Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Chemical Analysis of Water Samples
2.2.1. Chemicals
2.2.2. Preparation of the Internal Standard Solution
2.2.3. Solid-Phase Extraction (SPE) of Antibiotics from Water Samples
2.2.4. Measurement of Antibiotic Concentrations in Wastewater Samples
2.3. Isolation and Identification of VRE
2.4. Detection of Antimicrobial Susceptibility
2.5. PCR Detection of Resistance Genes and Virulence Factors
2.6. Pulsed-Field Gel Electrophoresis
3. Results and Discussion
3.1. Detecting the Presence of Selected Antibiotics in Water Samples
3.2. Determining PNECs for the Tested Antibiotics in Samples
3.3. Molecular Biological Characteristics of VRE in Wastewater and Surface Water
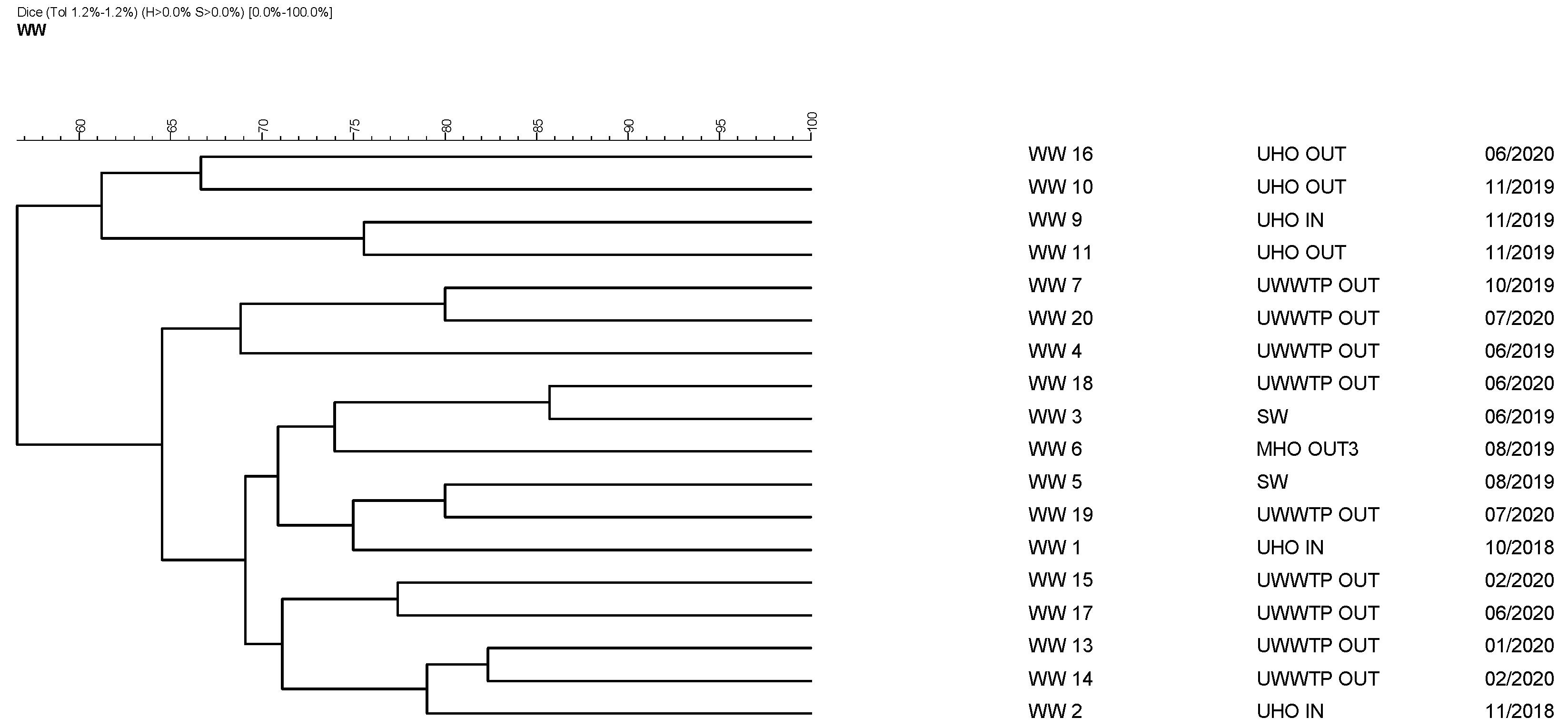
3.4. Pulsed-Field Gel Electrophoresis of 18 Isolates
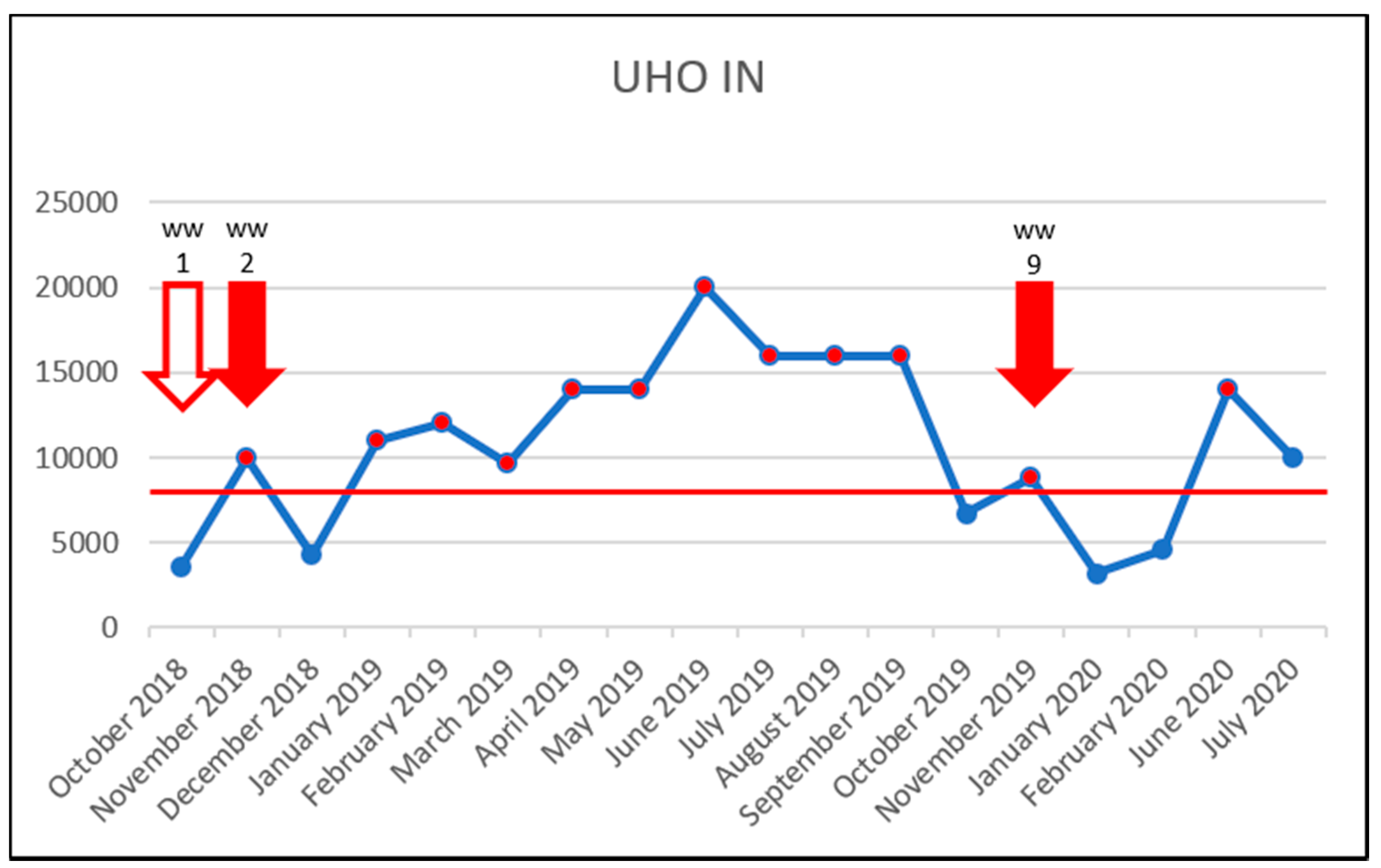
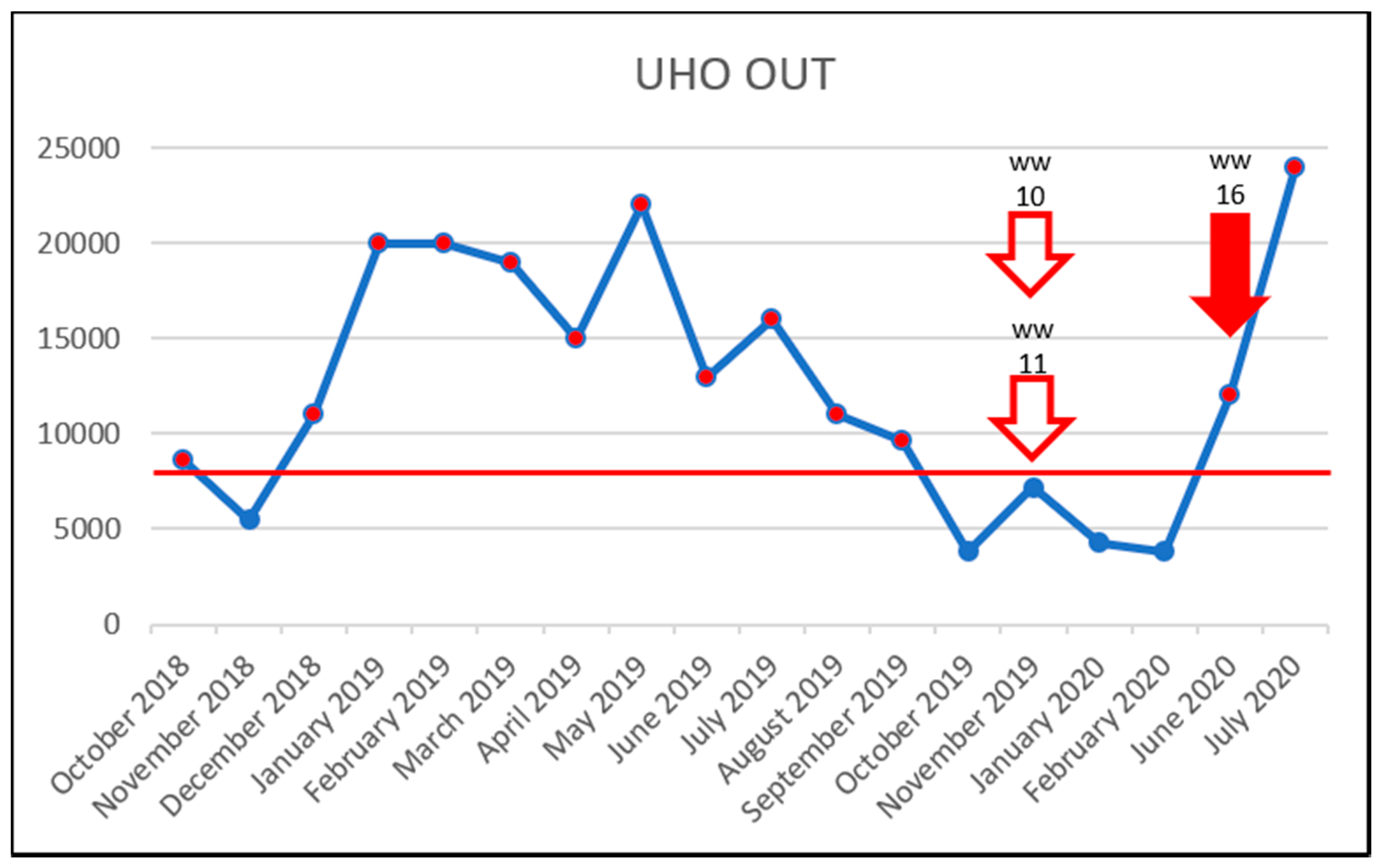
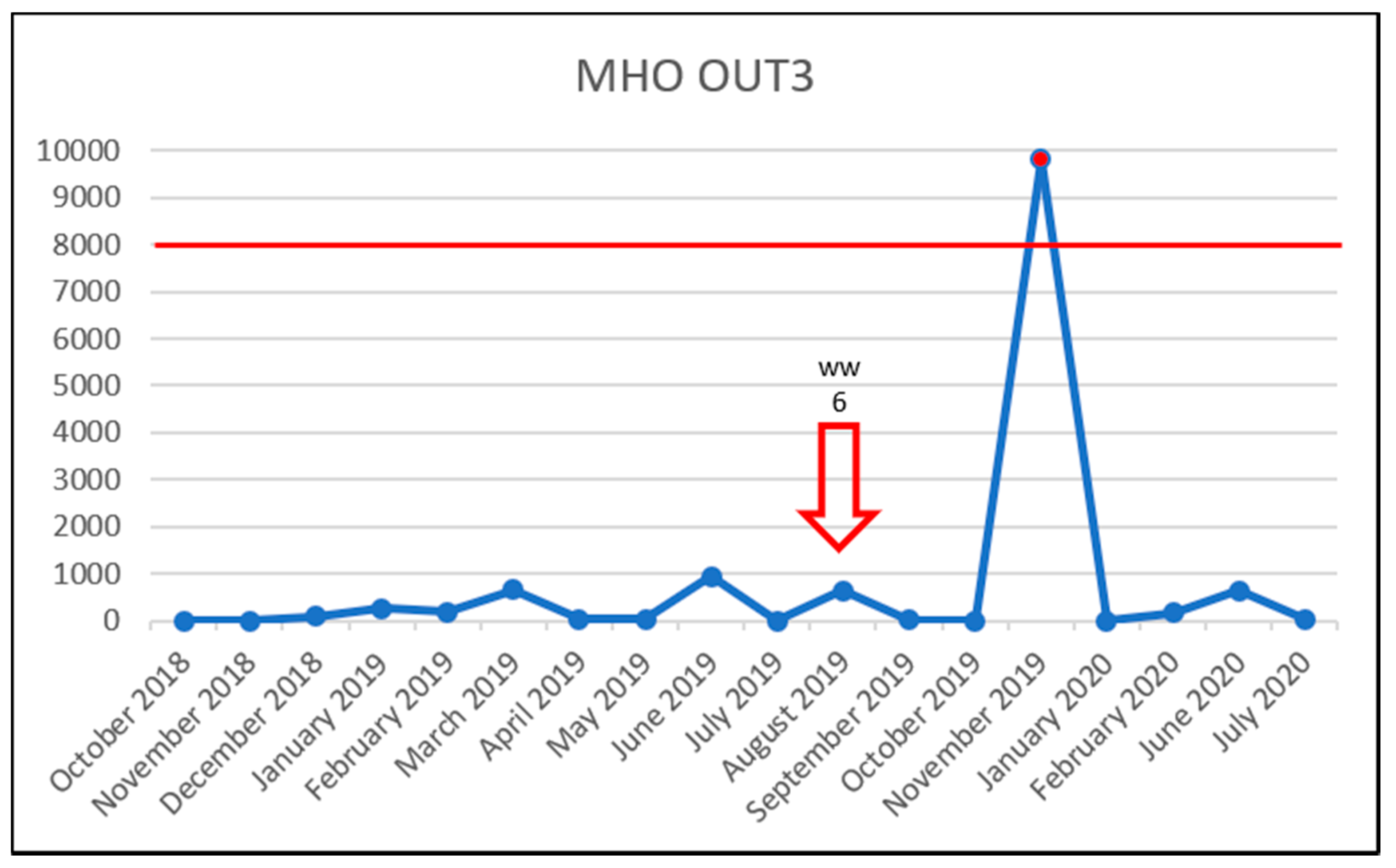
3.5. Effect of Vancomycin on VRE Prevalence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- One Health Initiative. Available online: http://www.onehealthinitiative.com (accessed on 2 November 2021).
- Černohorská, L. Rod Enterococcus. 1.8 Grampozitivní aerobní a fakultativně anaerobní koky. In A Kol: Lékařská Mikrobiologie Speciální; Votava, M., Ed.; Neptun: Brno, Czech Republic, 2006; pp. 127–128. [Google Scholar]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef] [Green Version]
- Yadav, G.; Thakuria, B.; Madan, M.; Agwan, V.; Pandey, A. Linezolid and Vancomycin Resistant Enterococci: A Therapeutic Problem. J. Clin. Diagn. Res. 2017, 11, 7–11. [Google Scholar] [CrossRef]
- Raza, T.; Ullah, S.R.; Mehmood, K.; Andleeb, S. Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar]
- Kirkizlar, T.A.; Akalin, H.; Kirkizlar, O.; Ozkalemkas, F.; Ozkocaman, V.; Kazak, E.; Ozakin, C.; Bulbul, E.N.; Ozboz, E.S.; Ali, R. Vancomycin-resistant enterococci infection and predisposing factors for infection and mortality in patients with acute leukaemia and febrile neutropenia. Leuk. Res. 2020, 99, 106463. [Google Scholar] [CrossRef]
- Prematunge, C.; MacDougall, C.; Johnstone, J.; Adomako, K.; Lam, F.; Robertson, J.; Garber, G. VRE and VSE Bacteremia Outcomes in the Era of Effective VRE Therapy: A Systematic Review and Meta-analysis. Infect. Control Hosp. Epidemiol. 2016, 37, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Hricová, K.; Štosová, T.; Kučová, P.; Fišerová, K.; Bardoň, J.; Kolář, M. Analysis of Vancomycin-Resistant Enterococci in Hemato-Oncological Patients. Antibiotics 2020, 9, 785. [Google Scholar] [CrossRef]
- Wist, V.; Morach, M.; Schneeberger, M.; Cernela, N.; Stevens, M.J.A.; Zurfluh, K.; Stephan, R.; Nüesch-Inderbinen, M. Phenotypic and Genotypic Traits of Vancomycin-Resistant Enterococci from Healthy Food-Producing Animals. Microorganisms 2020, 8, 261, Erratum in Microorganisms 2021, 9, 847. [Google Scholar] [CrossRef] [Green Version]
- Asgin, N.; Otlu, B. Antibiotic Resistance and Molecular Epidemiology of Vancomycin-Resistant Enterococci in a Tertiary Care Hospital in Turkey. Infect. Drug Resist. 2020, 13, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [Green Version]
- Hassoun-Kheir, N.; Stabholz, Y.; Kreft, J.-U.; de la Cruz, R.; Romalde, J.L.; Nesme, J.; Sørensen, S.J.; Smets, B.F.; Graham, D.; Paul, M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Sci. Total Environ. 2020, 743, 140804. [Google Scholar] [CrossRef]
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ștefan, M.G.; Loghin, F. Antibiotics in the environment: Causes and consequences. Med. Pharm. Rep. 2020, 93, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Pal, D. Antibiotic resistance and wastewater: Correlation, impact and critical human health challenges. J. Environ. Chem. Eng. 2018, 6, 52–58. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Vaz-Moreira, I.; Della Giustina, S.V.; Llorca, M.; Barceló, D.; Schubert, S.; Berendonk, T.U.; Michael-Kordatou, I.; Fatta-Kassinos, D.; Martinez, J.L.; et al. Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 2020, 140, 105733. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Sun, X.; Wu, J.; Ma, L.; Zhou, Y.; Lin, K.; Luo, Y.; Cui, C. Risk assessment of antibiotic resistance genes in the drinking water system. Sci. Total Environ. 2021, 800, 149650. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.; Galvin, S.; Boyle, F.; Hickey, P.; Mulligan, M.; Cormican, M. Enterococcus faecium of thevanAGenotype in Rural Drinking Water, Effluent, and the Aqueous Environment. Appl. Environ. Microbiol. 2011, 78, 596–598. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2017, 42, fux053. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Larsson, D.J. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef] [Green Version]
- Moore, B.; Perry, C.E.L.; Chard, S.T. A Survey by the sewage swab method of latent enteric infection in an urban area. J. Hyg. 1952, 50, 137–156. [Google Scholar] [CrossRef] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. Available online: http://www.eucast.org (accessed on 2 November 2021).
- Dutka-Malen, S.; Evers, S.; Courvalin, P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995, 33, 24–27. [Google Scholar] [CrossRef] [Green Version]
- Malhotra-Kumar, S.; Lammens, C.; Piessens, J.; Goossens, H. Multiplex PCR for Simultaneous Detection of Macrolide and Tetracycline Resistance Determinants in Streptococci. Antimicrob. Agents Chemother. 2005, 49, 4798–4800. [Google Scholar] [CrossRef] [Green Version]
- Ng, L.-K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Weigel, L.M.; Donlan, R.M.; Shin, D.H.; Jensen, B.; Clark, N.C.; McDougal, L.K.; Zhu, W.; Musser, K.A.; Thompson, J.; Kohlerschmidt, D.; et al. High-Level Vancomycin-Resistant Staphylococcus aureus Isolates Associated with a Polymicrobial Biofilm. Antimicrob. Agents Chemother. 2007, 51, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J.W. Multiplex PCR for Detection of Aminoglycoside Resistance Genes in Enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef] [Green Version]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a Multiplex PCR for the Detection of asa1, gelE, cylA, esp, and hyl Genes in Enterococci and Survey for Virulence Determinants among European Hospital Isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poeta, P.; Costa, D.; Igrejas, G.; Sáenz, Y.; Zarazaga, M.; Rodrigues, J.; Torres, C. Polymorphisms of the pbp5 gene and correlation with ampicillin resistance in Enterococcus faecium isolates of animal origin. J. Med. Microbiol. 2007, 56, 236–240. [Google Scholar] [CrossRef]
- Husickova, V.; Cekanova, L.; Chroma, M.; Htoutou-Sedlakova, M.; Hricova, K.; Kolář, M. Carriage of ESBL- and AmpC-positive Enterobacteriaceae in the gastrointestinal tract of community subjects and hospitalized patients in the Czech Republic. Biomed. Pap. 2012, 156, 348–353. [Google Scholar] [CrossRef] [Green Version]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Goessens, T.; Huysman, S.; De Troyer, N.; Deknock, A.; Goethals, P.; Lens, L.; Vanhaecke, L.; Croubels, S. Multi-class analysis of 46 antimicrobial drug residues in pond water using UHPLC-Orbitrap-HRMS and application to freshwater ponds in Flanders, Belgium. Talanta 2020, 220, 121326. [Google Scholar] [CrossRef]
- Guan, J.; Zhang, C.; Wang, Y.; Guo, Y.; Huang, P.; Zhao, L. Simultaneous determination of 12 pharmaceuticals in water samples by ultrasound-assisted dispersive liquid-liquid microextraction coupled with ultra-high performance liquid chromatography with tandem mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 8099–8109. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, L.; Liang, N.; Chen, H.; Hou, X. Simultaneous determination of seven residual pharmaceuticals in wastewater by solid-phase extraction and liquid chromatography coupled to tandem mass spectrometry with a switching ionization mode. Anal. Methods 2014, 6, 9045–9052. [Google Scholar] [CrossRef]
- Camacho-Muñoz, D.; Kasprzyk-Hordern, B.; Thomas, K.V. Enantioselective simultaneous analysis of selected pharmaceuticals in environmental samples by ultrahigh performance supercritical fluid based chromatography tandem mass spectrometry. Anal. Chim. Acta 2016, 934, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Zhang, T.; Xu, Z.; Fang, H.H.P. Rapid analysis of 21 antibiotics of multiple classes in municipal wastewater using ultra performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2009, 645, 64–72. [Google Scholar] [CrossRef]
- Gros, M.; Rodriguez-Mozaz, S.; Barceló, D. Rapid analysis of multiclass antibiotic residues and some of their metabolites in hospital, urban wastewater and river water by ultra-high-performance liquid chromatography coupled to quadrupole-linear ion trap tandem mass spectrometry. J. Chromatogr. A 2013, 1292, 173–188. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Ryu, H.-D.; Chung, E.G.; Kim, Y. Determination of 18 veterinary antibiotics in environmental water using high-performance liquid chromatography-q-orbitrap combined with on-line solid-phase extraction. J. Chromatogr. B 2018, 1084, 158–165. [Google Scholar] [CrossRef]
- Angeles, L.F.; Islam, S.; Aldstadt, J.; Saqeeb, K.N.; Alam, M.; Khan, A.; Johura, F.-T.; Ahmed, S.I.; Aga, D.S. Retrospective suspect screening reveals previously ignored antibiotics, antifungal compounds, and metabolites in Bangladesh surface waters. Sci. Total Environ. 2019, 712, 136285. [Google Scholar] [CrossRef]
- Kulkarni, P.; Olson, N.D.; Raspanti, G.A.; Goldstein, R.E.R.; Gibbs, S.G.; Sapkota, A.; Sapkota, A.R. Antibiotic Concentrations Decrease during Wastewater Treatment but Persist at Low Levels in Reclaimed Water. Int. J. Environ. Res. Public Healh 2017, 14, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craddock, H.A.; Panthi, S.; Rjoub, Y.; Lipchin, C.; Sapkota, A.; Sapkota, A.R. Antibiotic and herbicide concentrations in household greywater reuse systems and pond water used for food crop irrigation: West Bank, Palestinian Territories. Sci. Total Environ. 2019, 699, 134205. [Google Scholar] [CrossRef]
- Tylová, T.; Flieger, M.; Olšovská, J. Determination of antibiotics in influents and effluents of wastewater-treatment-plants in the Czech Republic—Development and application of the SPE and a UHPLC-ToFMS method. Anal. Methods 2013, 5, 2110–2118. [Google Scholar] [CrossRef]
- Rossmann, J.; Schubert, S.; Gurke, R.; Oertel, R.; Kirch, W. Simultaneous determination of most prescribed antibiotics in multiple urban wastewater by SPE-LC–MS/MS. J. Chromatogr. B 2014, 969, 162–170. [Google Scholar] [CrossRef]
- Panditi, V.R.; Batchu, S.R.; Gardinali, P.R. Online solid-phase extraction–liquid chromatography–electrospray–tandem mass spectrometry determination of multiple classes of antibiotics in environmental and treated waters. Anal. Bioanal. Chem. 2013, 405, 5953–5964. [Google Scholar] [CrossRef]
- Serra-Compte, A.; Pikkemaat, M.G.; Elferink, A.; Almeida, D.; Diogène, J.; Campillo, J.A.; Llorca, M.; Álvarez-Muñoz, D.; Barceló, D.; Rodríguez-Mozaz, S. Combining an effect-based methodology with chemical analysis for antibiotics determination in wastewater and receiving freshwater and marine environment. Environ. Pollut. 2020, 271, 116313. [Google Scholar] [CrossRef] [PubMed]
- Dinh, Q.T.; Alliot, F.; Moreau-Guigon, E.; Eurin, J.; Chevreuil, M.; Labadie, P. Measurement of trace levels of antibiotics in river water using on-line enrichment and triple-quadrupole LC–MS/MS. Talanta 2011, 85, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Dinh, Q.; Moreau-Guigon, E.; Labadie, P.; Alliot, F.; Teil, M.-J.; Blanchard, M.; Eurin, J.; Chevreuil, M. Fate of antibiotics from hospital and domestic sources in a sewage network. Sci. Total Environ. 2017, 575, 758–766. [Google Scholar] [CrossRef]
- Giebułtowicz, J.; Nałęcz-Jawecki, G.; Harnisz, M.; Kucharski, D.; Korzeniewska, E.; Płaza, G. Environmental Risk and Risk of Resistance Selection Due to Antimicrobials’ Occurrence in Two Polish Wastewater Treatment Plants and Receiving Surface Water. Molecules 2020, 25, 1470. [Google Scholar] [CrossRef] [Green Version]
- Giebułtowicz, J.; Tyski, S.; Wolinowska, R.; Grzybowska, W.; Zaręba, T.; Drobniewska, A.; Wroczyński, P.; Nałęcz-Jawecki, G. Occurrence of antimicrobial agents, drug-resistant bacteria, and genes in the sewage-impacted Vistula River (Poland). Environ. Sci. Pollut. Res. 2017, 25, 5788–5807. [Google Scholar] [CrossRef]
- Tran, N.H.; Hoang, L.; Nghiem, L.; Nguyen, N.M.H.; Ngo, H.H.; Guo, W.; Trinh, Q.T.; Mai, N.H.; Chen, H.; Nguyen, D.D.; et al. Occurrence and risk assessment of multiple classes of antibiotics in urban canals and lakes in Hanoi, Vietnam. Sci. Total Environ. 2019, 692, 157–174. [Google Scholar] [CrossRef]
- Omotola, E.O.; Olatunji, O.S. Quantification of selected pharmaceutical compounds in water using liquid chromatography-electrospray ionisation mass spectrometry (LC-ESI-MS). Heliyon 2020, 6, e05787. [Google Scholar] [CrossRef]
- Pascale, R.; Bianco, G.; Coviello, D.; Lafiosca, M.C.; Masi, S.; Mancini, I.M.; Bufo, S.A.; Scrano, L.; Caniani, D. Validation of a liquid chromatography coupled with tandem mass spectrometry method for the determination of drugs in wastewater using a three-phase solvent system. J. Sep. Sci. 2019, 43, 886–895. [Google Scholar] [CrossRef]
- Senta, I.; Krizman-Matasic, I.; Terzic, S.; Ahel, M. Comprehensive determination of macrolide antibiotics, their synthesis intermediates and transformation products in wastewater effluents and ambient waters by liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 2017, 1509, 60–68. [Google Scholar] [CrossRef]
- Araújo, C.; Torres, C.; Silva, N.; Carneiro, C.; Gonçalves, A.; Radhouani, H.; Correia, S.; Da Costa, P.M.; Paccheco, R.; Zarazaga, M.; et al. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 2010, 50, 605–609. [Google Scholar] [CrossRef]
- Goldstein, R.E.R.; Micallef, S.A.; Gibbs, S.G.; George, A.; Claye, E.; Sapkota, A.; Joseph, S.W.; Sapkota, A.R. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014, 466-467, 404–411. [Google Scholar] [CrossRef]
- Iversen, A.; Kühn, I.; Franklin, A.; Möllby, R. High Prevalence of Vancomycin-Resistant Enterococci in Swedish Sewage. Appl. Environ. Microbiol. 2002, 68, 2838–2842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caplin, J.L.; Hanlon, G.W.; Taylor, H.D. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ. Microbiol. 2008, 10, 885–892. [Google Scholar] [CrossRef]
- Sahlström, L.; Rehbinder, V.; Albihn, A.; Aspan, A.; Bengtsson, B. Vancomycin resistant enterococci (VRE) in Swedish sewage sludge. Acta Veter. Scand. 2009, 51, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
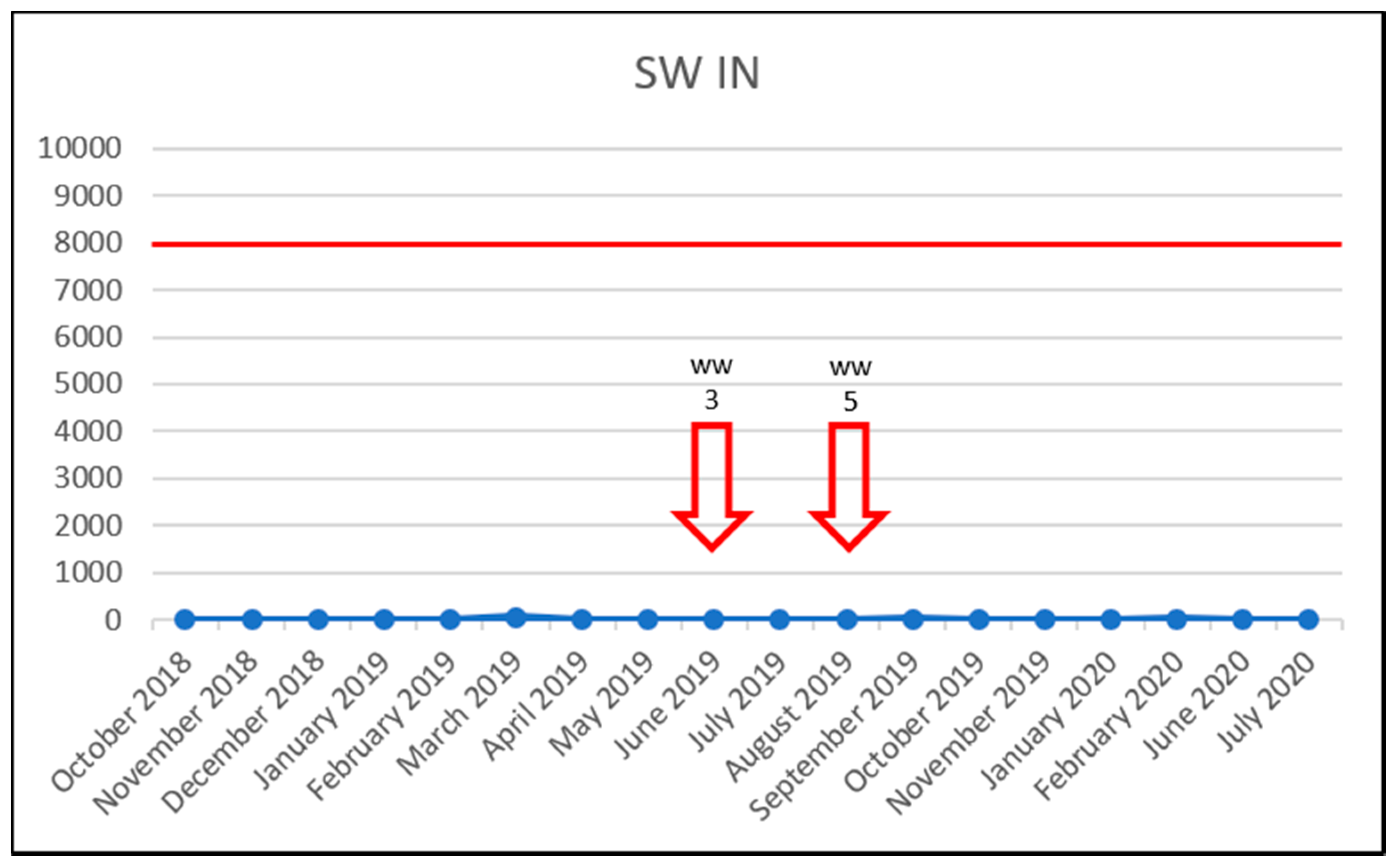
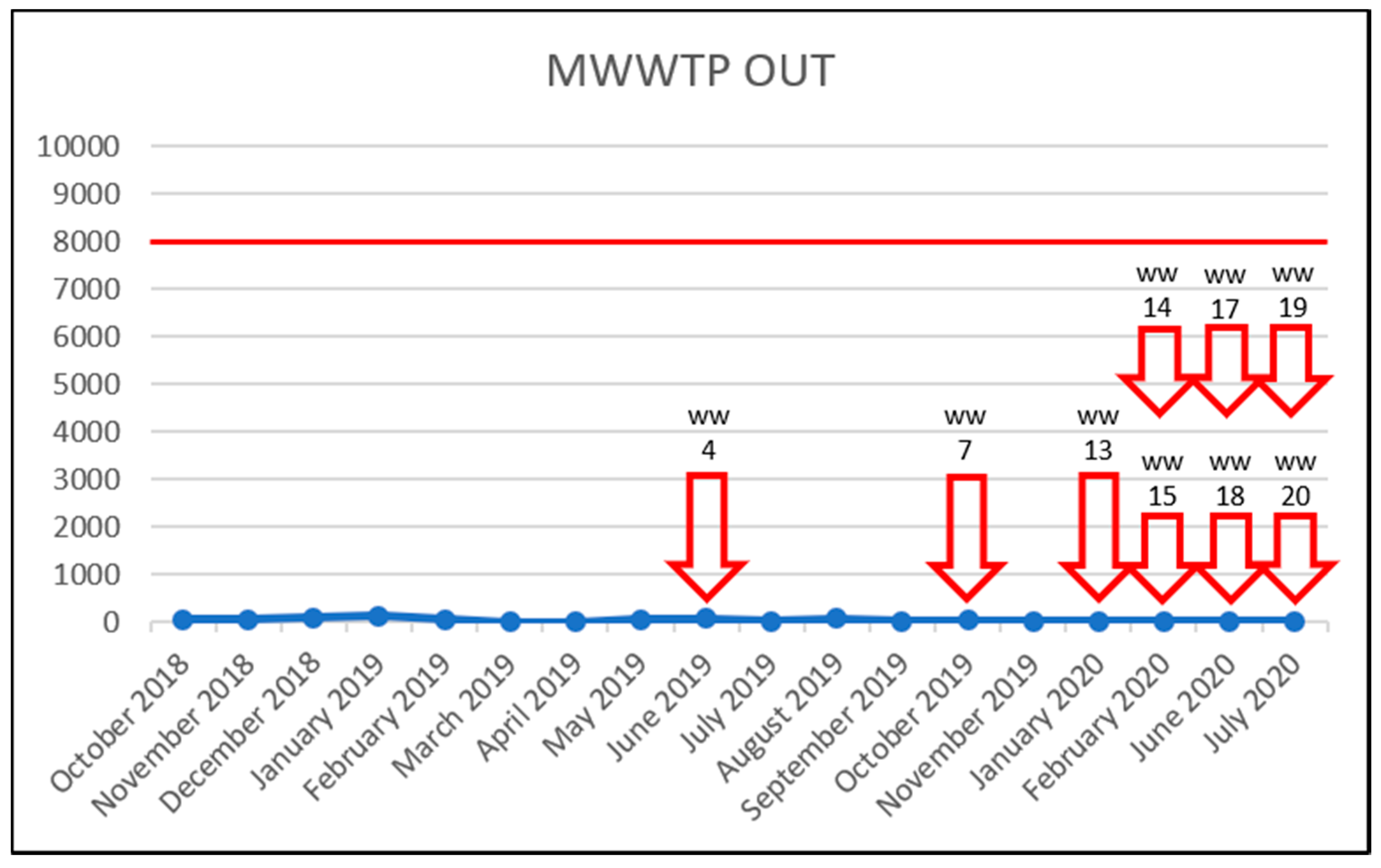
| Sample Identification | Acronym Explanation | Sample Type |
|---|---|---|
| MHO OUT1 | Military Hospital Olomouc—hospital sewer drain 1 | Raw sewage |
| MHO OUT2 | Military Hospital Olomouc—hospital sewer drain 2 | Raw sewage |
| MHO OUT3 | Military Hospital Olomouc—hospital sewer drain 3 | Raw sewage |
| UHO IN | University Hospital Olomouc—hospital wastewater treatment plant influent | Raw sewage |
| UHO OUT | University Hospital Olomouc—hospital wastewater treatment plant effluent | Treated sewage |
| MWWTP IN | Olomouc Municipal Wastewater Treatment Plant influent | Raw sewage |
| MWWTP OUT | Olomouc Municipal Wastewater Treatment Plant effluent | Treated sewage |
| SVI OUT | State Veterinary Institute wastewater treatment plant effluent | Treated sewage |
| SW IN | Surface water (Morava River)—before the Municipal Wastewater Treatment Plant influent | Surface water |
| SW OUT | Surface water (Morava River)—past the Municipal Wastewater Treatment Plant effluent | Surface water |
| Gene | Primer Name | Primer Sequence (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| pbp5 | PBP5 6F | AACCGGTGATCTTCTTGCG | 756 |
| PBP5 6R | TTGATTTCCGCTGTACCAGT | ||
| PBP5 11F | GGGCTTAAATGGCAAAAAGA | 694 | |
| PBP5 11R | ATTGATAATTTTGGTTGAGGTAT |
| Antibiotic | PNEC ng/L | Mean Concentration a,b (ng/L) | No. of Samples Exceeding PNEC c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw Waste-Water (Hospital) d | Raw Waste-Water (Municipal) | Treated Waste-Water (Hospital) | Treated Wastewater (Municipal) | Surface Water | Raw Waste-Water (Hospital) | Raw Waste-Water (Municipal) | Treated Waste-Water (Hospital) | Treated Waste-Water (Municipal) | Surface Water | Total | ||
| nitrofurantoin | 64,000 | 9300 (2) | - | - | - | - | - | - | - | - | - | - |
| chloramphenicol | 8000 | 890 (69) | 160 (18) | 240 (8) | 10 (4) | 16 (4) | - | - | - | - | - | - |
| linezolid | 8000 | 1200 (22) | 48 (9) | 980 (16) | 10 (10) | 3 (5) | - | - | - | - | - | - |
| ampicillin | 250 | 48,000 (5) | 92 (1) | 38 (1) | - | 9 (1) | 4 | - | - | - | - | 4 |
| clindamycin | 1000 | 1500 (66) | 81 (16) | 1200 (17) | 38 (18) | 2 (9) | 16 | - | 6 | - | - | 22 |
| tetracycline | 1000 | 2700 (5) | 37 (2) | 120 (4) | 8 (1) | 7 (4) | 1 | - | - | - | - | 1 |
| tigecycline | 1000 | 800 (6) | 280 (1) | - | 950 (1) | - | 2 | - | - | - | - | 2 |
| vancomycin | 8000 | 4700 (58) | 140 (17) | 13,000 (18) | 45 (16) | 25 (6) | 16 | - | 13 | - | - | 29 |
| erythromycin | 1000 | 67 (35) | 36 (18) | 52 (10) | 12 (13) | - | - | - | - | - | - | - |
| Laboratory Sample Identification | Sampling Site | Sample Type | Collection Date |
|---|---|---|---|
| WW1 | UHO IN | raw sewage | 10/2018 |
| WW2 | UHO IN | raw sewage | 11/2018 |
| WW3 | SW IN | surface water | 06/2019 |
| WW4 | UWWTP OUT | treated sewage | 06/2019 |
| WW5 | SW IN | surface water | 08/2019 |
| WW6 | MHO OUT3 | raw sewage | 08/2019 |
| WW7 | UWWTP OUT | treated sewage | 10/2019 |
| WW9 | UHO IN | raw sewage | 11/2019 |
| WW10 | UHO OUT | treated sewage | 11/2019 |
| WW11 | UHO OUT | treated sewage | 11/2019 |
| WW13 | UWWTP OUT | treated sewage | 01/2020 |
| WW14 | UWWTP OUT | treated sewage | 02/2020 |
| WW15 | UWWTP OUT | treated sewage | 02/2020 |
| WW16 | UHO OUT | treated sewage | 06/2020 |
| WW17 | UWWTP OUT | treated sewage | 06/2020 |
| WW18 | UWWTP OUT | treated sewage | 06/2020 |
| WW19 | UWWTP OUT | treated sewage | 07/2020 |
| WW20 | UWWTP OUT | treated sewage | 07/2020 |
| Laboratory Sample Identification | PEN | LNZ | AMP | TIG | CMP | TET | ERY | CLI | VAN | TEI | FUR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WW1 | 4 | 1.5 | 16 | 0.03 | 2 | 0.25 | 16 | 8 | 8 | 2 | 64 |
| WW2 | 4 | 2 | 16 | 0.03 | 2 | 16 | 16 | 8 | 32 | 32 | 512 |
| WW3 | 0.5 | 0.25 | 16 | 0.03 | 2 | 16 | 8 | 4 | 64 | 64 | 32 |
| WW4 | 2 | 1.5 | 16 | 0.03 | 1 | 16 | 8 | 4 | 64 | 32 | 32 |
| WW5 | 4 | 0.5 | 16 | 0.03 | 2 | 0.25 | 16 | 8 | 64 | 8 | 64 |
| WW6 | 4 | 0.25 | 16 | 0.03 | 2 | 16 | 16 | 8 | 64 | 8 | 64 |
| WW7 | 8 | 1 | 16 | 4 | 1 | 16 | 8 | 4 | 8 | 4 | 32 |
| WW9 | 2 | 1.5 | 16 | 0.5 | 2 | 0.5 | 4 | 4 | 32 | 2 | 64 |
| WW10 | 2 | 1.5 | 8 | 0.03 | 2 | 16 | 4 | 4 | 32 | 8 | 32 |
| WW11 | 2 | 1.5 | 16 | 0.03 | 2 | 0.5 | 4 | 4 | 8 | 0.5 | 32 |
| WW13 | 2 | 1 | 16 | 0.03 | 2 | 16 | 8 | 16 | 16 | 16 | 8 |
| WW14 | 2 | 1.5 | 16 | 0.03 | 2 | 16 | 8 | 2 | 16 | 16 | 16 |
| WW15 | 2 | 1.5 | 16 | 0.03 | 4 | 16 | 8 | 16 | 16 | 16 | 16 |
| WW16 | 2 | 2 | 16 | 0.03 | 4 | 16 | 8 | 4 | 16 | 32 | 256 |
| WW17 | 2 | 1 | 16 | 0.06 | 1 | 16 | 4 | 0.06 | 32 | 16 | 32 |
| WW18 | 2 | 0.03 | 16 | 0.03 | 4 | 0.5 | 8 | 4 | 16 | 8 | 256 |
| WW19 | 2 | 0.03 | 16 | 0.03 | 2 | 8 | 4 | 8 | 16 | 8 | 32 |
| WW20 | 2 | 0.03 | 16 | 0.03 | 4 | 0.25 | 4 | 8 | 64 | 8 | 64 |
| No. of Isolates | Presence of Gene(s) | MIC Range (µg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tet (K) | tet (L) | tet (M) | tet (O) | tet (S) | tet (X) | erm (A) | erm (B) | mef (A/E) | ERY | CLI | TET | |
| 3 | - | + | + | - | - | - | - | + | - | 4–8 | 4–16 | 16 |
| 4 | - | + | + | - | - | - | - | - | - | 4–16 | 0.06–8 | 0.25–16 |
| 5 | - | - | + | - | - | - | - | + | - | 4–16 | 4–16 | 0.25–16 |
| 1 | - | - | + | - | - | - | - | - | - | 8 | 4 | 16 |
| 5 | - | - | - | - | - | - | - | + | - | 4–16 | 4–8 | 0.25–16 |
| pbp5 Allele | Amino Acid Changes at Position | MIC Range of Ampicillin (µg/mL) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Isolates | 461 | 466 | 470 | 471 | 485 | 487 | 496 | 497 | 499 | 525 | 581 | 586 | 595 | 622 | 629 | 631 | 634 | ||
| X84860 | Q | - | H | V | M | Q | N | F | A | E | I | V | E | E | E | G | N | - | |
| A | 11 | K | S | Q | V | A | Q | K | F | T | D | I | V | E | E | V | G | N | 16 |
| B | 6 | Q | S | Q | V | A | Q | K | F | T | D | I | V | E | E | V | G | N | 16 |
| C | 1 | Q | D | Q | V | A | Q | K | F | I | D | I | V | E | E | V | G | N | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hricová, K.; Röderová, M.; Fryčák, P.; Pauk, V.; Kurka, O.; Mezerová, K.; Štosová, T.; Bardoň, J.; Milde, D.; Kučová, P.; et al. Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life 2021, 11, 1403. https://doi.org/10.3390/life11121403
Hricová K, Röderová M, Fryčák P, Pauk V, Kurka O, Mezerová K, Štosová T, Bardoň J, Milde D, Kučová P, et al. Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life. 2021; 11(12):1403. https://doi.org/10.3390/life11121403
Chicago/Turabian StyleHricová, Kristýna, Magdaléna Röderová, Petr Fryčák, Volodymyr Pauk, Ondřej Kurka, Kristýna Mezerová, Taťána Štosová, Jan Bardoň, David Milde, Pavla Kučová, and et al. 2021. "Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water" Life 11, no. 12: 1403. https://doi.org/10.3390/life11121403
APA StyleHricová, K., Röderová, M., Fryčák, P., Pauk, V., Kurka, O., Mezerová, K., Štosová, T., Bardoň, J., Milde, D., Kučová, P., & Kolář, M. (2021). Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life, 11(12), 1403. https://doi.org/10.3390/life11121403








