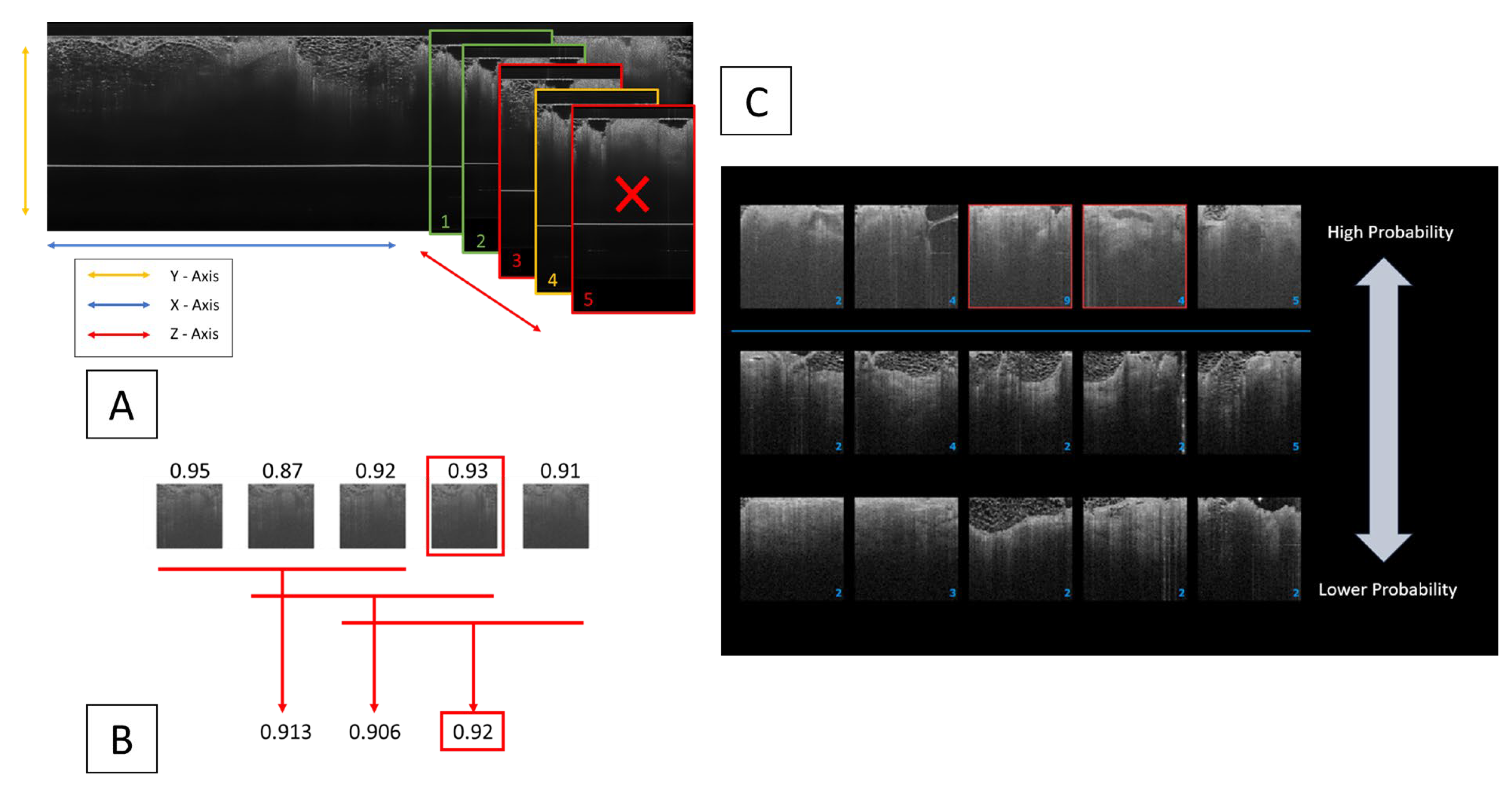
2.3.1. Clustering Algorithm Integration for Enhanced Diagnostic Precision
2.3.2. Key Thumbnail Selection for Clinician Review
3. Results
3.1. Patch-Wise Performance
3.2. Two-Tiered Confidence Threshold Analysis
- Sensitivity: 0.93
- Specificity: 0.98
- Precision (PPV): 0.41
- F1-Score: 0.78
- MCC: 0.61
- Sensitivity: 0.7
- Specificity: 1.0
- Precision (PPV): 0.79
- F1-Score: 0.87
- MCC: 0.74
3.3. Margin-Wise Analysis
- Evaluated Margins: 155 (31 positive)
- True Positives: 507 (92%)
- True Negatives: 1,894,239 (97.3%)
- False Positives: 53,225 (2.7%)
- Average Positive Patches per Margin: 347 (Positive margins: 882, Negative margins: 213)
- Evaluated Margins: 155 (31 positive)
- True Positives: 387 (70.2%)
- True Negatives: 1,825,709 (99.5%)
- False Positives: 9645 (0.5%)
- Average Positive Patches per Margin: 65 (Positive margins: 197, Negative margins: 32)
4. Discussion
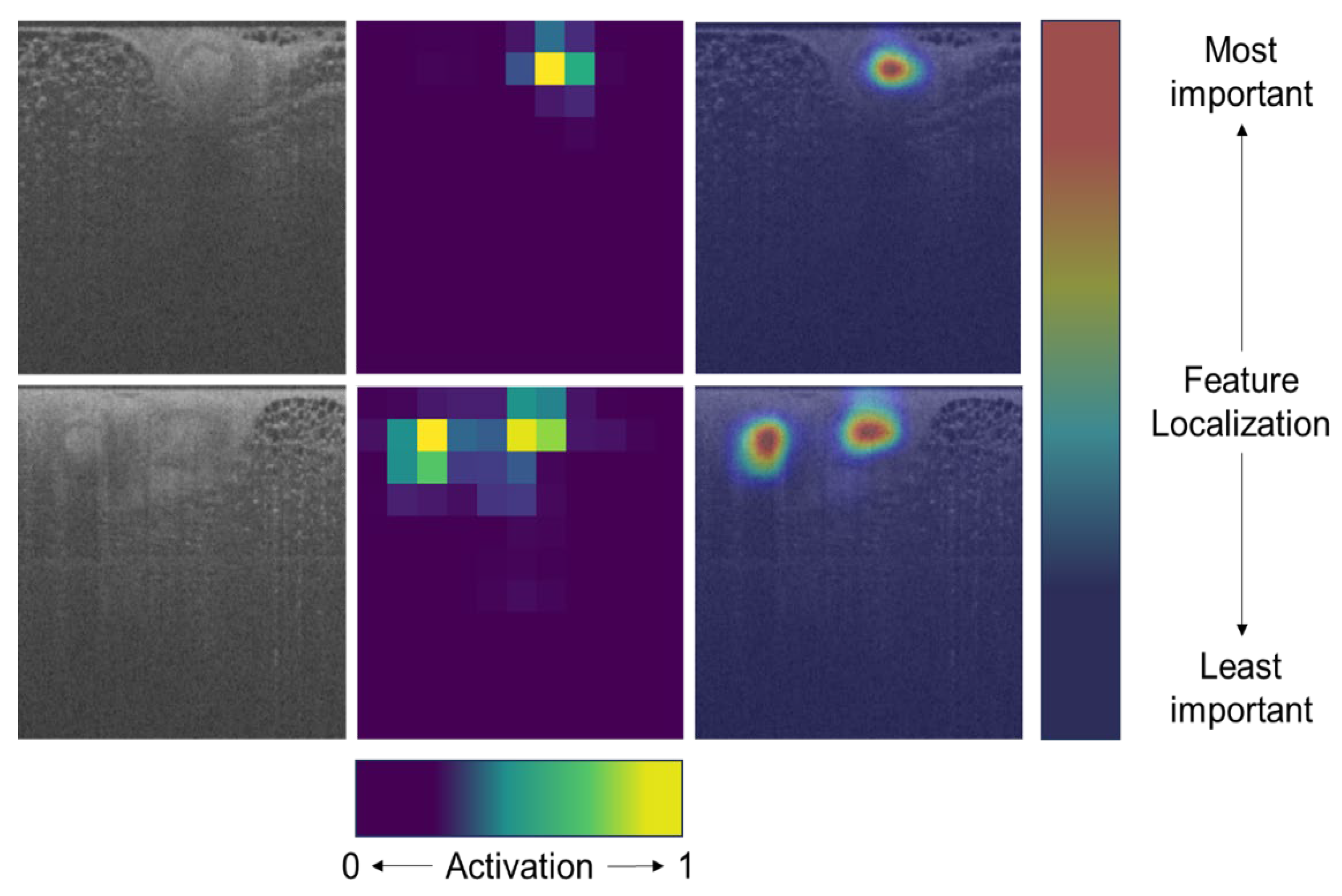
4.1. Interpreting Patch-Wise Results
4.2. Two-Tiered Confidence Threshold, Patch-Wise Performance
4.3. Enhancing Clinical Decision-Making: Integrating AI Model and User Interface for Optimal Margin Performance
4.4. Generalizability and Future Work
4.4.1. Generalizability
4.4.2. Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Breast Cancer Initiative Implementation Framework: Assessing, Strengthening and Scaling up of Services for the Early Detection and Management of Breast Cancer: Executive Summary. Available online: https://www.who.int/publications/i/item/9789240067134 (accessed on 7 December 2023).
- Gray, R.J.; Pockaj, B.A.; Garvey, E.; Blair, S. Intraoperative margin management in breast-conserving surgery: A systematic review of the literature. Ann. Surg. Oncol. 2018, 25, 18–27. [Google Scholar] [CrossRef]
- Alison, L.; Brar, M.S.; Bouchard-Fortier, A.; Leong, B.; Quan, M.L. Intraoperative margin assessment in wire-localized breast-conserving surgery for invasive cancer: A population-level comparison of techniques. Ann. Surg. Oncol. 2016, 23, 3290–3296. [Google Scholar]
- McCahill, L.E.; Single, R.M.; Bowles, E.J.A.; Feigelson, H.S.; James, T.A.; Barney, T.; Engel, J.M.; Onitilo, A.A. Variability in reexcision following breast conservation surgery. JAMA 2012, 307, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Jeevan, R.; Cromwell, D.A.; Trivella, M.; Lawrence, G.; Kearins, O.; Pereira, J.; Sheppard, C.; Caddy, C.M.; Van Der Meulen, J.H.P. Reoperation rates after breast conserving surgery for breast cancer among women in England: Retrospective study of hospital episode statistics. BMJ 2012, 345, e4505. [Google Scholar] [CrossRef] [PubMed]
- Wilke, L.G.; Czechura, T.; Wang, C.; Lapin, B.; Liederbach, E.; Winchester, D.P.; Yao, K. Repeat surgery after breast conservation for the treatment of stage 0 to II breast carcinoma: A report from the National Cancer Data Base, 2004–2010. JAMA Surg. 2014, 149, 1296–1305. [Google Scholar] [CrossRef]
- Landercasper, J.; Whitacre, E.; Degnim, A.C.; Al-Hamadani, M. Reasons for re-excision after lumpectomy for breast cancer: Insight from the American Society of Breast Surgeons Mastery SM database. Ann. Surg. Oncol. 2014, 21, 3185–3191. [Google Scholar] [CrossRef] [PubMed]
- Schulman, A.M.; Mirrielees, J.A.; Leverson, G.; Landercasper, J.; Greenberg, C.; Wilke, L.G. Reexcision surgery for breast cancer: An analysis of the American Society of Breast Surgeons (ASBrS) Mastery SM database following the SSO-ASTRO “no ink on tumor” guidelines. Ann. Surg. Oncol. 2017, 24, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, A.J.; Gemignani, M.L.; Pusic, A.; Sedrakyan, A. Association of breast conservation surgery for cancer with 90-day reoperation rates in New York state. JAMA Surg. 2016, 151, 648–655. [Google Scholar] [CrossRef]
- Eck, D.L.; Koonce, S.L.; Goldberg, R.F.; Bagaria, S.; Gibson, T.; Bowers, S.P.; McLaughlin, S.A. Breast surgery outcomes as quality measures according to the NSQIP database. Ann. Surg. Oncol. 2012, 19, 3212–3217. [Google Scholar] [CrossRef]
- Blair, S.L.; Thompson, K.; Rococco, J.; Malcarne, V.; Beitsch, P.D.; Ollila, D.W. Attaining negative margins in breast-conservation operations: Is there a consensus among breast surgeons? J. Am. Coll. Surg. 2009, 209, 608–613. [Google Scholar] [CrossRef]
- Simiyoshi, K.; Nohara, T.; Iwamoto, M.; Tanaka, S.; Kimura, K.; Takahashi, Y.; Kurisu, Y.; Tsuji, M.; Tanigawa, N. Usefulness of intraoperative touch smear cytology in breast-conserving surgery. Exp. Ther. Med. 2010, 1, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Klimberg, V.S. Accuracy of Intraoperative Gross Examination of Surgical Margin Status in Women Undergoing Partial Mastectomy for Breast Malignancy. Breast Dis. Year Book Q. 2005, 3, 258. [Google Scholar] [CrossRef]
- Chan, B.K.Y.; Wiseberg-Firtell, J.A.; Jois, R.H.; Jensen, K.; Audisio, R.A. Localization techniques for guided surgical excision of non-palpable breast lesions. Cochrane Database Syst. Rev. 2015, 12, CD009206. [Google Scholar] [CrossRef]
- Lange, M.; Reimer, T.; Hartmann, S.; Glass, Ä.; Stachs, A. The role of specimen radiography in breast-conserving therapy of ductal carcinoma in situ. Breast 2016, 26, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Ihrai, T.; Quaranta, D.; Fouche, Y.; Machiavello, J.-C.; Raoust, I.; Chapellier, C.; Maestro, C.; Marcy, M.; Ferrero, J.-M.; Flipo, B. Intraoperative radiological margin assessment in breast-conserving surgery. Eur. J. Surg. Oncol. 2014, 40, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Ha, R.; Friedlander, L.C.; Hibshoosh, H.; Hendon, C.; Feldman, S.; Ahn, S.; Schmidt, H.; Akens, M.K.; Fitzmaurice, M.; Wilson, B.C.; et al. Optical coherence tomography: A novel imaging method for post-lumpectomy breast margin assessment—A multi-reader study. Acad. Radiol. 2018, 25, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Savastru, D.; Chang, E.W.; Miclos, S.; Pitman, M.B.; Patel, A.; Iftimia, N. Detection of breast surgical margins with optical coherence tomography imaging: A concept evaluation study. J. Biomed. Opt. 2014, 19, 056001. [Google Scholar] [CrossRef]
- Nguyen, F.T.; Zysk, A.M.; Chaney, E.J.; Kotynek, J.G.; Oliphant, U.J.; Bellafiore, F.J.; Rowland, K.M.; Johnson, P.A.; Boppart, S.A. Intraoperative evaluation of breast tumor margins with optical coherence tomography. Cancer Res. 2009, 69, 8790–8796. [Google Scholar] [CrossRef]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical Coherence Tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Schmidt, H.; Connolly, C.; Jaffer, S.; Oza, T.; Weltz, C.R.; Port, E.R.; Corben, A. Evaluation of surgically excised breast tissue microstructure using wide-field optical coherence tomography. Breast J. 2020, 26, 917–923. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef]
- Khan, S.; Rahmani, H.; Shah, S.A.A.; Bennamoun, M. Applications of CNNs in Computer Vision. In A Guide to Convolutional Neural Networks for Computer Vision; Synthesis Lectures on Computer Vision; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, R.J.; Hughes, S. Real-Time Image Classification in Video Surveillance. J. Comput. Vis. Image Underst. 2022, 204, 103020. [Google Scholar]
- Zhao, Y.; Wang, X. Adapting Convolutional Neural Networks for Specialized Tasks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 2123–2134. [Google Scholar]
- Taylor, J. Efficient Training of Convolutional Networks in Data-Limited Regimes. Mach. Learn. Res. 2022, 23, 77–89. [Google Scholar]
- Murphy, K.; O’Connell, A. Edge Computing: A New Paradigm for Constrained Environments. Comput. Netw. 2023, 68, 456–469. [Google Scholar]
- Khan, M.A.; Gupta, A. Model Transparency and Compliance in Healthcare AI. Health Inform. J. 2021, 27, 1460458220985691. [Google Scholar]
- Nguyen, P.T. Comparative Study of CNN Architectures for Image Processing. Pattern Recognit. Lett. 2022, 150, 136–143. [Google Scholar]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD 1996, 96, 226–231. [Google Scholar]
- ISO/IEC TS 4213:2022; Information Technology—Artificial Intelligence—Assessment of Machine Learning Classification Performance. International Organization for Standardization (ISO): Geneva, Switzerland, 2022.
- Rempel, D.; Berkeley, A.; DiPasquale Sr, A.A.; Elmi, M.; Fine, R.E.; Lee, M.C.; O’Brien, B.; Wilke, L.G.; Thompson, A.M. A Prospective, Multicenter, Randomized, Double-Arm Trial to Determine the Impact of the Perimeter B-Series Optical Coherence Tomography and Artificial Intelligence System on Positive Margin Rates in Breast Conservation Surgery. J. Am. Coll. Surg. 2022, 235, S4. [Google Scholar] [CrossRef]
- Wide Field OCT + AI for Positive Margin Rates in Breast Conservation Surgery. (RCT). Available online: https://clinicaltrials.gov/study/NCT05113927?a=1 (accessed on 16 October 2023).
| Characteristic | Training and Validation (n = 151) | Testing (n = 29) |
|---|---|---|
| Age, years, mean (SD) | 63 (11.7) | 58.5 (9.1) |
| Race, n (%) | ||
| White | 116 (76.8%) | 20 (69%) |
| Black | 18 (11.9%) | 6 (20.7%) |
| Asian | 10 (6.6%) | 3 (10.3%) |
| Other | 6 (4%) | 0 (0%) |
| Not reported | 1 (0.7%) | 0 (0%) |
| Ethnicity, n (%) | ||
| Hispanic or Latino | 29 (19.2%) | 7 (24.1%) |
| Not Hispanic or Latino | 121 (80.1%) | 22 (75.9%) |
| Unknown | 1 (0.7%) | 0 (0%) |
| Characteristic | Training and Validation (n = 151) | Testing (n = 29) |
|---|---|---|
| Malignant Tumor type, n (%) | ||
| Invasive Ductal | 27 (17.9%) | 8 (27.6%) |
| Invasive Lobular | 4 (2.6%) | 0 (0%) |
| Ductal carcinoma in situ | 34 (22.5%) | 5 (17.2%) |
| Mixed | 77 (51%) | 15 (51.7%) |
| Benign (Not applicable for tumor type) | 5 (3.3%) | 1 (3.4%) |
| Other findings, n (%) | ||
| Lymphatic invasion | 6 (4.0%) | 1 (3.4%) |
| Atypical ductal hyperplasia | 23 (15.2%) | 7 (24.1%) |
| Lobular carcinoma in situ | 16 (10.6%) | 3 (10.3%) |
| Atypical lobular hyperplasia | 15 (9.9%) | 9 (31%) |
| Usual ductal hyperplasia | 26 (17.2%) | 12 (41.4%) |
| Duct Ectasia | 3 (2.0%) | 6 (20.7%) |
| Classification Threshold | Sensitivity (Recall) | Specificity | F1-Score | Matthew’s Correlation Coefficient (MCC) | Positive Predictive Value (PPV) (Precision) | Negative Predictive Value (NPV) | Positive Likelihood Ratio | Negative Likelihood Ratio |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 0.96 | 0.969 | 0.73 | 0.542 | 0.317 | 0.999 | 30.97 | 0.04 |
| 0.6 | 0.948 | 0.974 | 0.749 | 0.567 | 0.35 | 0.999 | 36.46 | 0.05 |
| 0.7 | 0.935 | 0.978 | 0.768 | 0.594 | 0.387 | 0.999 | 42.50 | 0.07 |
| 0.75 | 0.928 | 0.98 | 0.779 | 0.609 | 0.41 | 0.999 | 46.40 | 0.07 |
| 0.8 | 0.894 | 0.986 | 0.808 | 0.648 | 0.479 | 0.998 | 63.86 | 0.11 |
| 0.9 | 0.768 | 0.996 | 0.871 | 0.743 | 0.727 | 0.997 | 192.00 | 0.23 |
| 0.925 | 0.702 | 0.997 | 0.868 | 0.737 | 0.782 | 0.996 | 234.00 | 0.30 |
| 1 | 0 | 1 | 0 | 0 | 1 | 0 | - | 1.00 |
| Metric | 1st Confidence Threshold (0.75) | 2nd Confidence Threshold (0.925) |
|---|---|---|
| Number of Margins Evaluated | 155 | 155 |
| Number of Positive Margins | 31 | 31 |
| Positive Identification (Margins with Clusters/Key Thumbnails) | 30/27 | 26/26 |
| True Positive Patches (%) | 507 (92.0%) | 387 (70.2%) |
| False Negative Patches (%) | 44 (8.0%) | 164 (29.8%) |
| True Negative Patches (%) | 1,894,239 (97.3%) | 1,825,709 (99.5%) |
| False Positive Patches (%) | 53,225 (2.7%) | 9645 (0.5%) |
| Average Patches per Margin (Positive/Negative) | 882/213 | 197/32 |
| Discarded Single Patches (True Positive/True Negative) | 10/18,629 | 42/5234 |
| Clusters (Total/with True Positives) | 9135/154 | 1515/103 |
| True Positive Key Thumbnails | 91 | 74 |
| Average Clusters per Margin (Positive/Negative) | 147/37 | 33/4 |
| Scan Times (Seconds) (Total/Average Margin/Std Dev) | 1504.1/10.51/6.48 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levy, Y.; Rempel, D.; Nguyen, M.; Yassine, A.; Sanati-Burns, M.; Salgia, P.; Lim, B.; Butler, S.L.; Berkeley, A.; Bayram, E. The Fusion of Wide Field Optical Coherence Tomography and AI: Advancing Breast Cancer Surgical Margin Visualization. Life 2023, 13, 2340. https://doi.org/10.3390/life13122340
Levy Y, Rempel D, Nguyen M, Yassine A, Sanati-Burns M, Salgia P, Lim B, Butler SL, Berkeley A, Bayram E. The Fusion of Wide Field Optical Coherence Tomography and AI: Advancing Breast Cancer Surgical Margin Visualization. Life. 2023; 13(12):2340. https://doi.org/10.3390/life13122340
Chicago/Turabian StyleLevy, Yanir, David Rempel, Mark Nguyen, Ali Yassine, Maggie Sanati-Burns, Payal Salgia, Bryant Lim, Sarah L. Butler, Andrew Berkeley, and Ersin Bayram. 2023. "The Fusion of Wide Field Optical Coherence Tomography and AI: Advancing Breast Cancer Surgical Margin Visualization" Life 13, no. 12: 2340. https://doi.org/10.3390/life13122340
APA StyleLevy, Y., Rempel, D., Nguyen, M., Yassine, A., Sanati-Burns, M., Salgia, P., Lim, B., Butler, S. L., Berkeley, A., & Bayram, E. (2023). The Fusion of Wide Field Optical Coherence Tomography and AI: Advancing Breast Cancer Surgical Margin Visualization. Life, 13(12), 2340. https://doi.org/10.3390/life13122340







