The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Reporter Assays for In Vivo Verification of Targets
2.3. RNA Isolation, Northern Blot and Primer Extension Analysis
2.4. In Vitro Synthesis and Labelling of RNA
2.5. In Vitro Structure Probing and Footprinting
2.6. Expression and Purification of Protein All1871 and Western Blot
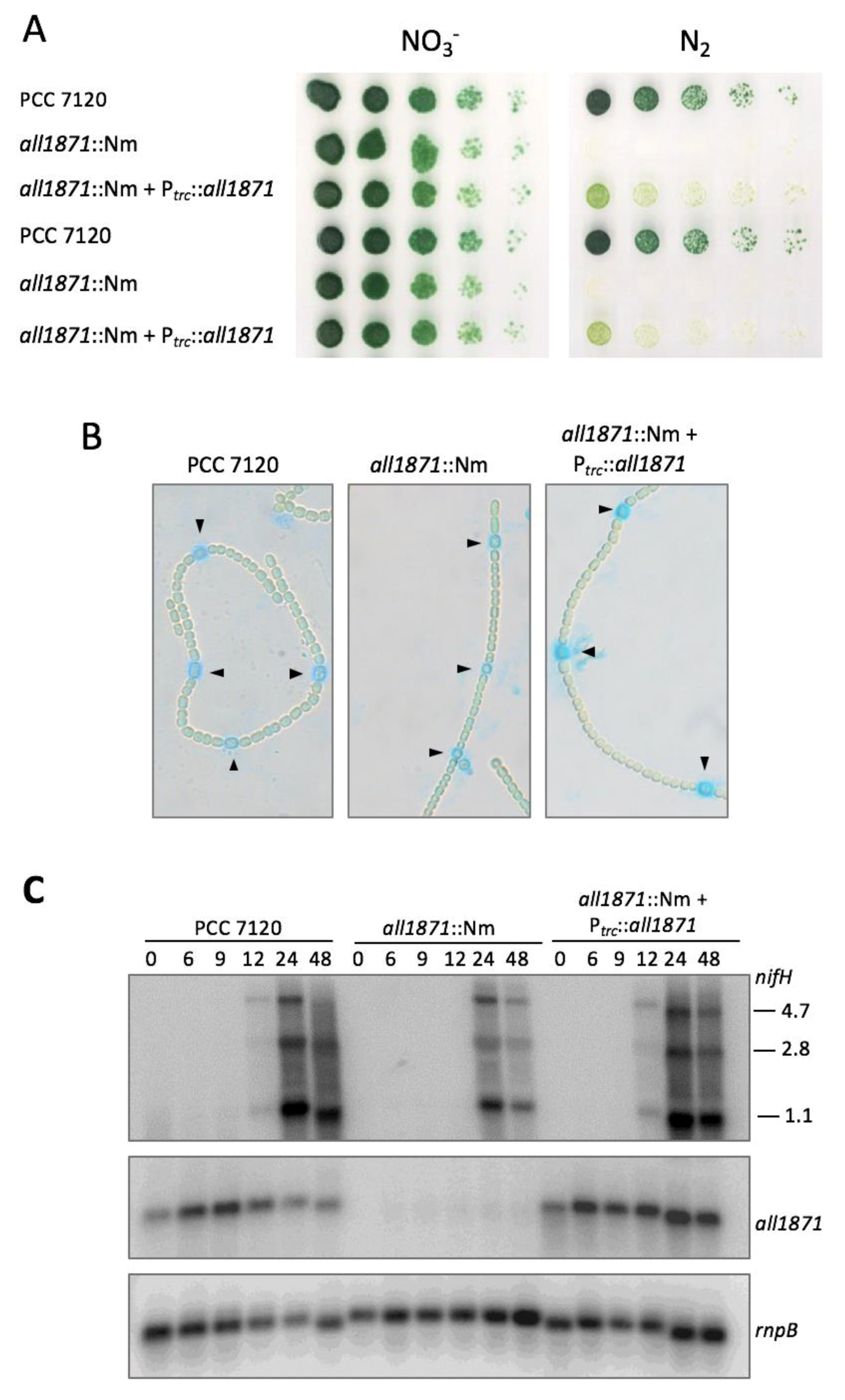
2.7. Generation and Complementation of all1871 Mutant Strain
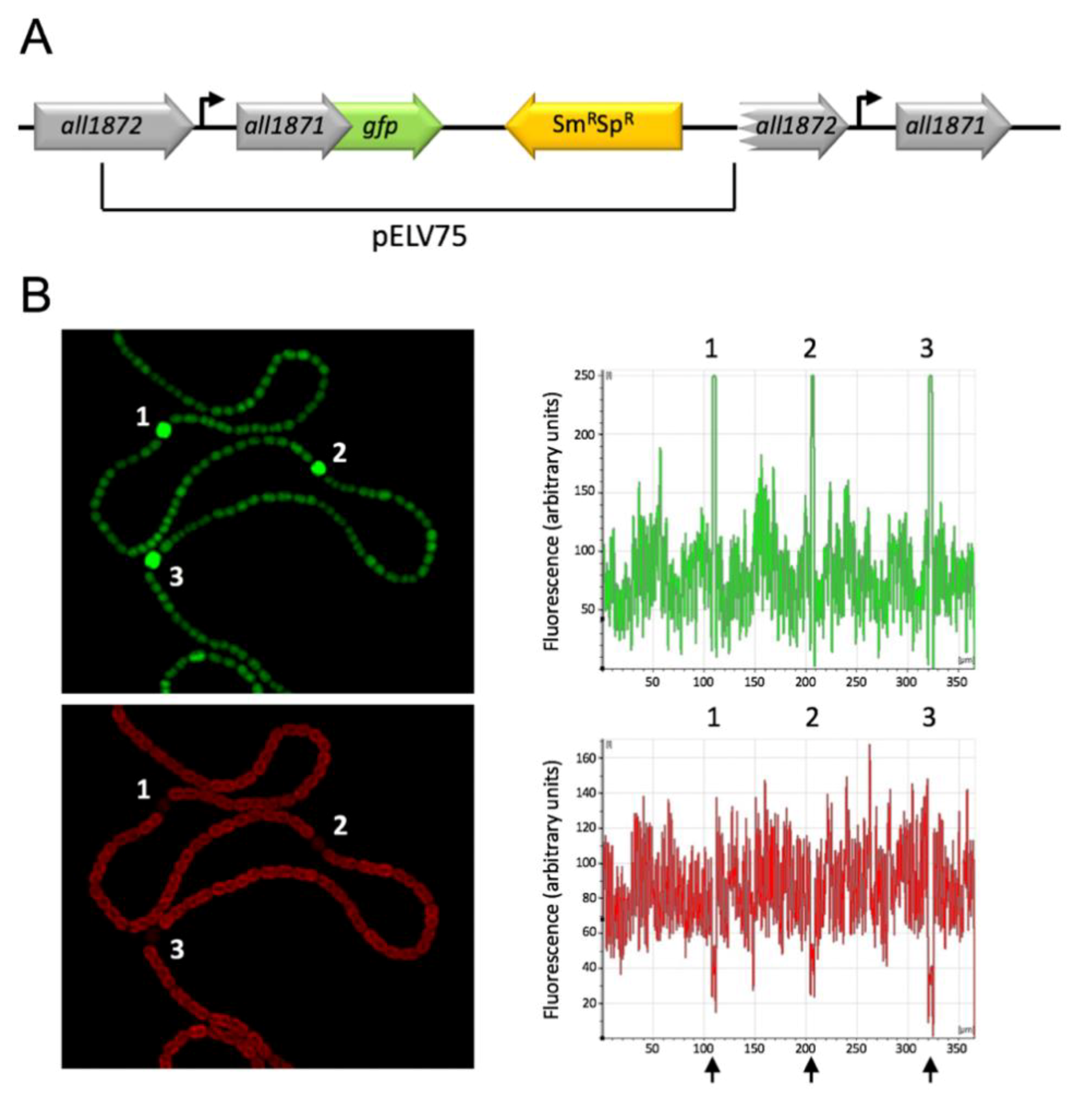
2.8. Construction of a Strain Bearing a Translational Fusion all1871-gfpmut2
2.9. Microscopy
3. Results
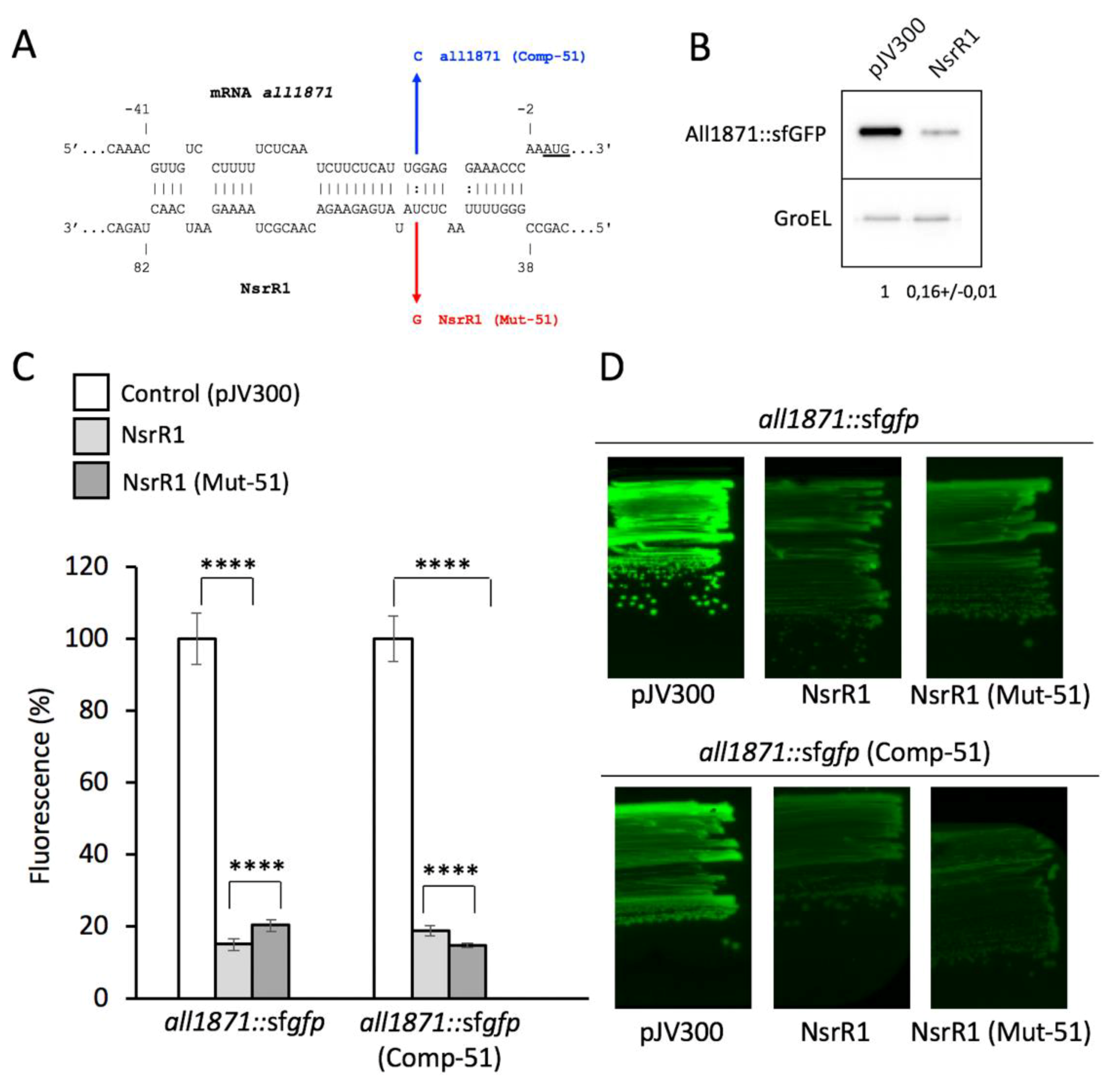
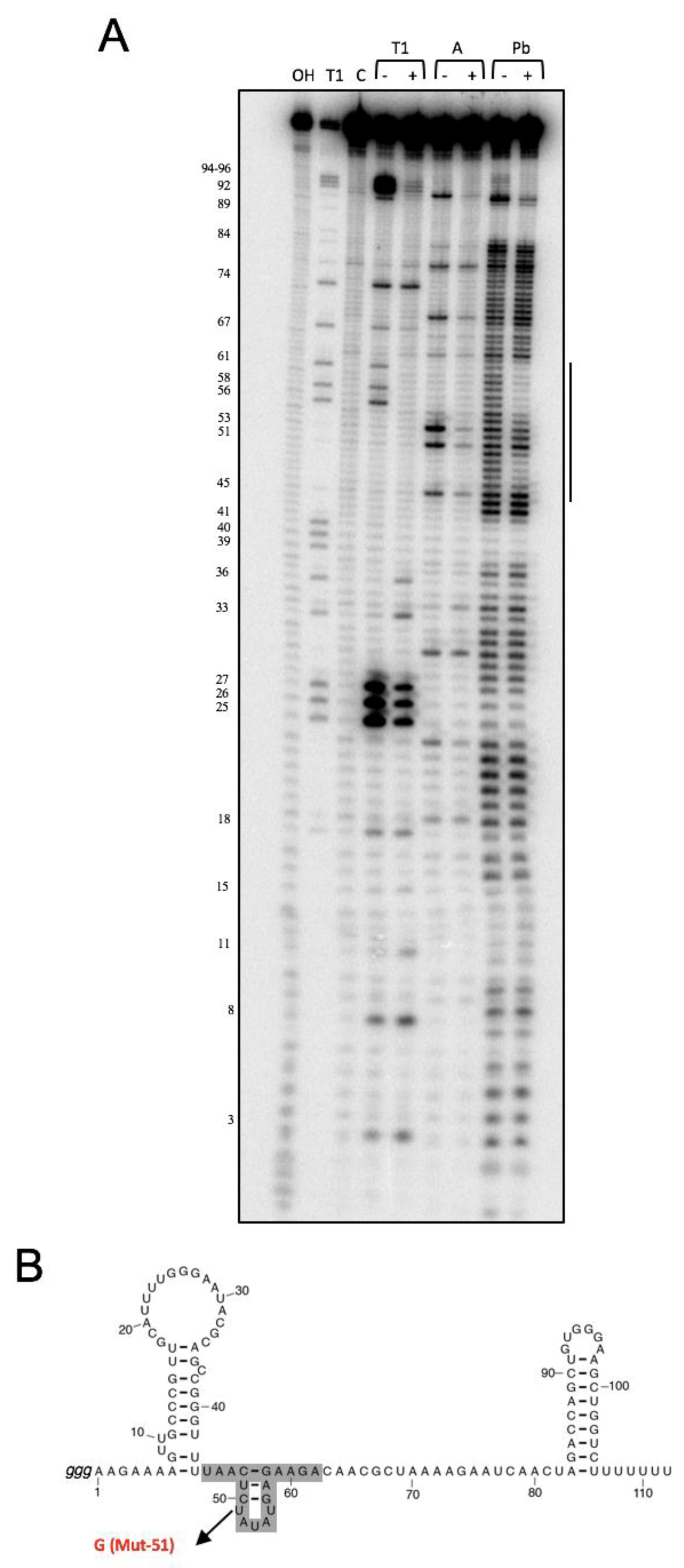
3.1. NsrR1 Interacts with all1871 mRNA 5′-UTR
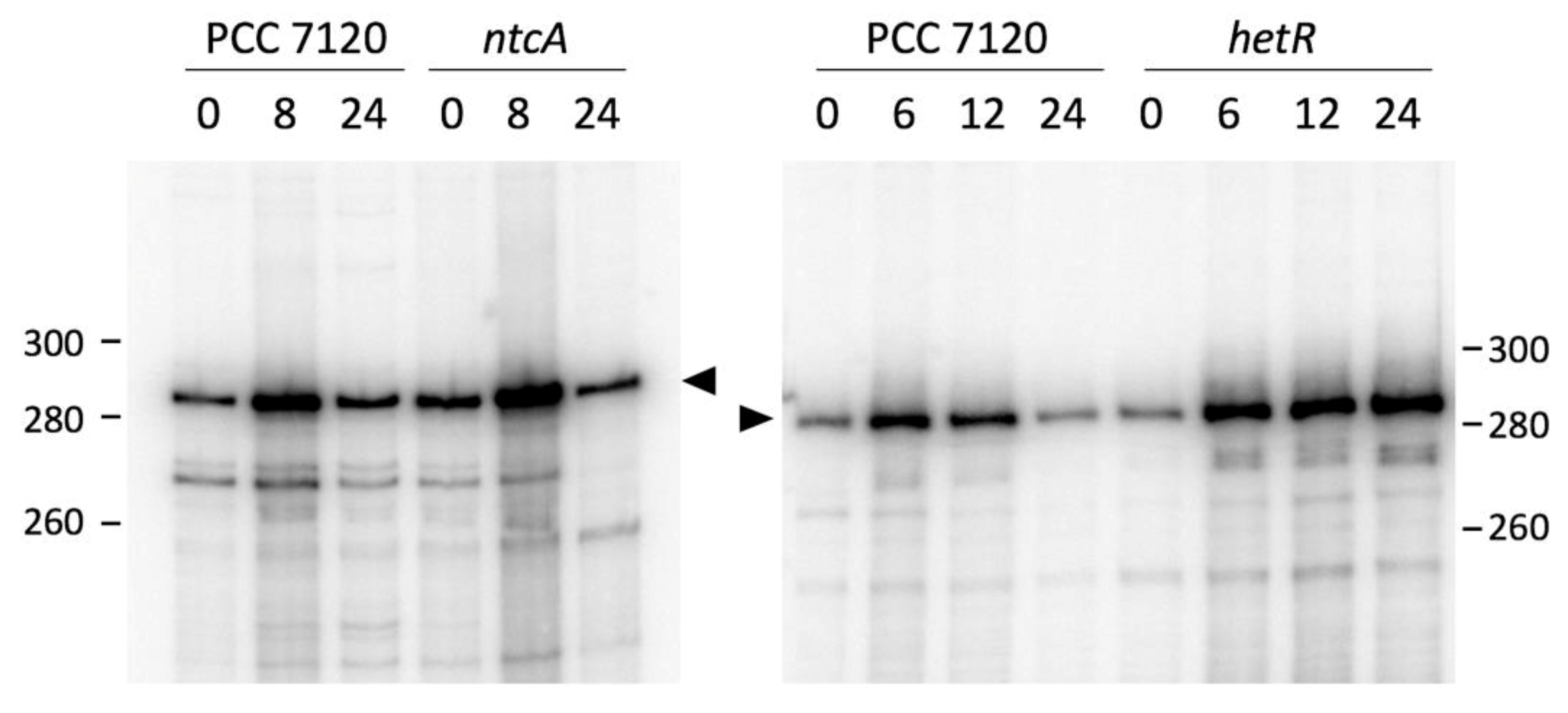
3.2. Expression of all1871 Is Regulated by Nitrogen Availability But Does Not Require NtcA or HetR
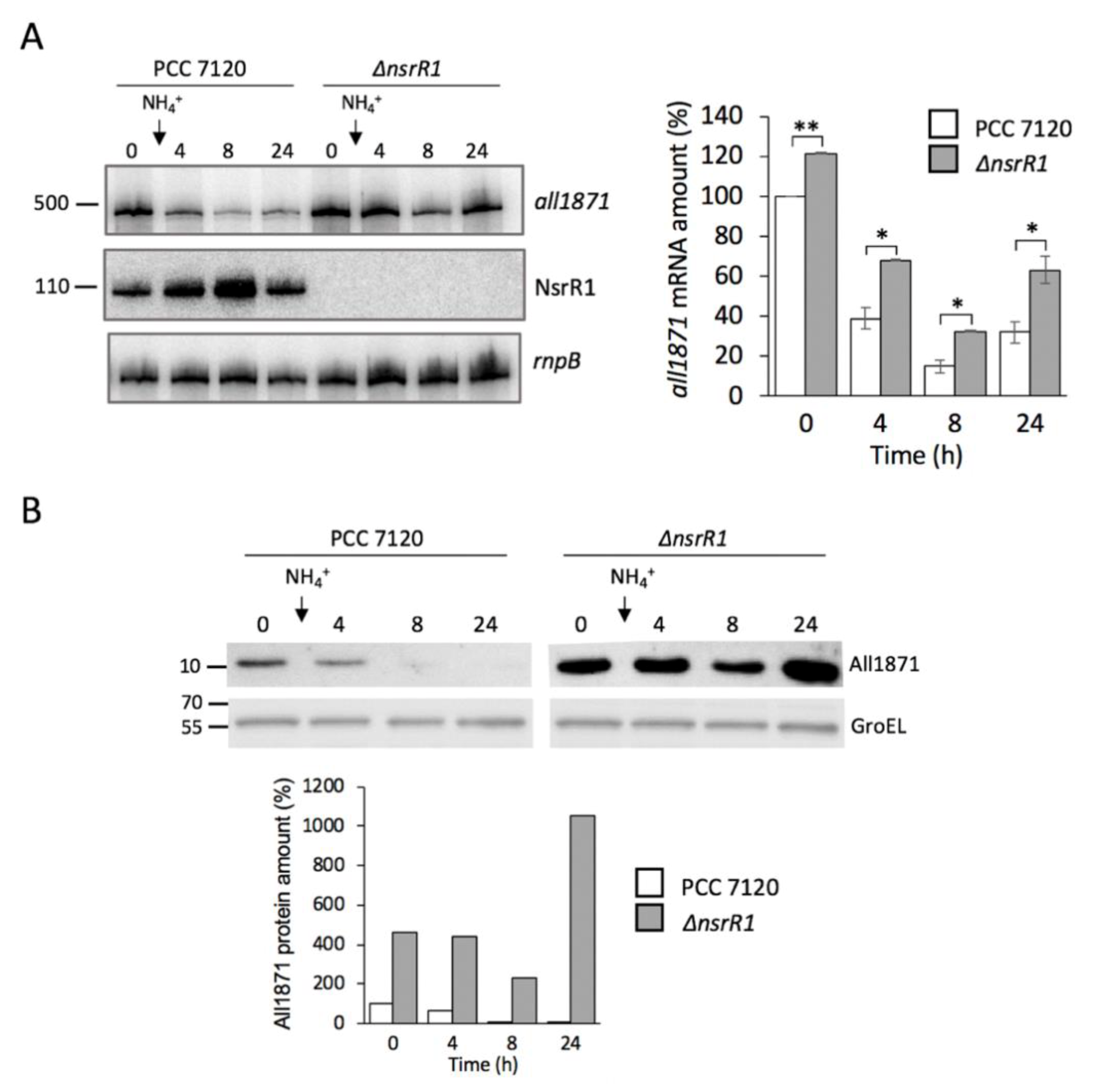
3.3. Expression of all1871 Is Reduced by NsrR1 in Nostoc sp. PCC 7120
3.4. All1871 Is Differentially Expressed in Heterocysts and Required for Diazotrophic Growth But Not for Heterocyst Differentiation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wagner, E.G.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef]
- Brosse, A.; Guillier, M. Bacterial small RNAs in mixed regulatory networks. Microbiol. Spectr. 2018, 6, RWR-0014-2017. [Google Scholar] [CrossRef]
- Nitzan, M.; Rehani, R.; Margalit, H. Integration of bacterial small RNAs in regulatory networks. Annu. Rev. Biophys. 2017, 46, 131–148. [Google Scholar] [CrossRef]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 2011, 108, 20130–20135. [Google Scholar] [CrossRef]
- Picossi, S.; Flores, E.; Herrero, A. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genom. 2014, 15, 22. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernández-Prieto, M.A.; Muro-Pastor, M.I.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef]
- Klähn, S.; Schaal, C.; Georg, J.; Baumgartner, D.; Knippen, G.; Hagemann, M.; Muro-Pastor, A.M.; Hess, W.R. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. USA 2015, 112, E6243–E6252. [Google Scholar] [CrossRef]
- Brenes-Álvarez, M.; Olmedo-Verd, E.; Vioque, A.; Muro-Pastor, A.M. Identification of conserved and potentially regulatory small RNAs in heterocystous cyanobacteria. Front. Microbiol. 2016, 7, 48. [Google Scholar] [CrossRef]
- Álvarez-Escribano, I.; Vioque, A.; Muro-Pastor, A.M. NsrR1, a nitrogen stress-repressed sRNA, contributes to the regulation of nblA in Nostoc sp. PCC 7120. Front. Microbiol. 2018, 9, 2267. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13, 1039–1047. [Google Scholar] [CrossRef]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000, 35, 1192–1201. [Google Scholar] [CrossRef]
- Flores, E.; Herrero, A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nature Rev. Microbiol. 2010, 8, 39–50. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Hess, W.R. Heterocyst differentiation: From single mutants to global approaches. Trends Microbiol. 2012, 20, 548–557. [Google Scholar] [CrossRef]
- Olmedo-Verd, E.; Brenes-Álvarez, M.; Vioque, A.; Muro-Pastor, A.M. A heterocyst-specific antisense RNA contributes to metabolic reprogramming in Nostoc sp. PCC 7120. Plant Cell Physiol. 2019, 60, 1646–1655. [Google Scholar] [CrossRef]
- Brenes-Álvarez, M.; Mitschke, J.; Olmedo-Verd, E.; Georg, J.; Hess, W.R.; Vioque, A.; Muro-Pastor, A.M. Elements of the heterocyst-specific transcriptome unravelled by co-expression analysis in Nostoc sp. PCC 7120. Environ. Microbiol. 2019, 21, 2544–2558. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain stories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar]
- Urban, J.H.; Vogel, J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007, 35, 1018–1037. [Google Scholar] [CrossRef]
- Corcoran, C.P.; Podkaminski, D.; Papenfort, K.; Urban, J.H.; Hinton, J.C.; Vogel, J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012, 84, 428–445. [Google Scholar] [CrossRef]
- Sittka, A.; Pfeiffer, V.; Tedin, K.; Vogel, J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007, 63, 193–217. [Google Scholar] [CrossRef]
- Wright, P.R.; Richter, A.S.; Papenfort, K.; Mann, M.; Vogel, J.; Hess, W.R.; Backofen, R.; Georg, J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, E3487–E3496. [Google Scholar] [CrossRef]
- Mohamed, A.; Jansson, C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 1989, 13, 693–700. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Valladares, A.; Flores, E.; Herrero, A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 1999, 181, 6664–6669. [Google Scholar] [CrossRef]
- Steglich, C.; Futschik, M.E.; Lindell, D.; Voβ, B.; Chisholm, S.W.; Hess, W.R. The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet. 2008, 4, e1000173. [Google Scholar] [CrossRef]
- Vioque, A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992, 20, 6331–6337. [Google Scholar] [CrossRef]
- Merino-Puerto, V.; Mariscal, V.; Mullineaux, C.W.; Herrero, A.; Flores, E. Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol. 2010, 75, 1159–1170. [Google Scholar] [CrossRef]
- Black, T.A.; Cai, Y.; Wolk, C.P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 1993, 9, 77–84. [Google Scholar] [CrossRef]
- Elhai, J.; Wolk, C.P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988, 167, 747–754. [Google Scholar]
- Cai, Y.; Wolk, C.P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1990, 172, 3138–3145. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Flores, E.; Herrero, A. NtcA-regulated heterocyst differentiation genes hetC and devB from Anabaena sp. strain PCC 7120 exhibit a similar tandem promoter arrangement. J. Bacteriol. 2009, 191, 5765–5774. [Google Scholar] [CrossRef][Green Version]
- Olmedo-Verd, E.; Muro-Pastor, A.M.; Flores, E.; Herrero, A. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 2006, 188, 6694–6699. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M. The heterocyst-specific NsiR1 small RNA is an early marker of cell differentiation in cyanobacterial filaments. mBio 2014, 5, e01079-14. [Google Scholar] [CrossRef]
- Olmedo-Verd, E.; Flores, E.; Herrero, A.; Muro-Pastor, A.M. HetR-dependent and -independent expression of heterocyst-related genes in an Anabaena strain overproducing the NtcA transcription factor. J. Bacteriol. 2005, 187, 1985–1991. [Google Scholar] [CrossRef]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Flaherty, B.L.; Van Nieuwerburgh, F.; Head, S.R.; Golden, J.W. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genom. 2011, 12, 332. [Google Scholar] [CrossRef]
- Frías, J.E.; Flores, E.; Herrero, A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 1994, 14, 823–832. [Google Scholar] [CrossRef]
- Brenes-Álvarez, M.; Vioque, A.; Muro-Pastor, A.M. The integrity of the cell wall and its remodeling during heterocyst differentiation are regulated by phylogenetically conserved small RNA Yfr1 in Nostoc sp. strain PCC 7120. mBio 2020, 11, e02599-19. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Hess, W.R. Regulatory RNA at the crossroads of carbon and nitrogen metabolism in photosynthetic cyanobacteria. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194477. [Google Scholar] [CrossRef]
- Kadowaki, T.; Nagayama, R.; Georg, J.; Nishiyama, Y.; Wilde, A.; Hess, W.R.; Hihara, Y. A feed-forward loop consisting of the response regulator RpaB and the small RNA PsrR1 controls light acclimation of photosystem I gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016, 57, 813–823. [Google Scholar] [CrossRef]
- Sandh, G.; Ramstrom, M.; Stensjö, K. Analysis of the early heterocyst Cys-proteome in the multicellular cyanobacterium Nostoc punctiforme reveals novel insights into the division of labor within diazotrophic filaments. BMC Genom. 2014, 15, 1064. [Google Scholar] [CrossRef]

| Strain | Nitrogenase Activity 1 (µmol Ethylene·h−1·mg Chl−1) | |
|---|---|---|
| Oxic Conditions | Anoxic Conditions | |
| Nostoc sp. PCC 7120 | 10.50 ± 3.49 | 10.64 ± 2.05 |
| all1871::Nm | 0.00 ± 0.00 | 0.00 ± 0.00 |
| all1871::Nm + Ptrc::all1871 | 3.66 ± 1.67 | 5.05 ± 3.41 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Escribano, I.; Brenes-Álvarez, M.; Olmedo-Verd, E.; Vioque, A.; Muro-Pastor, A.M. The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120. Life 2020, 10, 54. https://doi.org/10.3390/life10050054
Álvarez-Escribano I, Brenes-Álvarez M, Olmedo-Verd E, Vioque A, Muro-Pastor AM. The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120. Life. 2020; 10(5):54. https://doi.org/10.3390/life10050054
Chicago/Turabian StyleÁlvarez-Escribano, Isidro, Manuel Brenes-Álvarez, Elvira Olmedo-Verd, Agustín Vioque, and Alicia M. Muro-Pastor. 2020. "The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120" Life 10, no. 5: 54. https://doi.org/10.3390/life10050054
APA StyleÁlvarez-Escribano, I., Brenes-Álvarez, M., Olmedo-Verd, E., Vioque, A., & Muro-Pastor, A. M. (2020). The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120. Life, 10(5), 54. https://doi.org/10.3390/life10050054





