Profiling of Lymphovascular Space Invasion in Cervical Cancer Revealed PI3K/Akt Signaling Pathway Overactivation and Heterogenic Tumor-Immune Microenvironments
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Pathological Processing and LVSI Detection
2.3. RNA Sequencing and LVSI DEGs
2.4. Immune Pathways Enrichment Analysis with IMPAGT
2.5. Functional Analysis of Intersected LVSI DEGs with IMPAGT
2.6. Immune Microenvironment Estimation with Immune Deconvolution
3. Results
3.1. Clinicopathological Characteristics of the 21 Samples
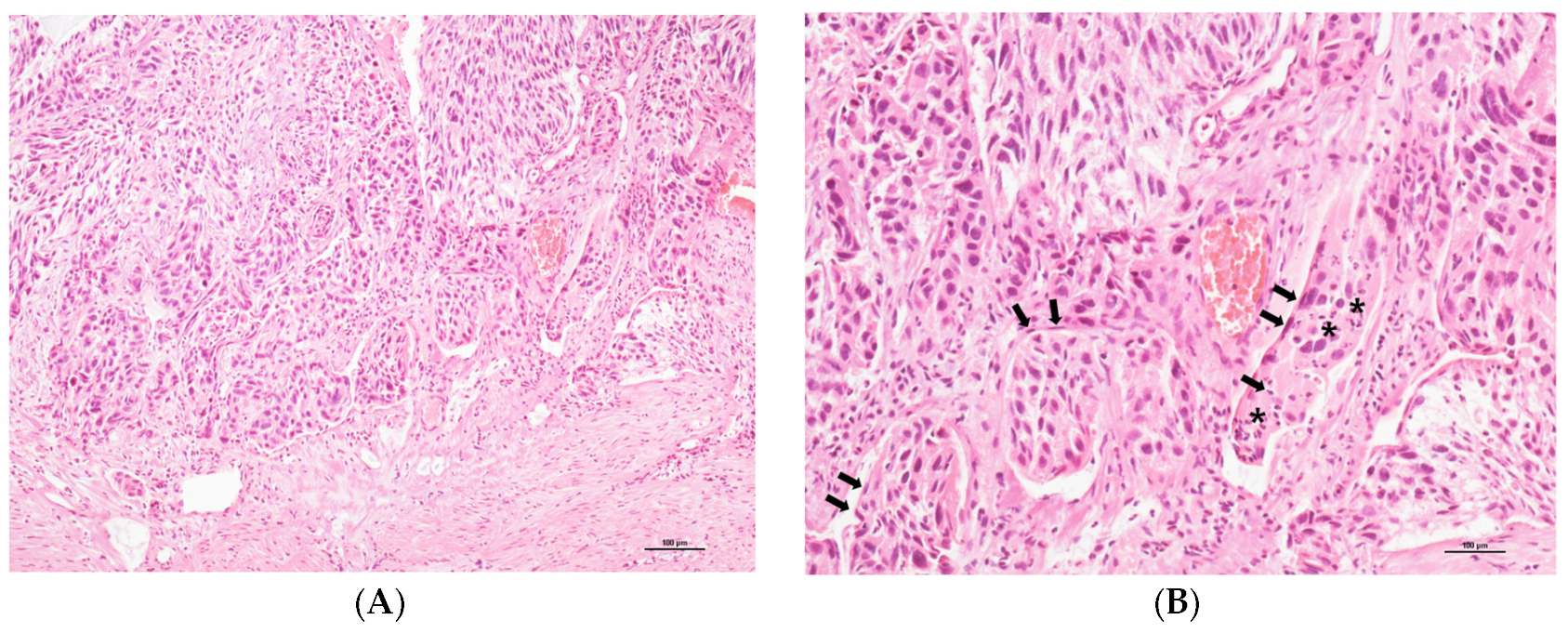
3.2. Representative LVSI Detection
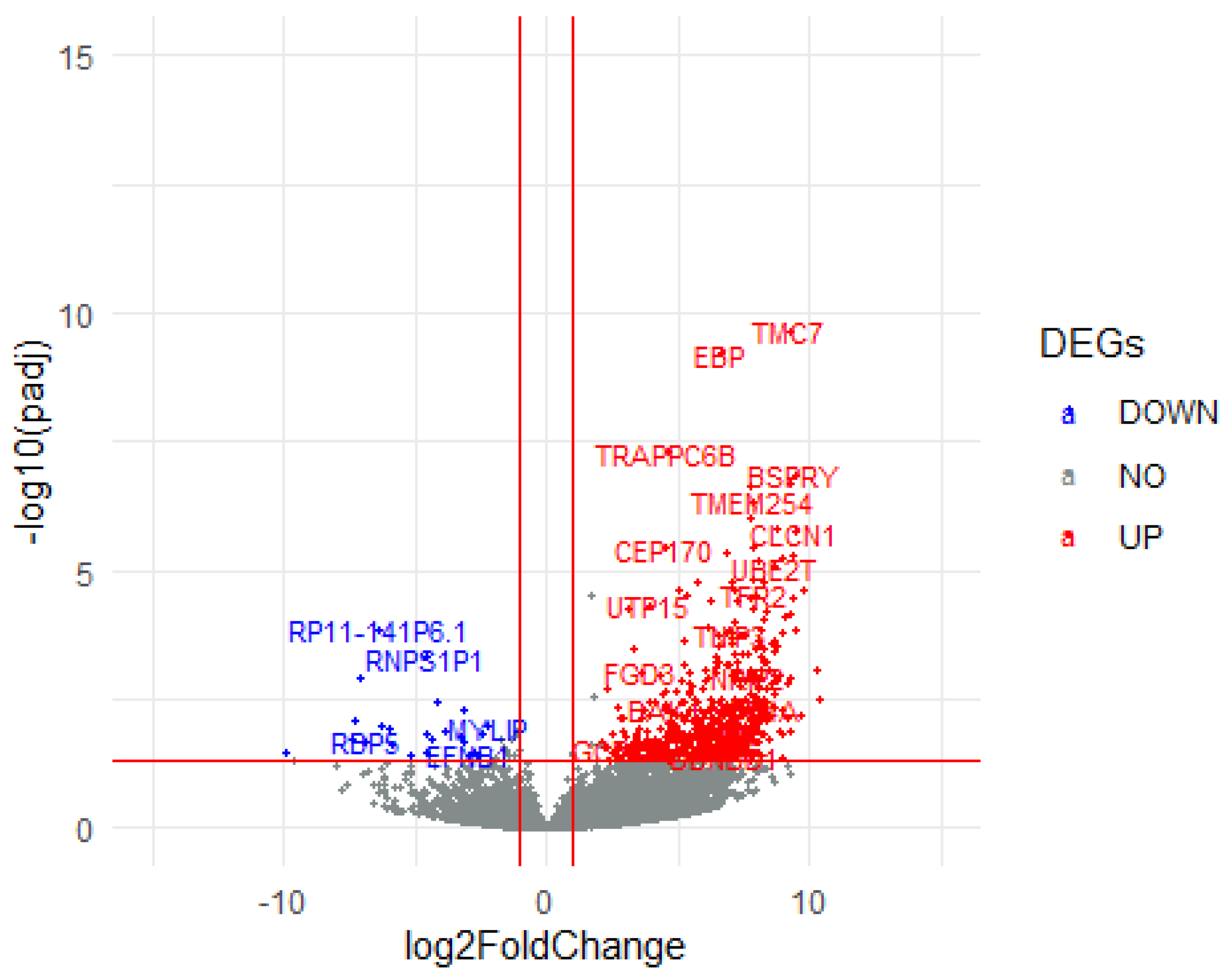
3.3. Differentially Expressed Genes (DEGs)
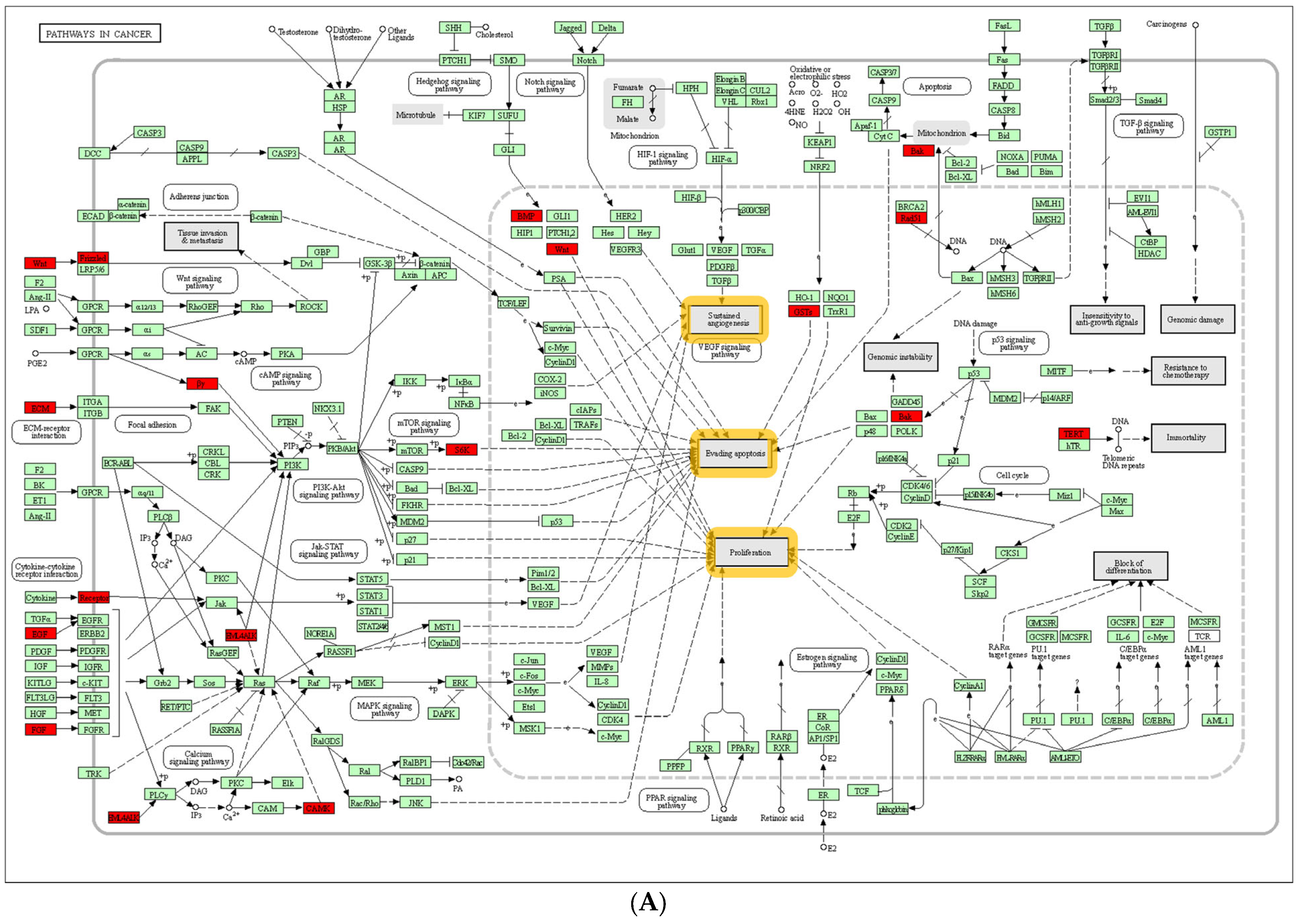
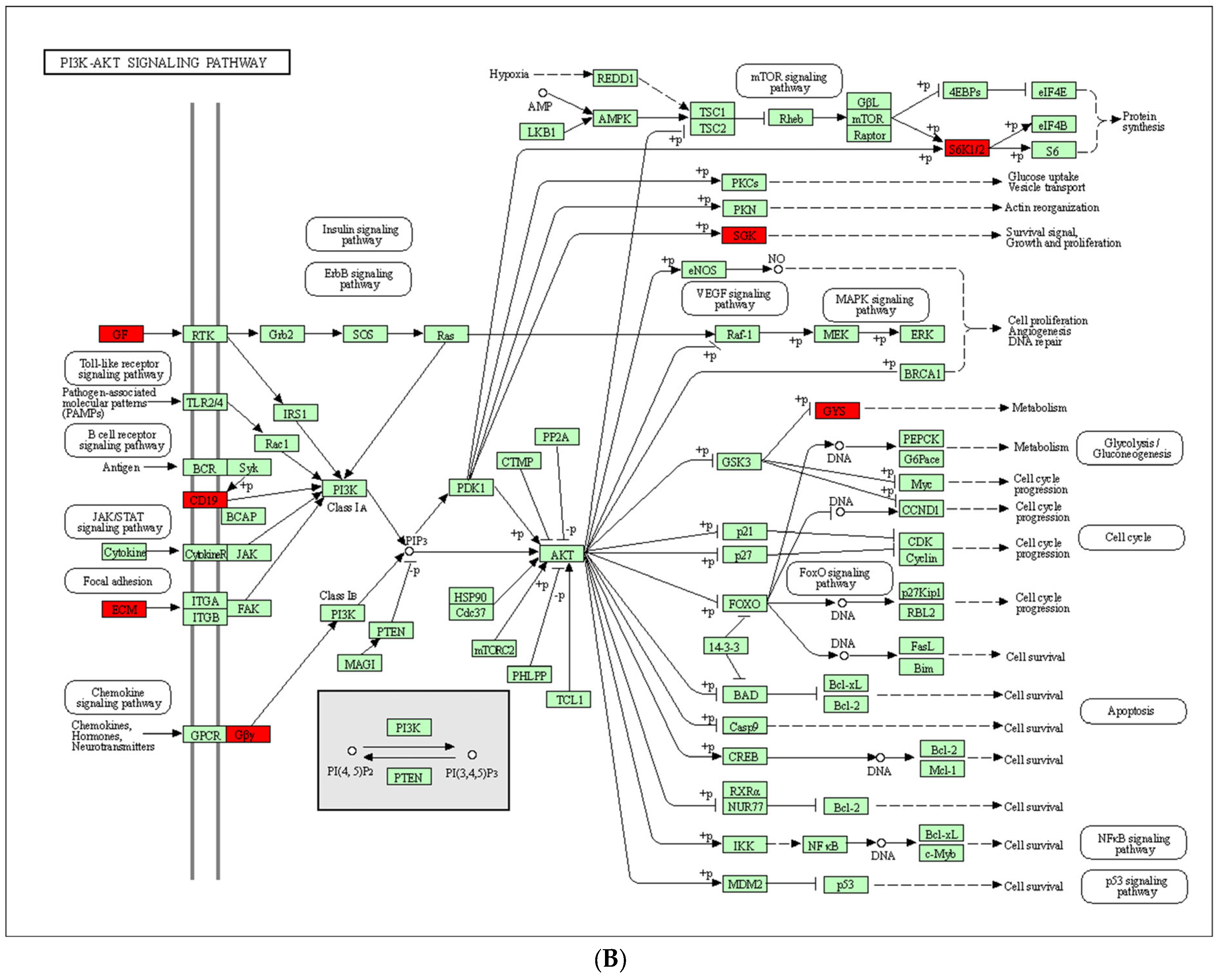
3.4. Intersection of LVSI DEGs with IMPAGT Revealed PI3K/Akt Signaling Pathway Overactivation
3.4.1. Functional Analysis Revealed 67 LVSI-IMPAGT DEGs Are Related to Tumoral Angiogenesis in CC
3.4.2. Immune Deconvolution Exposed Heterogeneity in the Immune Composition of the LVSI+ Group with CC
3.4.3. Regulatory T Cells as Primary Immune Markers Related to the LVSI+ Group within the Heterogenic Immune Composition of LVSI+ in CC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sabel, M.S. Reading the Pathology Report. In Essentials of Breast Surgery; Elsevier: Amsterdam, The Netherlands, 2009; pp. 123–131. [Google Scholar] [CrossRef]
- Tortorella, L.; Restaino, S.; Zannoni, G.F.; Vizzielli, G.; Chiantera, V.; Cappuccio, S.; Gioè, A.; La Fera, E.; Dinoi, G.; Angelico, G.; et al. Substantial Lymph-Vascular Space Invasion (Lvsi) as Predictor of Distant Relapse and Poor Prognosis in Low-Risk Early-Stage Endometrial Cancer. J. Gynecol. Oncol. 2021, 32, e11. [Google Scholar] [CrossRef] [PubMed]
- Schade, G.R.; Wright, J.L.; Lin, D.W. Prognostic Significance of Positive Surgical Margins and Other Implications of Pathology Report. In Prostate Cancer: Science and Clinical Practice, 2nd ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 295–306. [Google Scholar] [CrossRef]
- Oliver-Perez, M.R.; Padilla-Iserte, P.; Arencibia-Sanchez, O.; Martin-Arriscado, C.; Muruzabal, J.C.; Diaz-Feijóo, B.; Cabrera, S.; Coronado, P.; Martín-Salamanca, M.B.; Pantoja-Garrido, M.; et al. Lymphovascular Space Invasion in Early-Stage Endometrial Cancer (LySEC): Patterns of Recurrence and Predictors. A Multicentre Retrospective Cohort Study of the Spain Gynecologic Oncology Group. Cancers 2023, 15, 2612. [Google Scholar] [CrossRef] [PubMed]
- Aleskandarany, M.A.; Sonbul, S.N.; Mukherjee, A.; Rakha, E.A. Molecular Mechanisms Underlying Lymphovascular Invasion in Invasive Breast Cancer. Pathobiology 2015, 82, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Reig, B.; Moy, L.; Sigmund, E.E.; Heacock, L. Biomarkers, Prognosis, and Prediction Factors; Elsevier: Amsterdam, The Netherlands, 2022; ISBN 9780323797023. [Google Scholar]
- Alexander-Sefre, F.; Singh, N.; Ayhan, A.; Salveson, H.B.; Wilbanks, G.; Jacobs, I.J. Detection of Tumour Lymphovascular Space Invasion Using Dual Cytokeratin and CD31 Immunohistochemistry. J. Clin. Pathol. 2003, 56, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, H.; Koumori, K.; Watanabe, H.; Kano, K.; Shimoda, Y.; Aoyama, T.; Yamada, T.; Hiroshi, T.; Yamamoto, N.; Cho, H.; et al. The Clinical Significance of Lymphovascular Invasion in Gastric Cancer. In Vivo 2020, 34, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Narayan, K.; Lin, M.Y.; Kondalsamy-Chennakesavan, S.; Mukhopadhyay, A. Lymphovascular Space Invasion (LVSI)-Based Prognostic Clusters in Endometrial Cancer Patients Treated with Primary Surgery and Adjuvant Radiotherapy. Indian J. Gynecol. Oncol. 2021, 19, 63. [Google Scholar] [CrossRef]
- Ørtoft, G.; Lausten-Thomsen, L.; Høgdall, C.; Hansen, E.S.; Dueholm, M. Lymph-Vascular Space Invasion (LVSI) as a Strong and Independent Predictor for Non-Locoregional Recurrences in Endometrial Cancer: A Danish Gynecological Cancer Group Study. J. Gynecol. Oncol. 2019, 30, e84. [Google Scholar] [CrossRef]
- Liu, Y.L.; Saraf, A.; Lee, S.M.; Zhong, X.; Hibshoosh, H.; Kalinsky, K.; Connolly, E.P. Lymphovascular Invasion Is an Independent Predictor of Survival in Breast Cancer after Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 2016, 157, 555–564. [Google Scholar] [CrossRef]
- Song, Y.J.; Shin, S.H.; Cho, J.S.; Park, M.H.; Yoon, J.H.; Jegal, Y.J. The Role of Lymphovascular Invasion as a Prognostic Factor in Patients with Lymph Node-Positive Operable Invasive Breast Cancer. J. Breast Cancer 2011, 14, 198–203. [Google Scholar] [CrossRef]
- Creasman, W.T.; Kohler, M.F. Is Lymph Vascular Space Involvement an Independent Prognostic Factor in Early Cervical Cancer? Gynecol. Oncol. 2004, 92, 525–529. [Google Scholar] [CrossRef]
- Morice, P.; Piovesan, P.; Rey, A.; Atallah, D.; Haie-Meder, C.; Pautier, P.; Sideris, L.; Pomel, C.; Duvillard, P.; Castaigne, D. Prognostic Value of Lymphovascular Space Invasion Determined with Hematoxylin-Eosin Staining in Early Stage Cervical Carcinoma: Results of a Multivariate Analysis. Ann. Oncol. 2003, 14, 1511–1517. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, C.; Yu, Y.; Guo, Y.; Sun, H. Prediction of Lymphovascular Space Invasion Using a Combination of Tenascin-C, Cox-2, and PET/CT Radiomics in Patients with Early-Stage Cervical Squamous Cell Carcinoma. BMC Cancer 2021, 21, 866. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Wang, Y.; Li, D.; Xia, X.; Tan, Q.; Xiong, X.; Li, Z. Preoperative Prediction of Lymphovascular Space Invasion in Cervical Cancer With Radiomics –Based Nomogram. Front. Oncol. 2021, 11, 637794. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Chong, G.O.; Park, J.Y.; Chung, D.; Lee, Y.H.; Lee, H.J.; Hong, D.G.; Han, H.S.; Lee, Y.S. Tumor Budding in Cervical Cancer as a Prognostic Factor and Its Possible Role as an Additional Intermediate-Risk Factor. Gynecol. Oncol. 2020, 159, 157–163. [Google Scholar] [CrossRef] [PubMed]
- He, X. Bioinformatics Analysis of the Primary Molecular Mechanism in Lymphatic Vascular Space Invasion and Parametrial Invasion of Cervical Cancer. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Santoro, A.; Inzani, F.; Angelico, G.; Arciuolo, D.; Bragantini, E.; Travaglino, A.; Valente, M.; D’Alessandris, N.; Scaglione, G.; Sfregola, S.; et al. Recent Advances in Cervical Cancer Management: A Review on Novel Prognostic Factors in Primary and Recurrent Tumors. Cancers 2023, 15, 1137. [Google Scholar] [CrossRef]
- Zhang, W.; He, W.; Shi, Y.; Zhao, J.; Liu, S.; Zhang, F.; Yang, J.; Xie, C.; Zhang, Y. Aberrant TIMELESS Expression Is Associated with Poor Clinical Survival and Lymph Node Metastasis in Early-Stage Cervical Carcinoma. Int. J. Oncol. 2017, 50, 173–184. [Google Scholar] [CrossRef]
- Pol, F.J.M.; Zusterzeel, P.L.M.; Van Ham, M.A.P.C.; Kuijpers, D.A.T.; Bulten, J.; Massuger, L.F.A.G. Satellite Lymphovascular Space Invasion: An Independent Risk Factor in Early Stage Cervical Cancer. Gynecol. Oncol. 2015, 138, 579–584. [Google Scholar] [CrossRef]
- Kariri, Y.A.; Aleskandarany, M.A.; Joseph, C.; Kurozumi, S.; Mohammed, O.J.; Toss, M.S.; Green, A.R.; Rakha, E.A. Molecular Complexity of Lymphovascular Invasion: The Role of Cell Migration in Breast Cancer as a Prototype. Pathobiology 2020, 87, 218–231. [Google Scholar] [CrossRef]
- Asaoka, M.; Patnaik, S.K.; Zhang, F.; Ishikawa, T.; Takabe, K. Lymphovascular Invasion in Breast Cancer Is Associated with Gene Expression Signatures of Cell Proliferation but Not Lymphangiogenesis or Immune Response. Breast Cancer Res. Treat. 2020, 181, 309–322. [Google Scholar] [CrossRef]
- Jiang, H.H.; Zhang, Z.Y.; Wang, X.Y.; Tang, X.; Liu, H.L.; Wang, A.L.; Li, H.G.; Tang, E.J.; Lin, M. Bin Prognostic Significance of Lymphovascular Invasion in Colorectal Cancer and Its Association with Genomic Alterations. World J. Gastroenterol. 2019, 25, 2489–2502. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zheng, M.; Zhou, Q.M.; Zhang, M.Y.; Jia, W.H.; Yun, J.P.; Wang, H.Y. Identification of a Gene-Expression Signature for Predicting Lymph Node Metastasis in Patients with Early Stage Cervical Carcinoma. Cancer 2011, 117, 3363–3373. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. In Bioinformatics the Impact of Accurate Quantification on Proteomic and Genetic Analysis and Research; Apple Academic Press: New York, NY, USA, 2014; pp. 41–74. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Choi, Y.; Park, N.J.Y.; Le, T.M.; Lee, E.; Lee, D.; Nguyen, H.D.T.; Cho, J.; Park, J.Y.; Han, H.S.; Chong, G.O. Immune Pathway and Gene Database (IMPAGT) Revealed the Immune Dysregulation Dynamics and Overactivation of the PI3K/Akt Pathway in Tumor Buddings of Cervical Cancer. Curr. Issues Mol. Biol. 2022, 44, 5139–5152. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A Novel Biological Module-Centric Algorithm to Functionally Analyze Large Gene Lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; List, M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2020, 2120, 223–232. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous Enumeration of Cancer and Immune Cell Types from Bulk Tumor Gene Expression Data. Elife 2017, 6, e26476. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. XCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Dai, L.J.; Wu, S.Y.; Xiao, Y.; Ma, D.; Jiang, Y.Z.; Shao, Z.M. Spatial Architecture of the Immune Microenvironment Orchestrates Tumor Immunity and Therapeutic Response. J. Hematol. Oncol. 2021, 14, 98. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/MTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef]
- Garrido, M.P.; Torres, I.; Vega, M.; Romero, C. Angiogenesis in Gynecological Cancers: Role of Neurotrophins. Front. Oncol. 2019, 9, 913. [Google Scholar] [CrossRef]
- Jiang, B.H.; Liu, L.Z. PI3K/PTEN Signaling in Tumorigenesis and Angiogenesis. Biochim. Biophys. Acta Proteins Proteom. 2008, 1784, 150–158. [Google Scholar] [CrossRef]
- Okkenhaug, K.; Graupera, M.; Vanhaesebroeck, B. Targeting PI3K in Cancer: Impact on Tumor Cells, Their Protective Stroma, Angiogenesis, and Immunotherapy. Cancer Discov. 2016, 6, 1090–1105. [Google Scholar] [CrossRef]
- Soler, A.; Serra, H.; Pearce, W.; Angulo, A.; Guillermet-Guibert, J.; Friedman, L.S.; Viñals, F.; Gerhardt, H.; Casanovas, O.; Graupera, M.; et al. Inhibition of the P110α Isoform of PI 3-Kinase Stimulates Nonfunctional Tumor Angiogenesis. J. Exp. Med. 2013, 210, 1937–1945. [Google Scholar] [CrossRef]
- Soler, A.; Angulo-Urarte, A.; Graupera, M. PI3K at the Crossroads of Tumor Angiogenesis Signaling Pathways. Mol. Cell. Oncol. 2015, 2, e975624. [Google Scholar] [CrossRef]
- Graupera, M.; Guillermet-Guibert, J.; Foukas, L.C.; Phng, L.K.; Cain, R.J.; Salpekar, A.; Pearce, W.; Meek, S.; Millan, J.; Cutillas, P.R.; et al. Angiogenesis Selectively Requires the P110α Isoform of PI3K to Control Endothelial Cell Migration. Nature 2008, 453, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Brackenbury, W.J. Membrane Potential and Cancer Progression. Front. Physiol. 2013, 4, 185. [Google Scholar] [CrossRef] [PubMed]
- Djamgoz, M.B.A.; Coombes, R.C.; Schwab, A. Ion Transport and Cancer: From Initiation to Metastasis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Fiorio Pla, A.; Munaron, L. Functional Properties of Ion Channels and Transporters in Tumour Vascularization. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130103. [Google Scholar] [CrossRef]
- Andersen, A.P.; Moreira, J.M.A.; Pedersen, S.F. Interactions of Ion Transporters and Channels with Cancer Cell Metabolism and the Tumour Microenvironment. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130098. [Google Scholar] [CrossRef]
- Arcangeli, A.; Crociani, O.; Bencini, L. Interaction of Tumour Cells with Their Microenvironment: Ion Channels and Cell Adhesion Molecules. A Focus on Pancreatic Cancer. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130101. [Google Scholar] [CrossRef][Green Version]
- Prevarskaya, N.; Ouadid-Ahidouch, H.; Skryma, R.; Shuba, Y. Remodelling of Ca2+ Transport in Cancer: How It Contributes to Cancer Hallmarks? Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130097. [Google Scholar] [CrossRef]
- Moccia, F.; Negri, S.; Shekha, M.; Faris, P.; Guerra, G. Endothelial Ca2+ Signaling, Angiogenesis and Vasculogenesis: Just What It Takes to Make a Blood Vessel. Int. J. Mol. Sci. 2019, 20, 3962. [Google Scholar] [CrossRef]
- Bong, A.H.L.; Monteith, G.R. Calcium Signaling and the Therapeutic Targeting of Cancer Cells. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1786–1794. [Google Scholar] [CrossRef]
- Martial, S. Involvement of Ion Channels and Transporters in Carcinoma Angiogenesis and Metastasis. Am. J. Physiol. Cell Physiol. 2016, 310, C710–C727. [Google Scholar] [CrossRef]
- Jones, C.A.; Hazlehurst, L.A. Role of Calcium Homeostasis in Modulating Emt in Cancer. Biomedicines 2021, 9, 1200. [Google Scholar] [CrossRef]
- Li, C.; Wu, H.; Guo, L.; Liu, D.; Yang, S.; Li, S.; Hua, K. Single-Cell Transcriptomics Reveals Cellular Heterogeneity and Molecular Stratification of Cervical Cancer. Commun. Biol. 2022, 5, 1208. [Google Scholar] [CrossRef]
- Litwin, T.R.; Irvin, S.R.; Chornock, R.L.; Sahasrabuddhe, V.V.; Stanley, M.; Wentzensen, N. Infiltrating T-Cell Markers in Cervical Carcinogenesis: A Systematic Review and Meta-Analysis. Br. J. Cancer 2021, 124, 831–841. [Google Scholar] [CrossRef]
- Facciabene, A.; Motz, G.T.; Coukos, G. T-Regulatory Cells: Key Players in Tumor Immune Escape and Angiogenesis. Cancer Res. 2012, 72, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Visser, J.; Nijman, H.W.; Hoogenboom, B.N.; Jager, P.; Van Baarle, D.; Schuuring, E.; Abdulahad, W.; Miedema, F.; Van Der Zee, A.G.; Daemen, T. Frequencies and Role of Regulatory T Cells in Patients with (Pre)Malignant Cervical Neoplasia. Clin. Exp. Immunol. 2007, 150, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Galli, F.; Aguilera, J.V.; Palermo, B.; Markovic, S.N.; Nisticò, P.; Signore, A. Relevance of Immune Cell and Tumor Microenvironment Imaging in the New Era of Immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lazarus, J.; Steele, N.G.; Yan, W.; Lee, H.J.; Nwosu, Z.C.; Halbrook, C.J.; Menjivar, R.E.; Kemp, S.B.; Sirihorachai, V.R.; et al. Regulatory T-Cell Depletion Alters the Tumor Microenvironment and Accelerates Pancreatic Carcinogenesis. Cancer Discov. 2020, 10, 422–439. [Google Scholar] [CrossRef]
- Pacella, I.; Piconese, S. Immunometabolic Checkpoints of Treg Dynamics: Adaptation to Microenvironmental Opportunities and Challenges. Front. Immunol. 2019, 10, 1889. [Google Scholar] [CrossRef]
- Hori, S. Regulatory T Cell Plasticity: Beyond the Controversies. Trends Immunol. 2011, 32, 295–300. [Google Scholar] [CrossRef]
- Barua, S.; Fang, P.; Sharma, A.; Fujimoto, J.; Wistuba, I.; Rao, A.U.K.; Lin, S.H. Spatial Interaction of Tumor Cells and Regulatory T Cells Correlates with Survival in Non-Small Cell Lung Cancer. Lung Cancer 2018, 117, 73–79. [Google Scholar] [CrossRef]
- Lužnik, Z.; Anchouche, S.; Dana, R.; Yin, J. Regulatory T Cells in Angiogenesis. J. Immunol. 2020, 205, 2557–2565. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W. Molecular Classification of Endometrial Cancer and the 2023 FIGO Staging: Exploring the Challenges and Opportunities for Pathologists. Cancers 2023, 15, 4101. [Google Scholar] [CrossRef] [PubMed]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Berek, J.S.; et al. FIGO Staging of Endometrial Cancer: 2023. Int. J. Gynecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef]
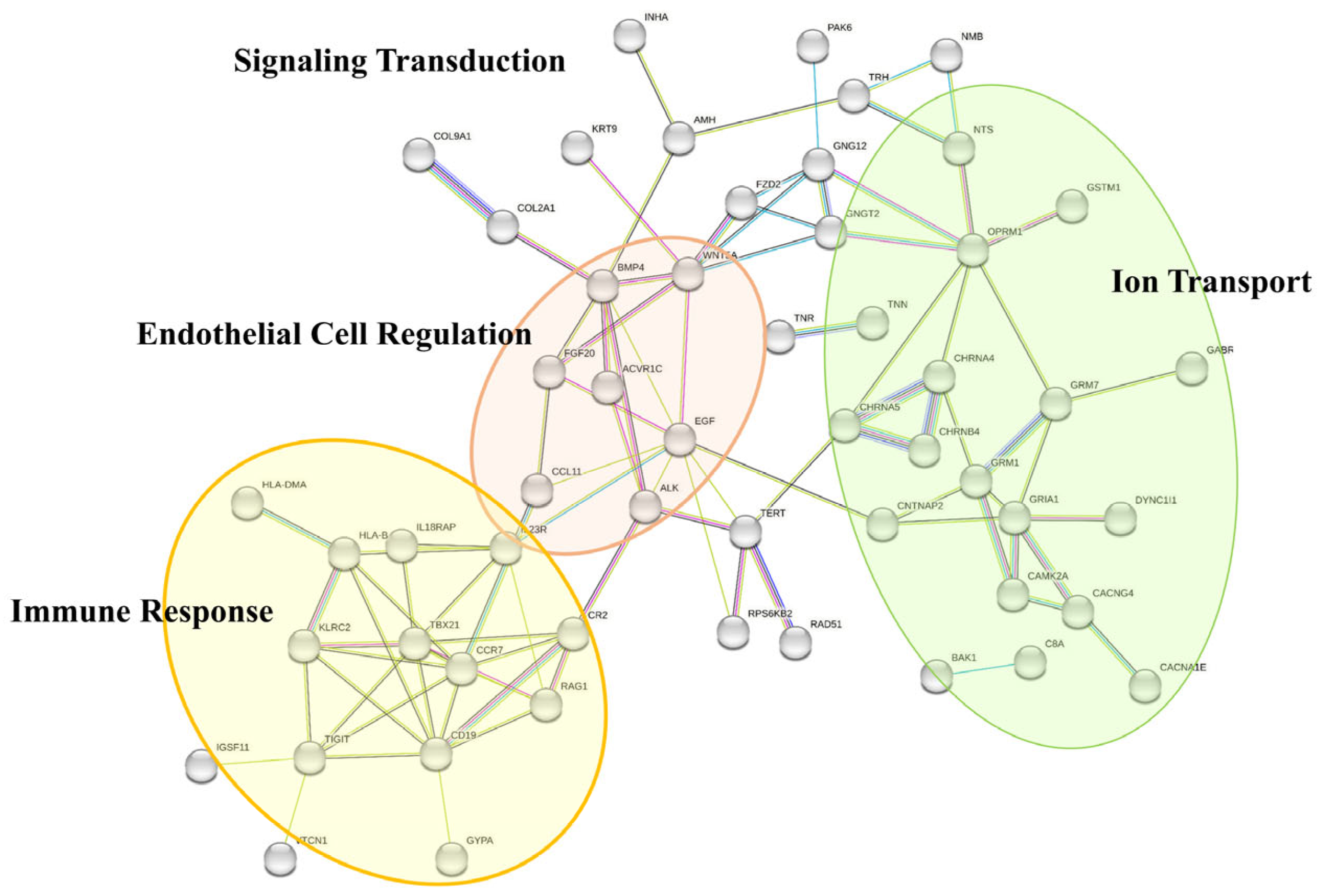

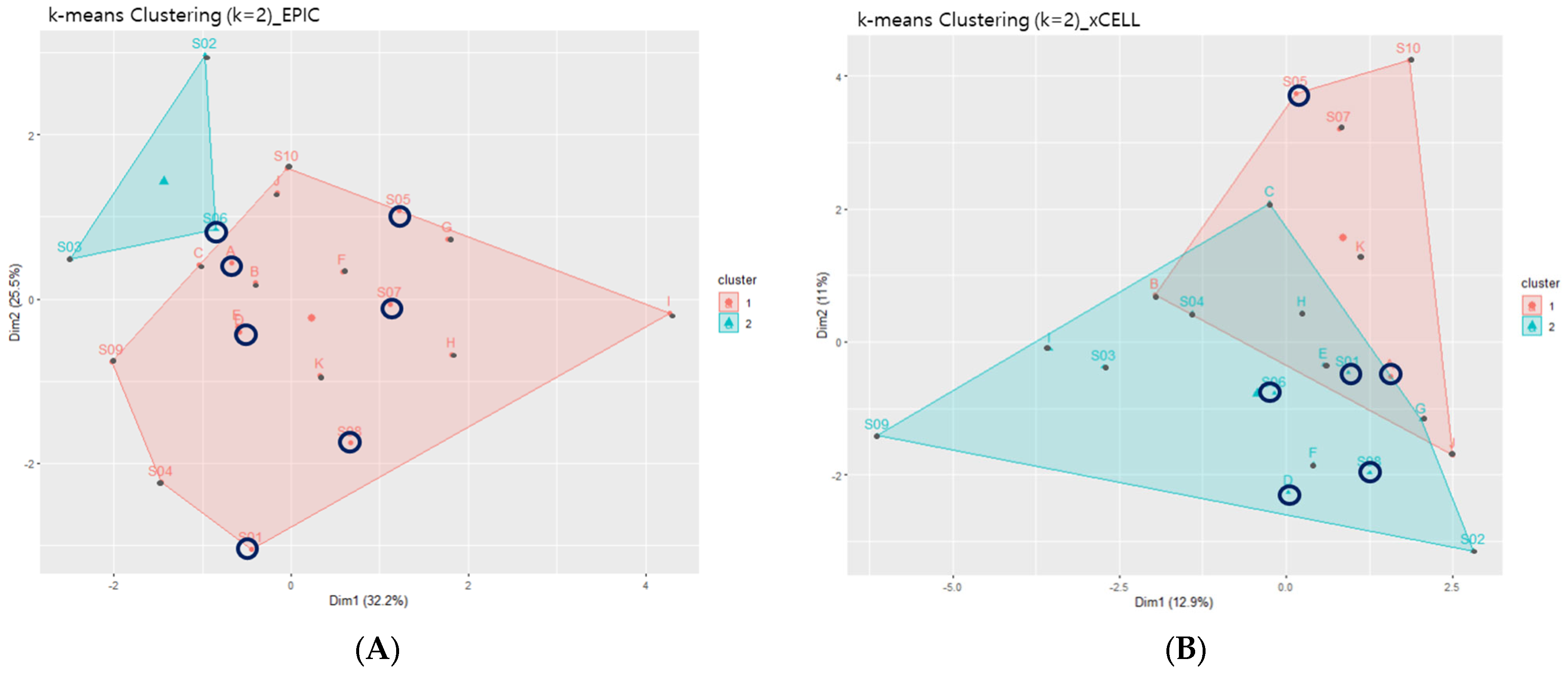
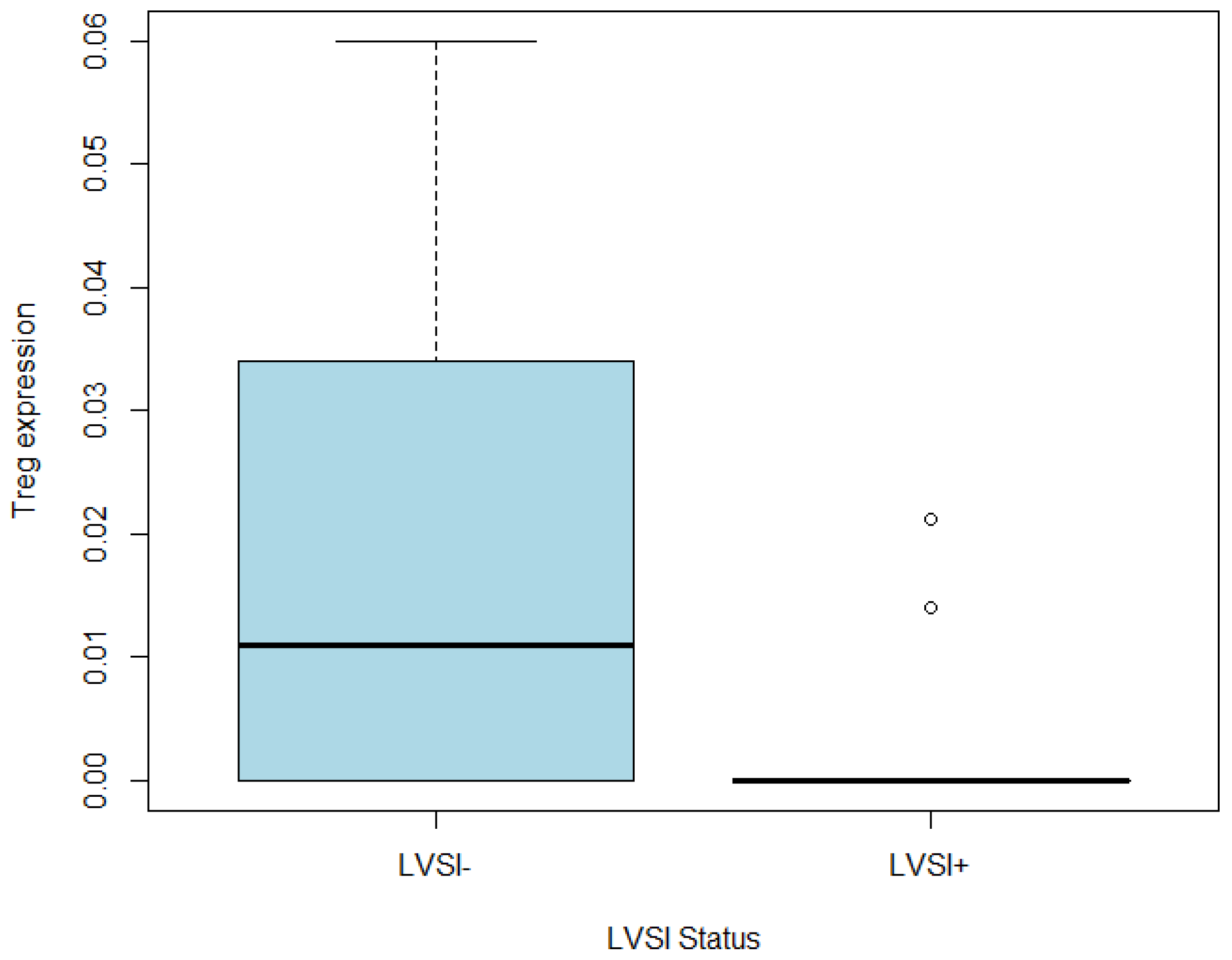
| Variables | All (n = 21) | LVSI− (n = 6) | LVSI+ (n = 15) | p-Value |
|---|---|---|---|---|
| Age (years ± SD) | 47.4 ± 11.0 | 46.8 ± 9.2 | 47.6 ± 11.3 | 0.880 |
| FIGO stage (n, %) | 0.586 | |||
| IB1 | 11 (52.4) | 4 (66.7) | 7 (46.7) | |
| IB2 | 2 (9.5) | 1 (16.7) | 1 (6.7) | |
| IIA2 | 4 (19.0) | 1 (16.7) | 3 (20.0) | |
| IIB | 4 (19.0) | 0 | 4 (26.7) | |
| Histology | 0.862 | |||
| Squamous cell carcinoma | 15 (71.4) | 5 (83.3) | 10 (66.7) | |
| Adenocarcinoma | 5 (23.8) | 1 (16.7) | 4 (26.7) | |
| Adenosquamous carcinoma | 1 (4.8) | 0 | 1 (6.7) | |
| Primary tumor size (cm ± SD) | 3.3 ± 1.6 | 2.7 ± 1.0 | 3.5 ± 2.0 | 0.367 |
| Deep stromal invasion (n, %) | 20 (95.2) | 5 (83.3) | 15 (100.0) | 0.286 |
| Parametrial invasion (n, %) | 9 (42.9) | 1 (16.7) | 8 (53.3) | 0.178 |
| Positive vaginal margin (n, %) | 2 (9.5) | 1 (16.7) | 1 (6.7) | 0.500 |
| Lymph node metastasis (n, %) | 11 (52.4) | 3 (50.0) | 8 (53.3) | 1.000 |
| Lymphadenectomy | 0.084 | |||
| Pelvic lymphadenectomy | 4 (19.0) | 3 (50.0) | 1 (6.7) | |
| Pelvic and low paraaortic lymphadenectomy | 7 (33.3) | 1 (16.7) | 6 (40.0) | |
| Pelvic and extended lymphadenectomy | 10 (47.6) | 2 (33.3) | 8 (53.3) | |
| Adjuvant therapy | 0.835 | |||
| Chemotherapy | 5 (23.8) | 1 (16.7) | 4 (26.7) | |
| Radiotherapy | 2 (9.5) | 0 | 2 (13.3) | |
| Concurrent chemotherapy | 10 (47.6) | 2 (33.3) | 8 (53.3) | |
| Recurrence (n, %) | 6 (28.6) | 4 (66.7) | 2 (13.3) | 0.031 |
| KEGG ID | IMPAGT Pathway | Number of DEGs |
|---|---|---|
| Hsa 05200 | Pathway in Cancer | 16 |
| Hsa 04121 | PI3K/Akt Signaling Pathway | 13 |
| Hsa 04060 | Cytokine-Cytokine Receptor Interaction | 9 |
| Hsa 04514 | Cell Adhesion Molecules | 7 |
| Hsa 04640 | Hematopoietic Cell Lineage | 5 |
| Hsa 04010 | MAPK Signaling Pathway | 5 |
| Group | GO: BP ID | BP Terms | p-Value |
|---|---|---|---|
| Tumor | GO: 0007165 | Signal Transduction | <0.0001 |
| GO: 0051092 | Positive Regulation of NF-kappa B Transcription Factor Activity | <0.001 | |
| GO: 0034220 | Ion Transmembrane Transport | 0.001 | |
| GO: 0001938 | Positive Regulation of Endothelial Cell Proliferation | 0.002 | |
| GO: 0030334 | Regulation of Cell Migration | 0.007 | |
| GO: 0007267 | Cell–Cell Signaling | 0.008 | |
| GO: 0006468 | Protein Phosphorylation | 0.001 | |
| GO: 0043410 | Positive Regulation of MAPK Cascade | 0.018 | |
| GO: 0010595 | Positive Regulation of Endothelial Cell Migration | 0.022 | |
| GO: 0032092 | Positive Regulation of Protein Binding | 0.028 | |
| GO: 0006816 | Calcium Ion Transport | 0.037 | |
| Immune | GO: 0006955 | Immune Response | <0.0001 |
| GO: 0001916 | Positive Regulation of T Cell-Mediated Cytotoxicity | 0.007 | |
| GO: 0050778 | Positive Regulation of Immune Response | 0.007 | |
| GO: 0030333 | Antigen Processing and Presentation | 0.012 | |
| GO: 0002250 | Adaptive Immune Response | 0.028 | |
| GO: 0030183 | B Cell Differentiation | 0.038 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.; Ando, Y.; Lee, D.; Kim, N.Y.; Lee, O.E.M.; Cho, J.; Seo, I.; Chong, G.O.; Park, N.J.-Y. Profiling of Lymphovascular Space Invasion in Cervical Cancer Revealed PI3K/Akt Signaling Pathway Overactivation and Heterogenic Tumor-Immune Microenvironments. Life 2023, 13, 2342. https://doi.org/10.3390/life13122342
Choi Y, Ando Y, Lee D, Kim NY, Lee OEM, Cho J, Seo I, Chong GO, Park NJ-Y. Profiling of Lymphovascular Space Invasion in Cervical Cancer Revealed PI3K/Akt Signaling Pathway Overactivation and Heterogenic Tumor-Immune Microenvironments. Life. 2023; 13(12):2342. https://doi.org/10.3390/life13122342
Chicago/Turabian StyleChoi, Yeseul, Yu Ando, Donghyeon Lee, Na Young Kim, Olive E. M. Lee, Junghwan Cho, Incheol Seo, Gun Oh Chong, and Nora Jee-Young Park. 2023. "Profiling of Lymphovascular Space Invasion in Cervical Cancer Revealed PI3K/Akt Signaling Pathway Overactivation and Heterogenic Tumor-Immune Microenvironments" Life 13, no. 12: 2342. https://doi.org/10.3390/life13122342
APA StyleChoi, Y., Ando, Y., Lee, D., Kim, N. Y., Lee, O. E. M., Cho, J., Seo, I., Chong, G. O., & Park, N. J.-Y. (2023). Profiling of Lymphovascular Space Invasion in Cervical Cancer Revealed PI3K/Akt Signaling Pathway Overactivation and Heterogenic Tumor-Immune Microenvironments. Life, 13(12), 2342. https://doi.org/10.3390/life13122342






