On the Role and Production of Polyhydroxybutyrate (PHB) in the Cyanobacterium Synechocystis sp. PCC 6803
Abstract
1. Introduction
2. Materials and Methods
2.1. Cyanobacterial Cultivation Conditions
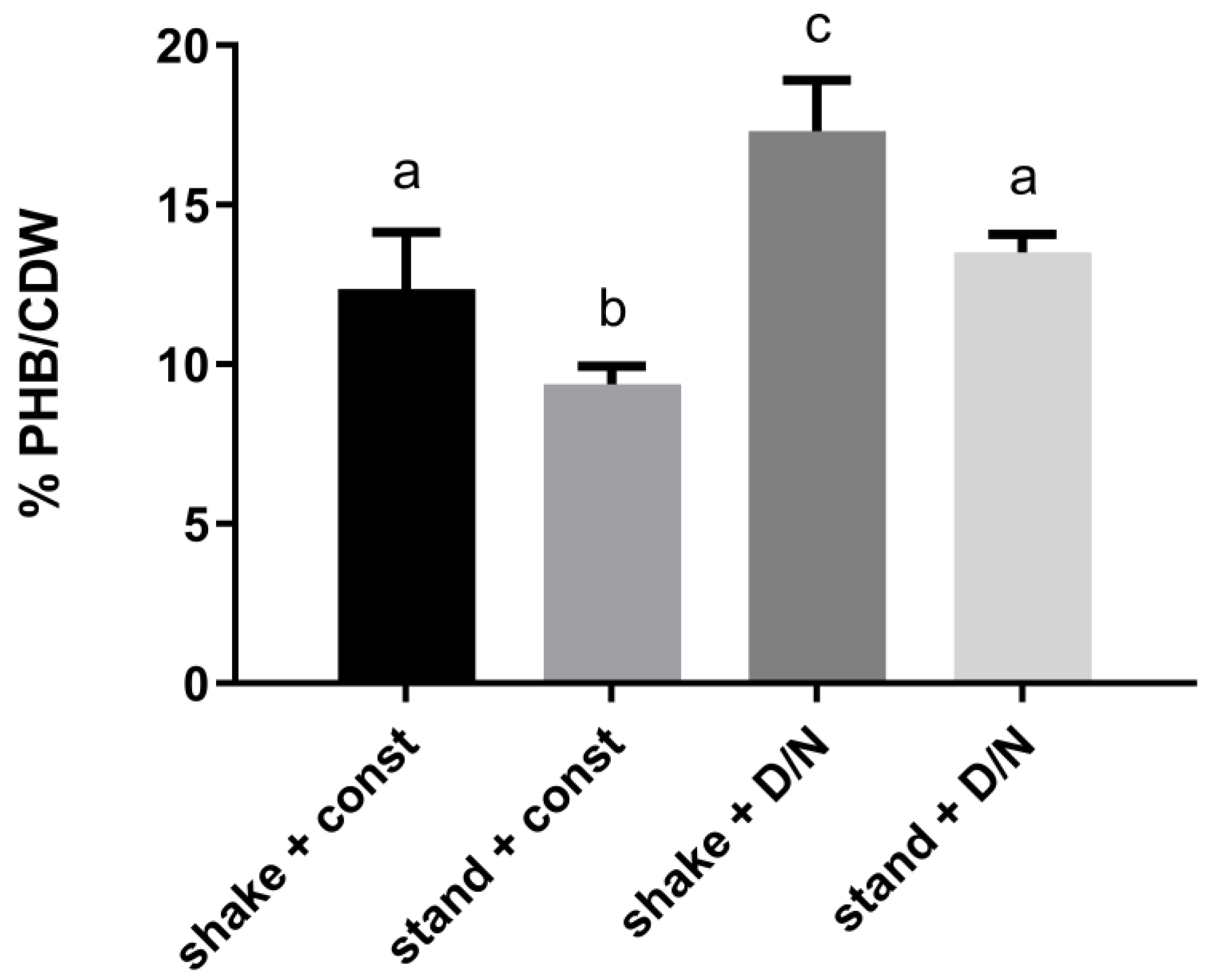
2.2. Physical Stress Conditions
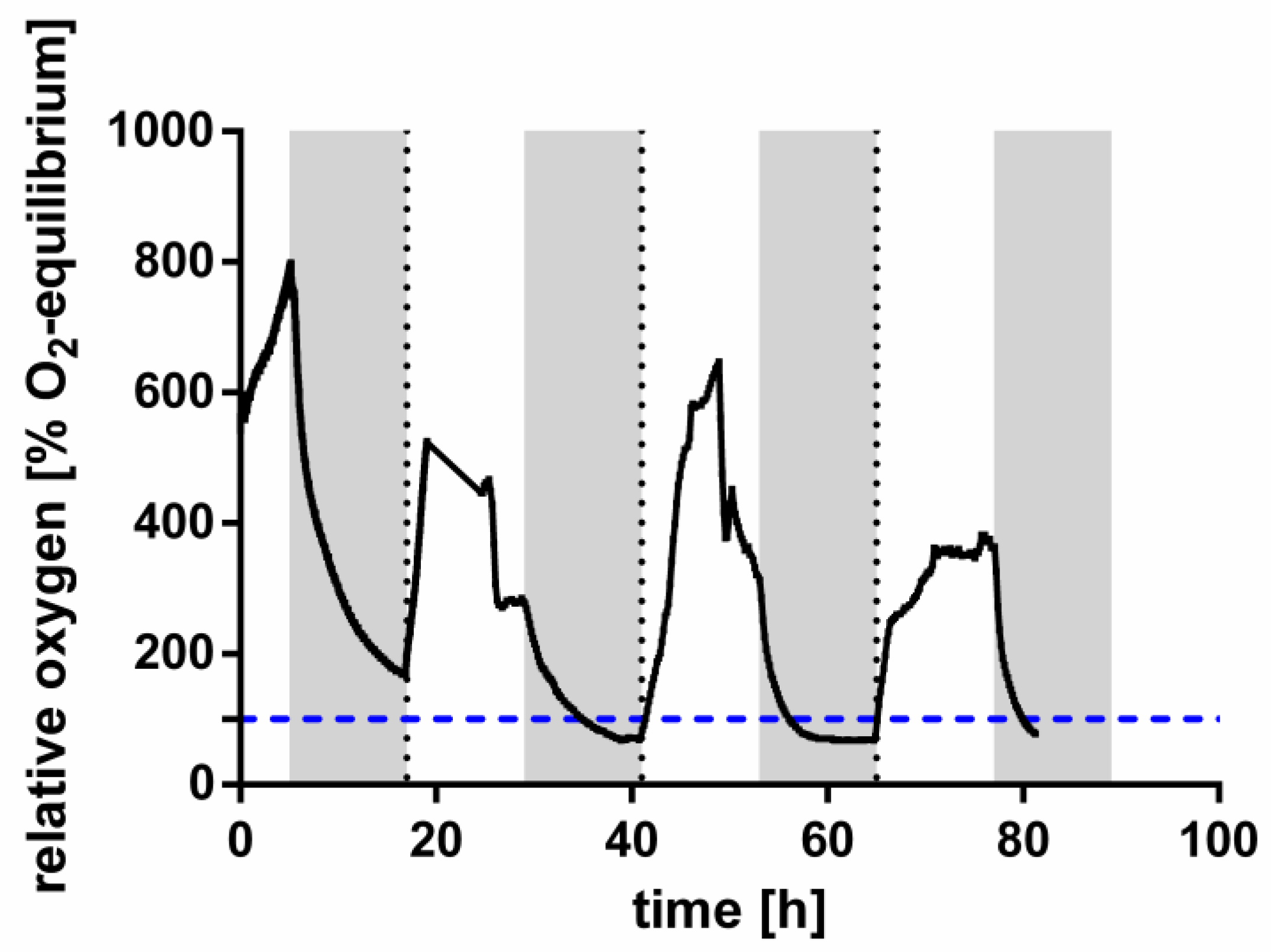
2.3. Oxygen Measurements
2.4. Microscopy and Staining Procedures
2.5. Electron Microscopy
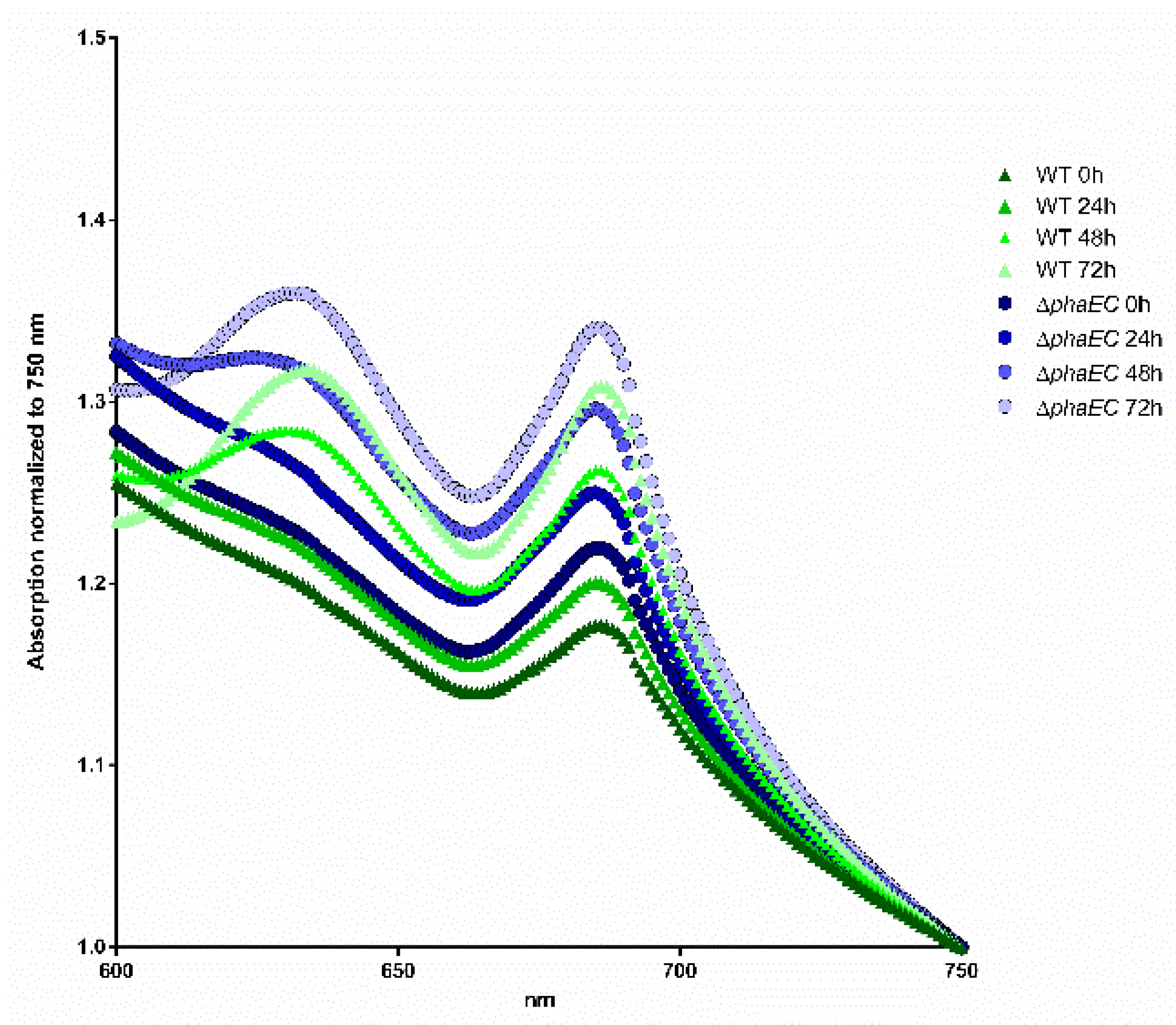
2.6. Spectral Analysis
2.7. PAM
2.8. PHB Quantification
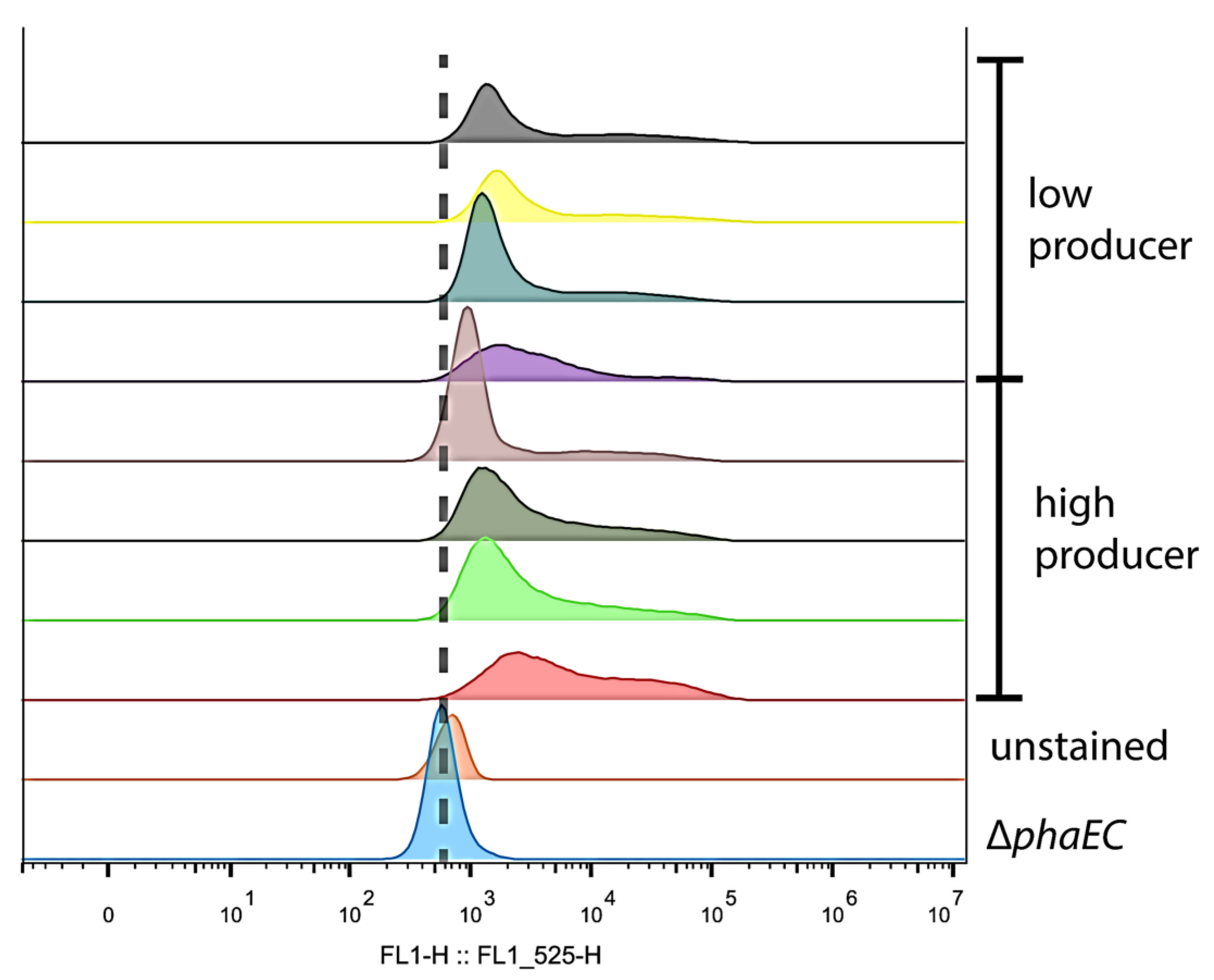
2.9. FACS and Flow Cytometry
3. Results
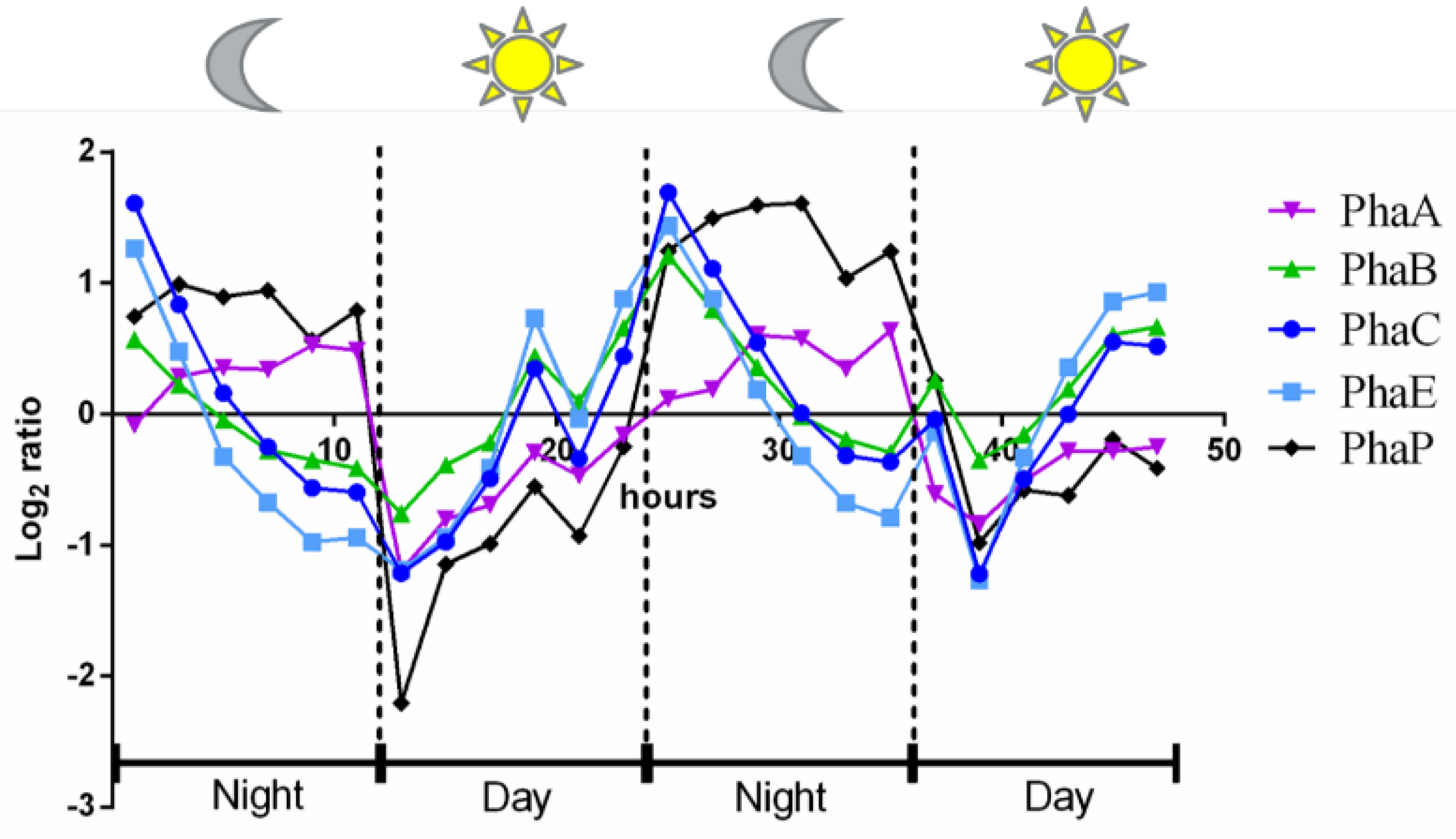
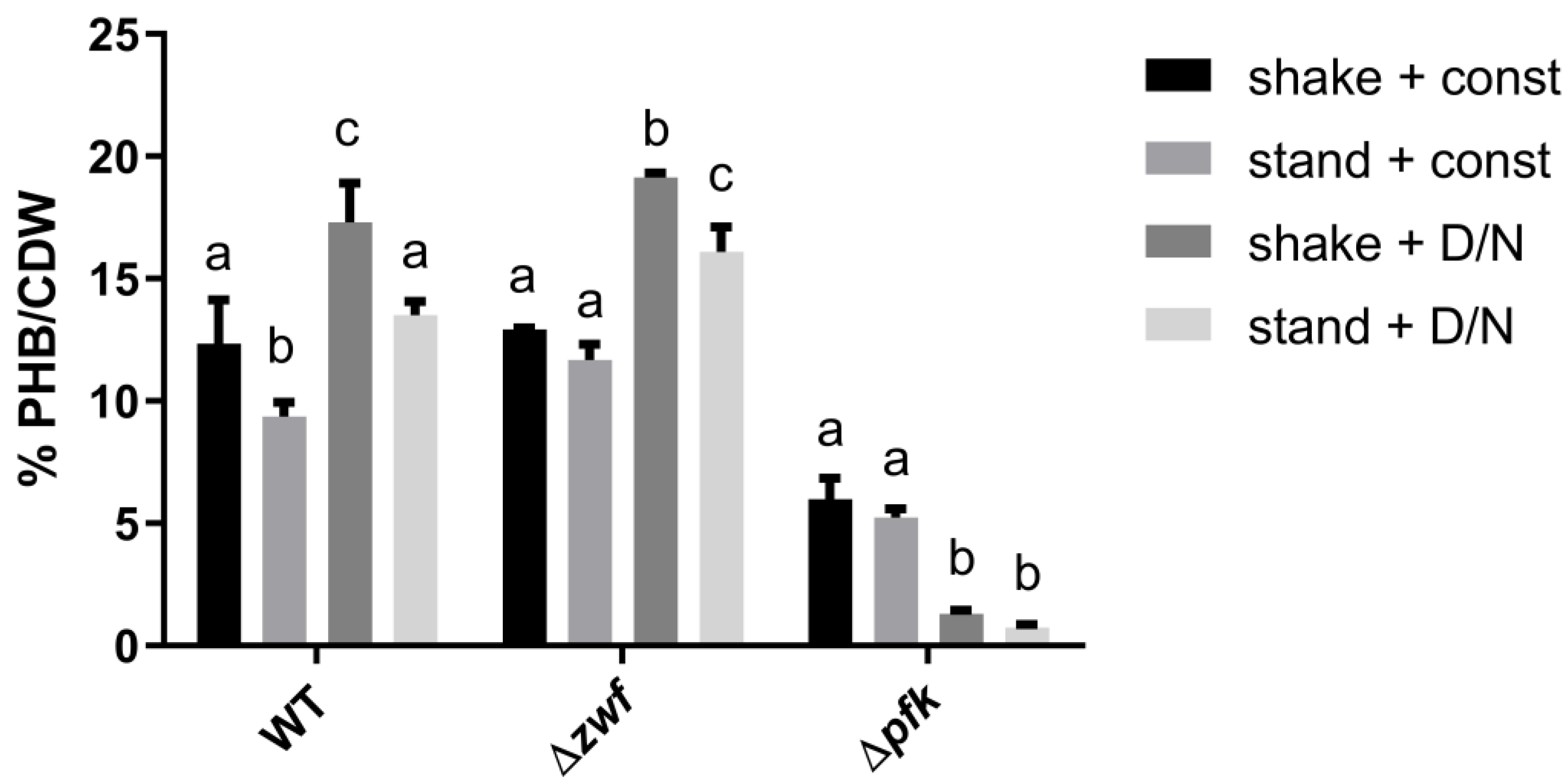
3.1. The Influence of Environmental Conditions and Central Pathways on PHB Production
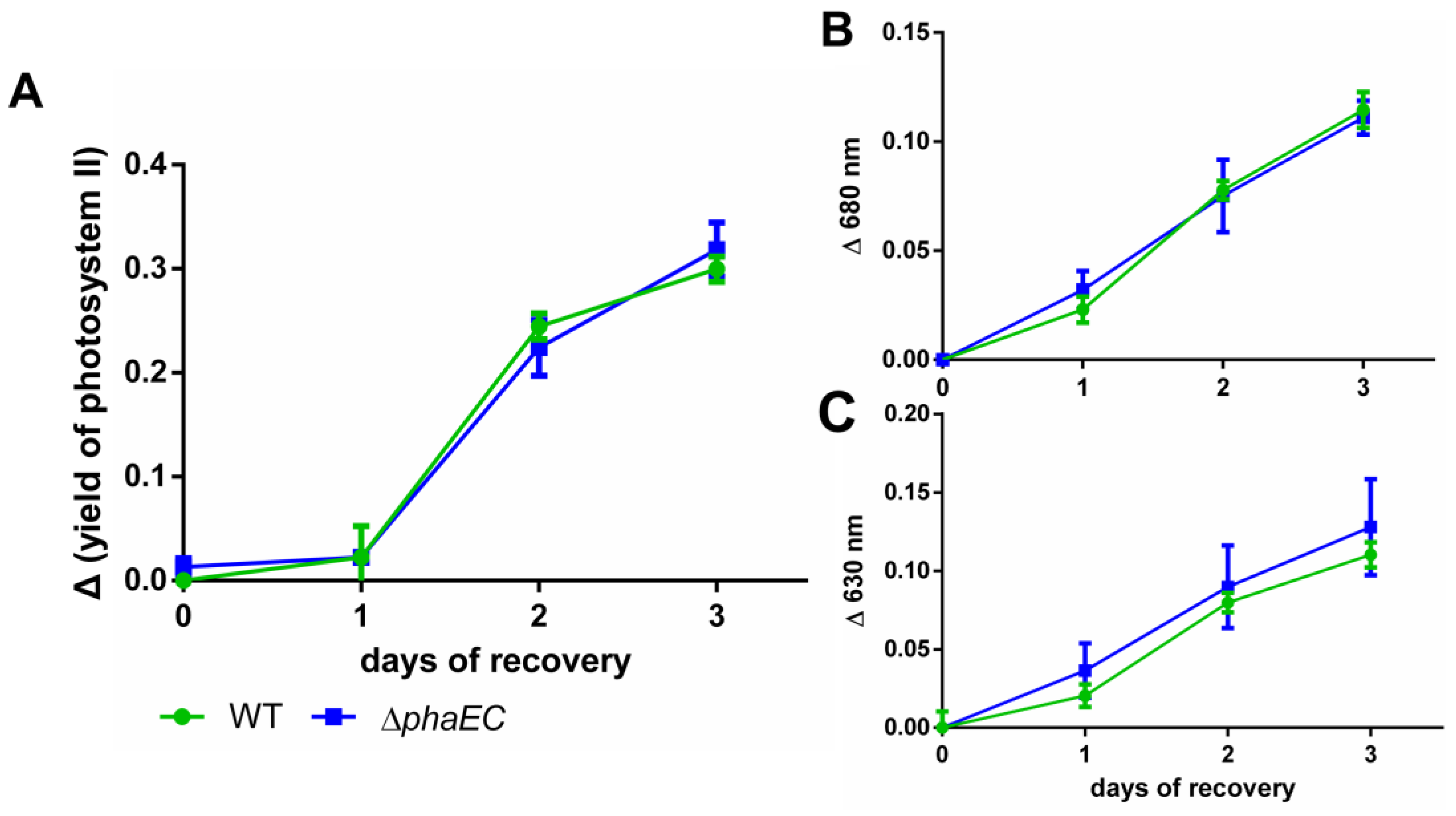
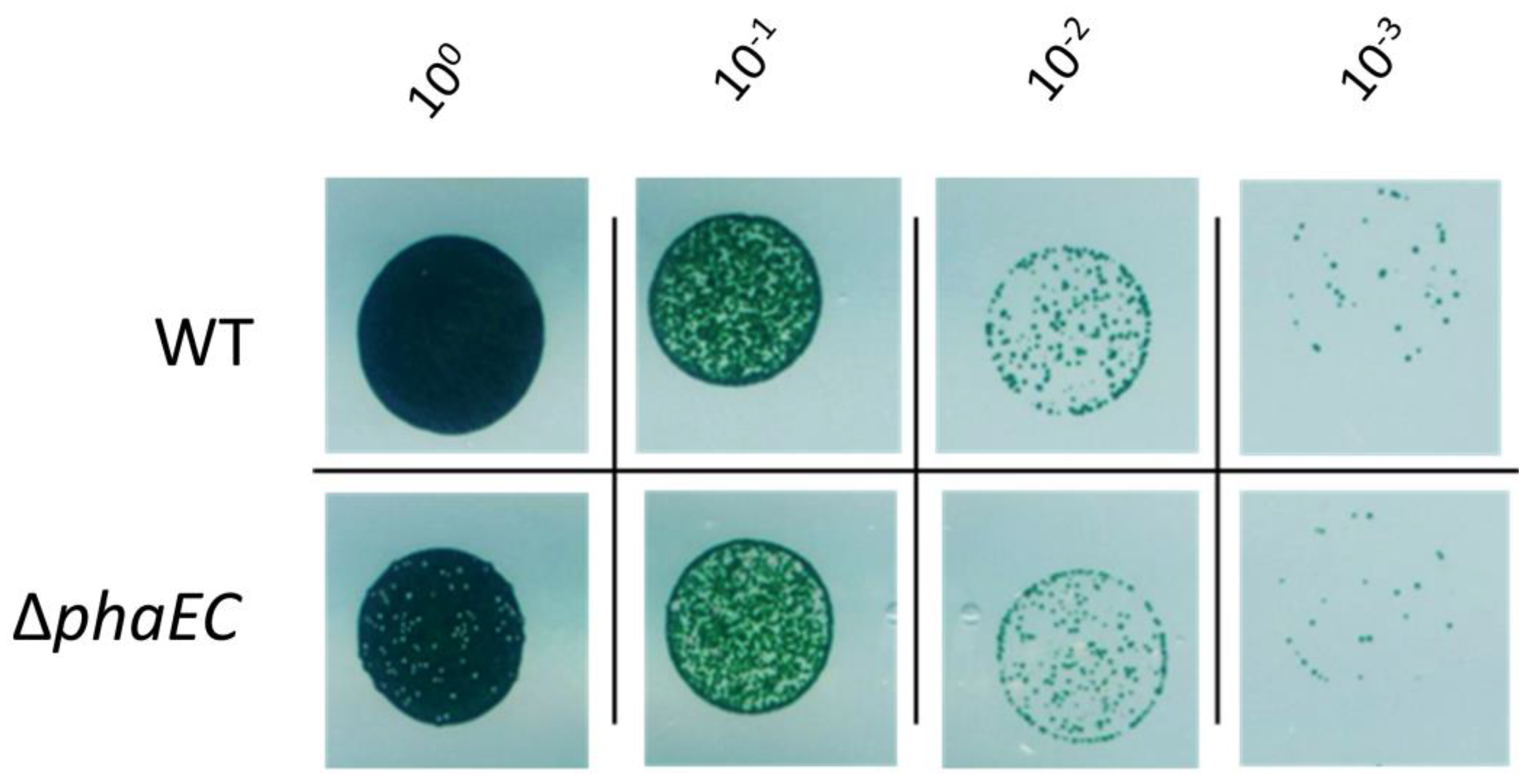
3.2. Recovery from Nitrogen Starvation
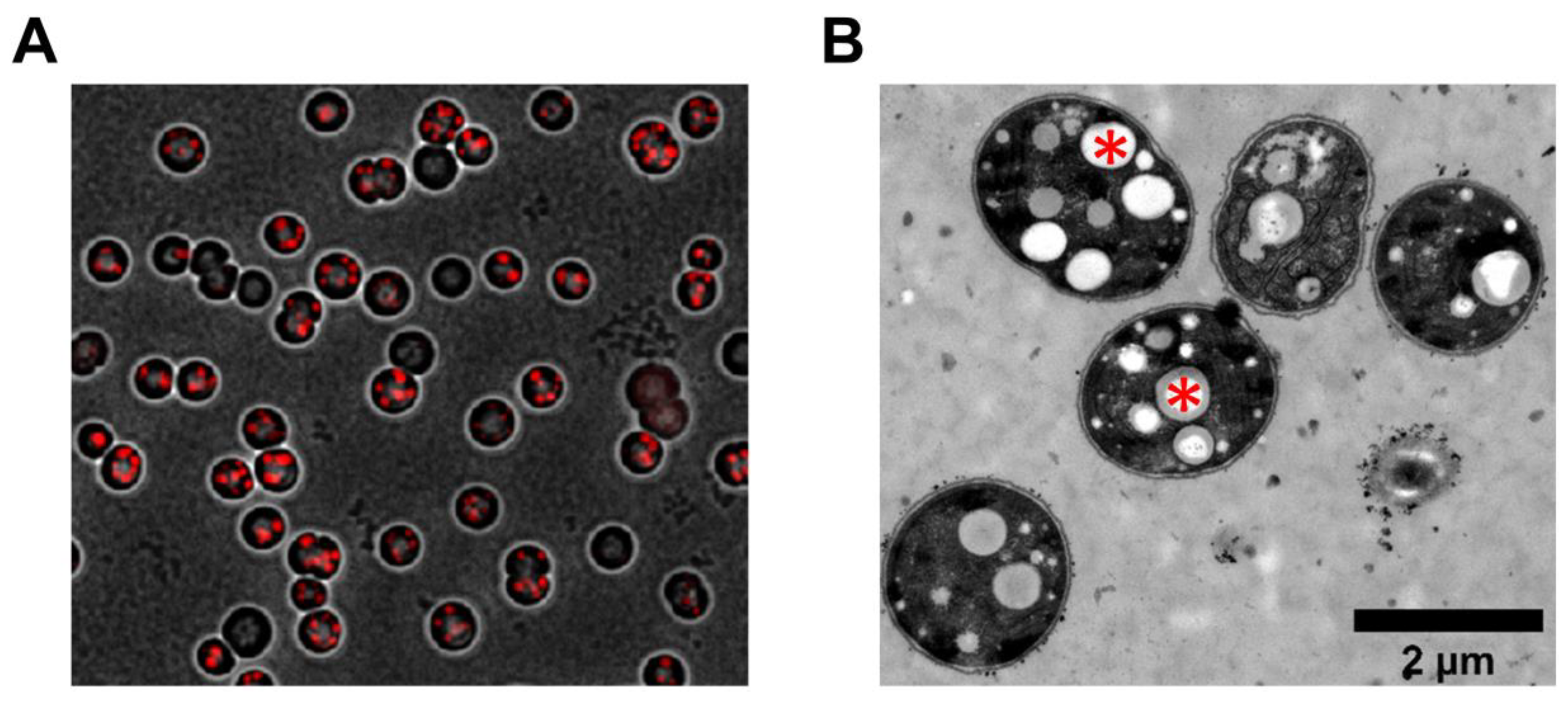
3.3. Heterogeneity of PHB Production
4. Discussion
4.1. Heterogeneity of PHB Content
4.2. Physiological Function of PHB
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Strain | Relevant Marker of Genotype | Reference |
|---|---|---|
| Synechocystis sp. PCC 6803 | - | Pasteur culture collection |
| Δzwf | Slr1843::cmR | Chen et al. 2016 |
| ΔpfkB1/2 | sll1196::kmR, sll0745::spR | Chen et al. 2016 |
| ΔphaEC | slr1829,slr1830::kmR | Klotz et al. 2016 |
References
- Herrero, A.; Flores, E. The Cyanobacteria: Molecular Biology, Genomics and Evolution; Caister Academic Press: Norfolk, UK, 2008. [Google Scholar]
- Allen, M.M. Cyanobacterial cell inclusions. Annu. Rev. Microbiol. 1984, 38. [Google Scholar] [CrossRef] [PubMed]
- Carr, N.G. The occurrence of poly-beta-hydroxybutyrate in the blue-green alga, Chlorogloea fritschii. Biochim. Biophys. Acta 1966, 120, 308–310. [Google Scholar] [CrossRef]
- Damrow, R.; Maldener, I.; Zilliges, Y. The Multiple Functions of Common Microbial Carbon Polymers, Glycogen and PHB, during Stress Responses in the Non-Diazotrophic Cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2016, 7, 966. [Google Scholar] [CrossRef] [PubMed]
- Klotz, A.; Georg, J.; Bučinská, L.; Watanabe, S.; Reimann, V.; Januszewski, W.; Sobotka, R.; Jendrossek, D.; Hess, W.R.; Forchhammer, K. Awakening of a Dormant Cyanobacterium from Nitrogen Chlorosis Reveals a Genetically Determined Program. Curr. Biol. 2016, 26, 2862–2872. [Google Scholar] [CrossRef]
- Hein, S.; Tran, H.; Steinbuchel, A. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch. Microbiol. 1998, 170, 162–170. [Google Scholar] [CrossRef]
- Wu, G.F.; Wu, Q.Y.; Shen, Z.Y. Accumulation of poly-beta-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresour. Technol. 2001, 76, 85–90. [Google Scholar] [CrossRef]
- Anderson, A.J.; Dawes, E.A. Occurrence, Metabolism, Metabolic Role, and Industrial Uses of Bacterial Polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar] [CrossRef]
- Jendrossek, D.; Pfeiffer, D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ. Microbiol. 2014, 16, 2357–2373. [Google Scholar] [CrossRef]
- Kadouri, D.; Jurkevitch, E.; Okon, Y. Involvement of the reserve material poly-beta-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl. Environ. Microbiol. 2003, 69, 3244–3250. [Google Scholar] [CrossRef]
- Batista, M.B.; Teixeira, C.S.; Sfeir, M.Z.T.; Alves, L.P.S.; Valdameri, G.; de Oliveira Pedrosa, F.; Sassaki, G.L.; Steffens, M.B.R.; de Souza, E.M.; Dixon, R.; et al. PHB Biosynthesis Counteracts Redox Stress in Herbaspirillum seropedicae. Front. Microbiol. 2018, 9, 472. [Google Scholar] [CrossRef]
- Van Gemerden, H. On the ATP generation by Chromatium in darkness. Arch. Mikrobiol. 1968, 64, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Hauf, W.; Schlebusch, M.; Hüge, J.; Kopka, J.; Hagemann, M.; Forchhammer, K. Metabolic Changes in Synechocystis PCC6803 upon Nitrogen-Starvation: Excess NADPH Sustains Polyhydroxybutyrate Accumulation. Metabolites 2013, 3, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Balaji, S.; Gopi, K.; Muthuvelan, B. A review on production of poly β hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Res. 2013, 2, 278–285. [Google Scholar] [CrossRef]
- Knöttner, S.; Drosg, B.; Ellersdorfer, M.; Meixner, K.; Fritz, I. Photoautotrophic production of poly-hydroxybutyrate—First detailed cost estimations. Algal Res. 2019, 41, 101558. [Google Scholar]
- Dutt, V.; Srivastava, S. Novel quantitative insights into carbon sources for synthesis of poly hydroxybutyrate in Synechocystis PCC 6803. Photosynth. Res. 2018, 136, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Doello, S.; Gutekunst, K.; Forchhammer, K. PHB is Produced from Glycogen Turn-over during Nitrogen Starvation in Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 2019, 20, 1942. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.; Lukas, M. Cyanobacterial Polyhydroxyalkanoate Production: Status Quo and Quo Vadis? Curr. Biotechnol. 2015, 4, 464–480. [Google Scholar]
- Rippka, R.; Deruelles, J.; aterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiol. Soc. 1979, 111. [Google Scholar] [CrossRef]
- Schlebusch, M.; Forchhammer, K. Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2010, 76, 6101–6107. [Google Scholar] [CrossRef]
- Fiedler, G.; Arnold, M.; Hannus, S.; Maldener, I. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 1998, 27, 1193–1202. [Google Scholar] [CrossRef]
- Schreiber, U.; Endo, T.; Mi, H.; Asada, K. Quenching Analysis of Chlorophyll Fluorescence by the Saturation Pulse Method: Particular Aspects Relating to the Study of Eukaryotic Algae and Cyanobacteria. Plant Cell Physiol. 1995, 36, 873–882. [Google Scholar] [CrossRef]
- Panda, B.; Mallick, N. Enhanced poly-beta-hydroxybutyrate accumulation in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. Lett. Appl. Microbiol. 2007, 44, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Veening, J.W.; Smits, W.K.; Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.J.E.; Halliday, J.A.; Blankschien, M.D.; Burns, P.A.; Yatagai, F.; Herman, C. Transcriptional Infidelity Promotes Heritable Phenotypic Change in a Bistable Gene Network. PLoS Biol. 2009, 7, e1000044. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F. Individual-level bet hedging in the bacterium Sinorhizobium meliloti. Curr. Biol. 2010, 20, 1740–1744. [Google Scholar] [CrossRef]
- Saha, R.; Liu, D.; Hoynes-O’Connor, A.; Liberton, M.; Yu, J.; Bhattacharyya-Pakrasi, M.; Balassy, A.; Zhang, F.; Moon, T.S.; Maranas, C.D.; et al. Diurnal Regulation of Cellular Processes in the Cyanobacterium Synechocystis sp. Strain PCC 6803: Insights from Transcriptomic, Fluxomic, and Physiological Analyses. mBio 2016, 7, e00464-16. [Google Scholar] [CrossRef]
- Ueda, S.; Kawamura, Y.; Iijima, H.; Nakajima, M.; Shirai, T.; Okamoto, M.; Kondo, A.; Hirai, M.Y.; Osanai, T. Anionic metabolite biosynthesis enhanced by potassium under dark, anaerobic conditions in cyanobacteria. Sci. Rep. 2016, 6, 32354. [Google Scholar] [CrossRef]
- Smith, A.J. Modes of cyanobacterial carbon metabolism. Ann. Inst. Pasteur/Microbiol. 1983, 134B, 93–113. [Google Scholar] [CrossRef]
- Stal, L.J.; Moezelaar, R. Fermentation in cyanobacteria1. FEMS Microbiol. Rev. 1997, 21, 179–211. [Google Scholar] [CrossRef]
- Makowka, A.; Nichelmann, L.; Schulze, D.; Spengler, K.; Wittmann, C.; Forchhammer, K.; Gutekunst, K. Glycolytic Shunts Replenish the Calvin-Benson-Bassham Cycle as Anaplerotic Reactions in Cyanobacteria. Mol. Plant 2020, 13, 471–482. [Google Scholar] [CrossRef]
- Heyer, H.; Krumbein, W.E. Excretion of Fermentation Products in Dark and Anaerobically Incubated Cyanobacteria. Arch. Microbiol. 1991, 155, 284–287. [Google Scholar] [CrossRef]
- Thiel, K.; Patrikainen, P.; Nagy, C.; Fitzpatrick, D.; Pope, N.; Aro, E.-M.; Kallio, P. Redirecting photosynthetic electron flux in the cyanobacterium Synechocystis sp. PCC 6803 by the deletion of flavodiiron protein Flv3. Microb. Cell Fact. 2019, 18, 189. [Google Scholar] [CrossRef] [PubMed]

| Abiotic Stress | Treatment | Effect |
|---|---|---|
| (1) | ||
| Cold | 4 °C over night 1 h at −20°C; 14 h at −20 °C | No difference |
| Heat | 45 min at 40 °C; 45 min at 40°C and 20 min at 50°C | No difference All cells were dead |
| Physical Force | Centrifugation for 30 min at 20,000 g; 1 h at 25,000 g 3 × 5 min at 4 m/s glass-bead milling in Ribolyser; 3 × 5 min at 7 m/s Ribolyser | No difference No difference All cells were dead |
| Darkness | 1 day; 2 days, 5 days; 8 days; 10 days; 15 days | No difference |
| Low Light | 14 days at 5 µE | No difference |
| Alternating Light | 12/12 light/dark | No difference |
| Drought | 30 min at 30 °C SpeedVac | All cells were dead |
| Nitrogen Starvation | 3 weeks 10 weeks | No difference ∆phaEC showed weak growth advantage |
| High Light | 1 day incubation at 500 µE | No difference |
| (2) | ||
| Buffered Medium | BG11 agar plates containing 300 µL, 1 mL, 3 mL TES buffer | No difference |
| Salt | Recovery at BG11 agar plates with: 100, 150, 300 mM NaCl 2 × BG11 salts | No difference No difference |
| Carbon Availability | 0, 10, 50, 150 mM bicarbonate (added to BG11 agar plates) | No difference |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koch, M.; Berendzen, K.W.; Forchhammer, K. On the Role and Production of Polyhydroxybutyrate (PHB) in the Cyanobacterium Synechocystis sp. PCC 6803. Life 2020, 10, 47. https://doi.org/10.3390/life10040047
Koch M, Berendzen KW, Forchhammer K. On the Role and Production of Polyhydroxybutyrate (PHB) in the Cyanobacterium Synechocystis sp. PCC 6803. Life. 2020; 10(4):47. https://doi.org/10.3390/life10040047
Chicago/Turabian StyleKoch, Moritz, Kenneth W. Berendzen, and Karl Forchhammer. 2020. "On the Role and Production of Polyhydroxybutyrate (PHB) in the Cyanobacterium Synechocystis sp. PCC 6803" Life 10, no. 4: 47. https://doi.org/10.3390/life10040047
APA StyleKoch, M., Berendzen, K. W., & Forchhammer, K. (2020). On the Role and Production of Polyhydroxybutyrate (PHB) in the Cyanobacterium Synechocystis sp. PCC 6803. Life, 10(4), 47. https://doi.org/10.3390/life10040047






