Evolutionary Multilabel Classification Algorithm Based on Cultural Algorithm
Abstract
1. Introduction
2. Bayesian Multilabel Classification and Cultural Algorithms
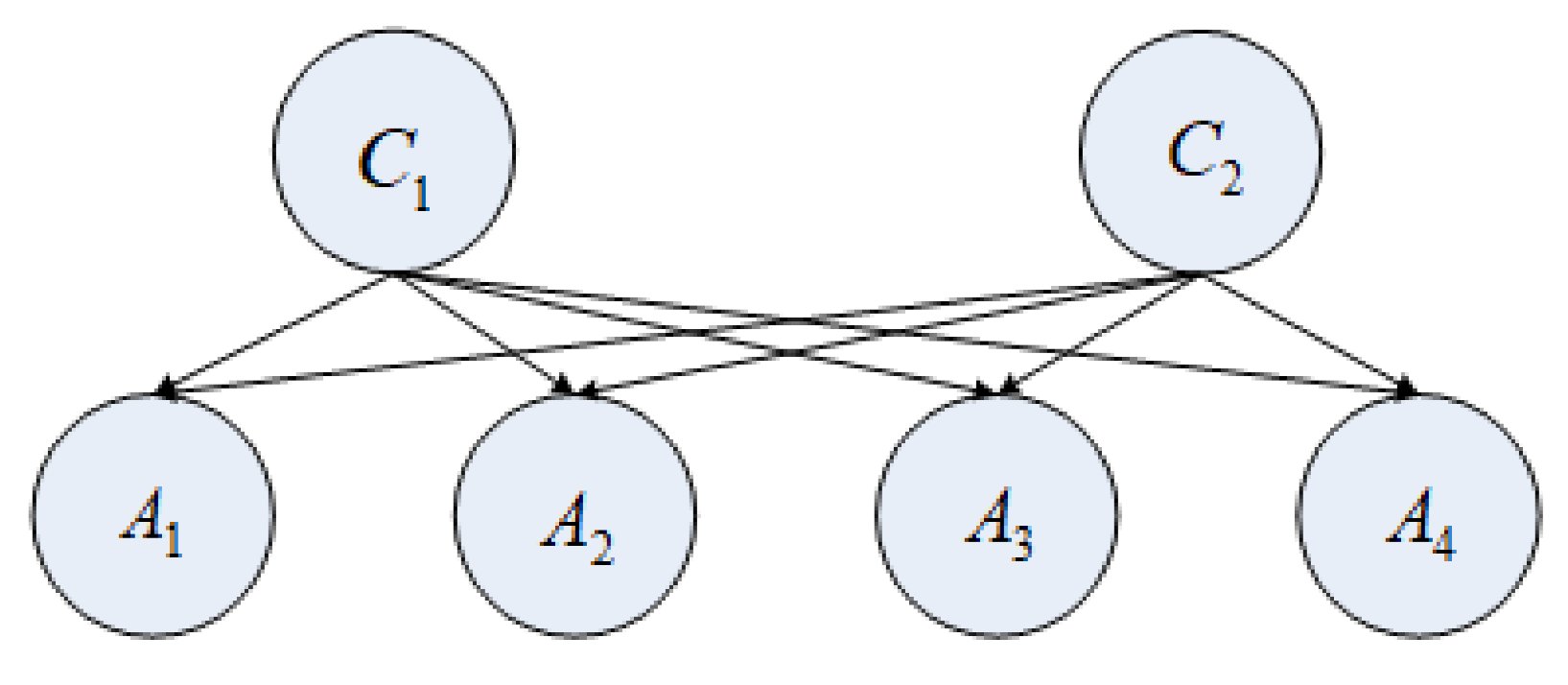
2.1. Bayesian Multilabel Classification
- All attribute nodes exhibit an equal level of importance for the selection of a class node.
- The attribute nodes (A1, A2, A3, and A4) are mutually independent and completely unrelated to each other.
- Assume that the class nodes and are unrelated and independent.
2.2. Cultural Algorithms
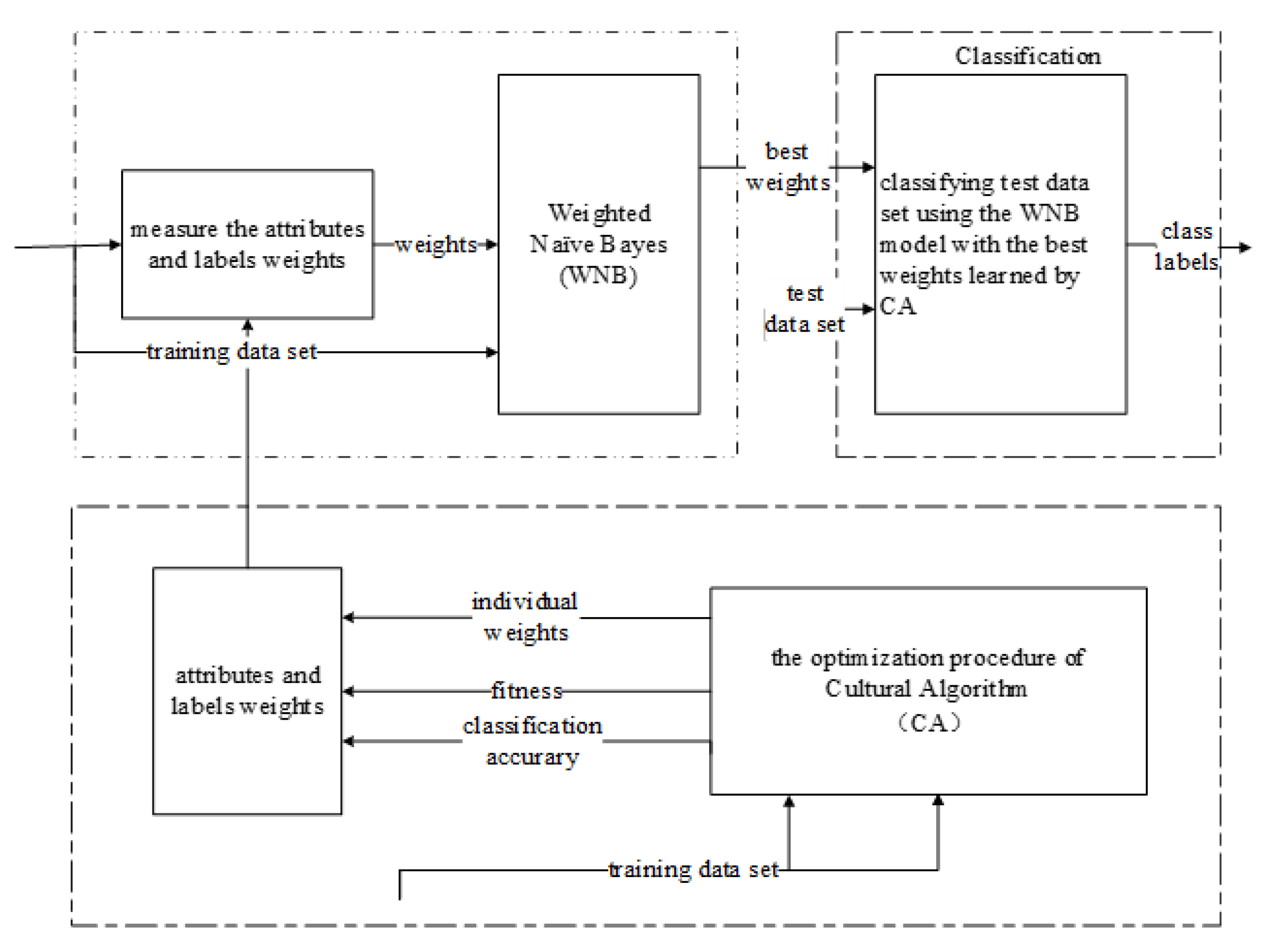
3. Cultural-Algorithm-Based Evolutionary Multilabel Classification Algorithm
3.1. Weighted Bayes Multilabel Classification Algorithm
3.1.1. Correction of the Prior Probability Formula
3.1.2. Correction of the Conditional Probability Formula
3.1.3. Correction of the Posterior Probability Formula
3.2. Improved Cultural Algorithm
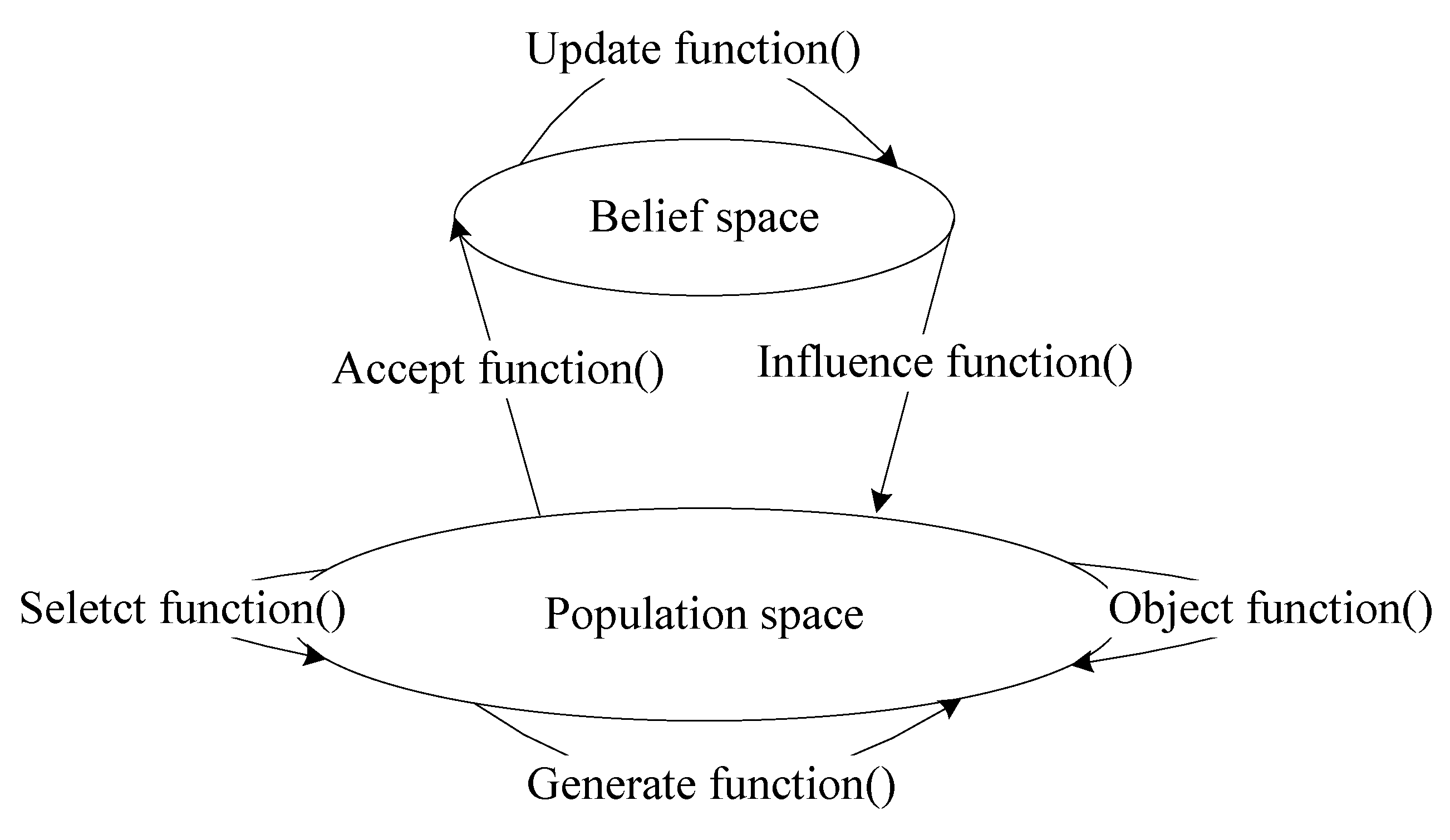
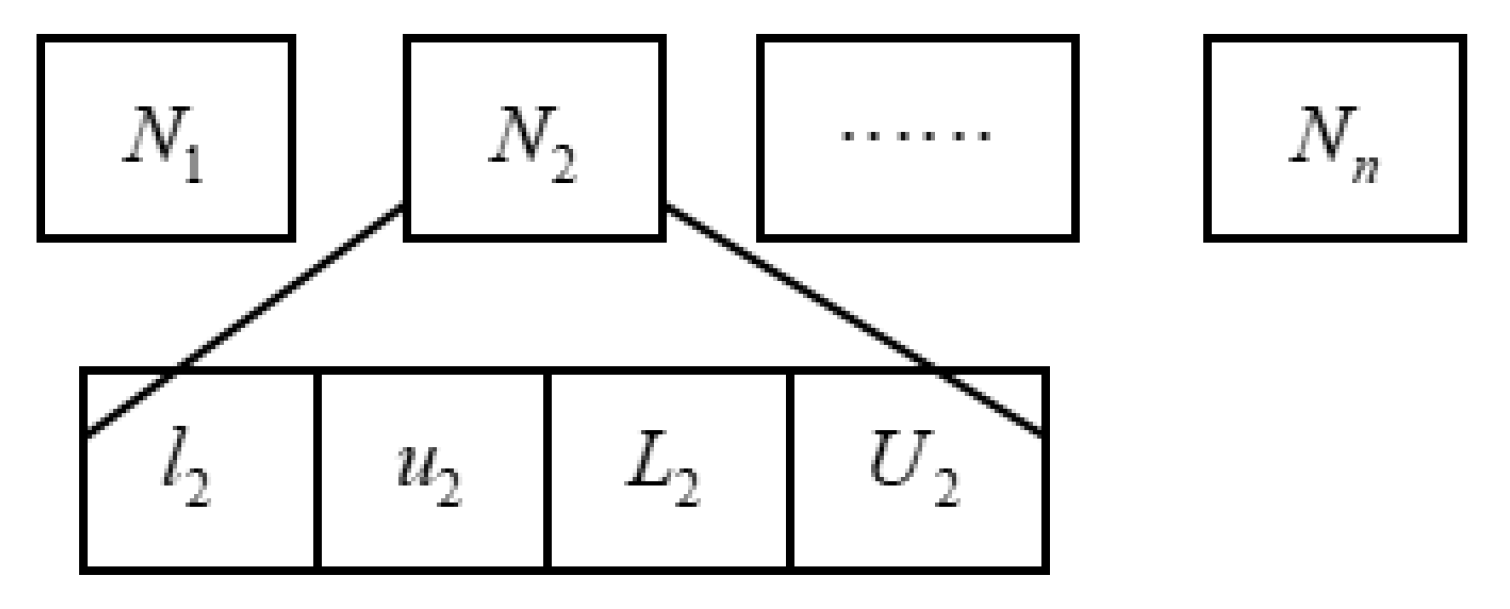
3.2.1. Definition and Update Rules of the Belief Space
3.2.2. Fitness Function
3.2.3. Influence Function
3.2.4. Selection Function
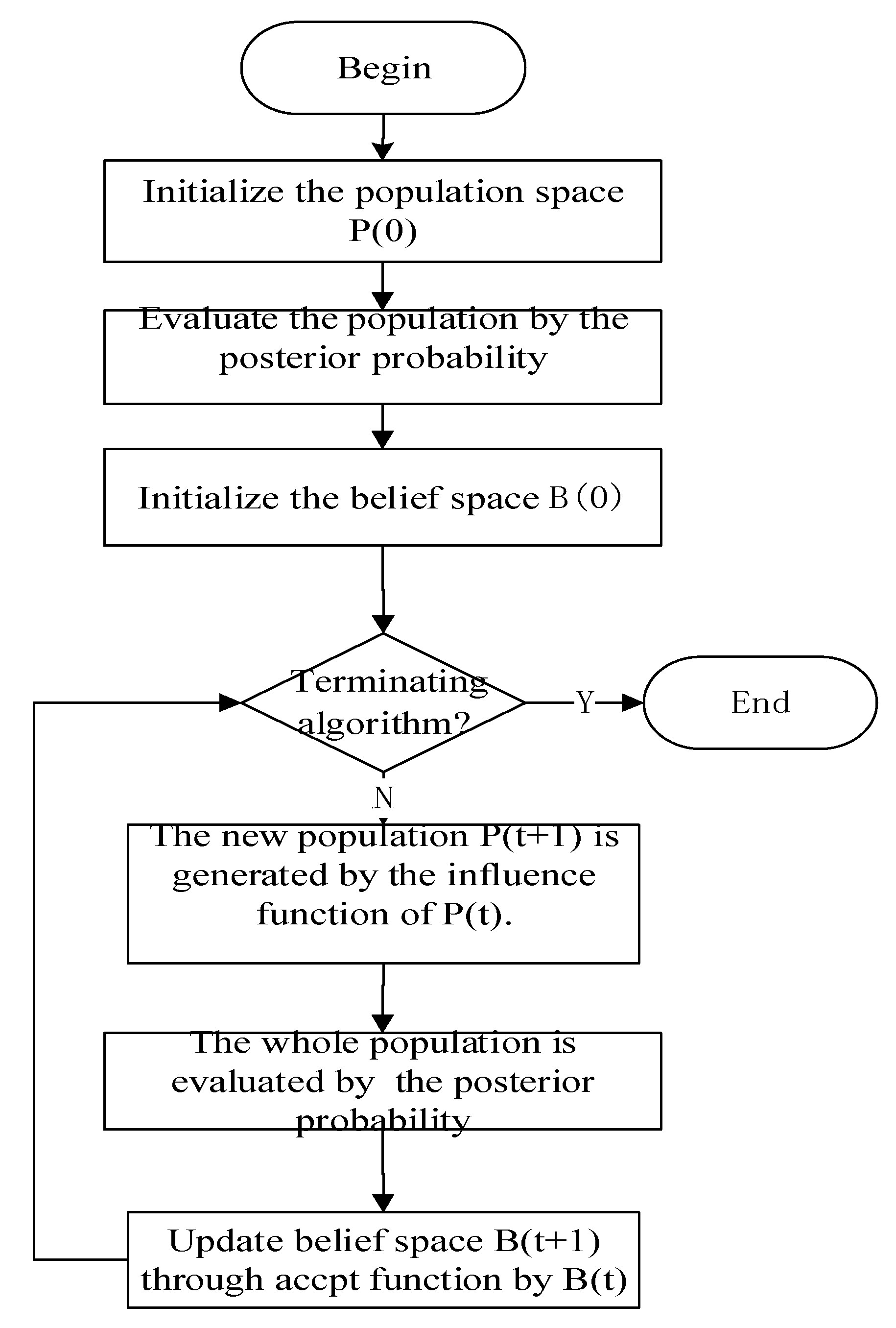
3.3. CA-Based Evolutionary Multilabel Classification Algorithm
4. Experimental Results and Analysis
4.1. Experimental Datasets
4.2. Classification Evaluation Criteria
4.3. Classification Prediction Methods
4.4. Analysis of the Results of the NBMLC Experiment
4.5. Analysis of Results of the CA-WNB Experiment
Gaussian and Cauchy Distribution
- (1)
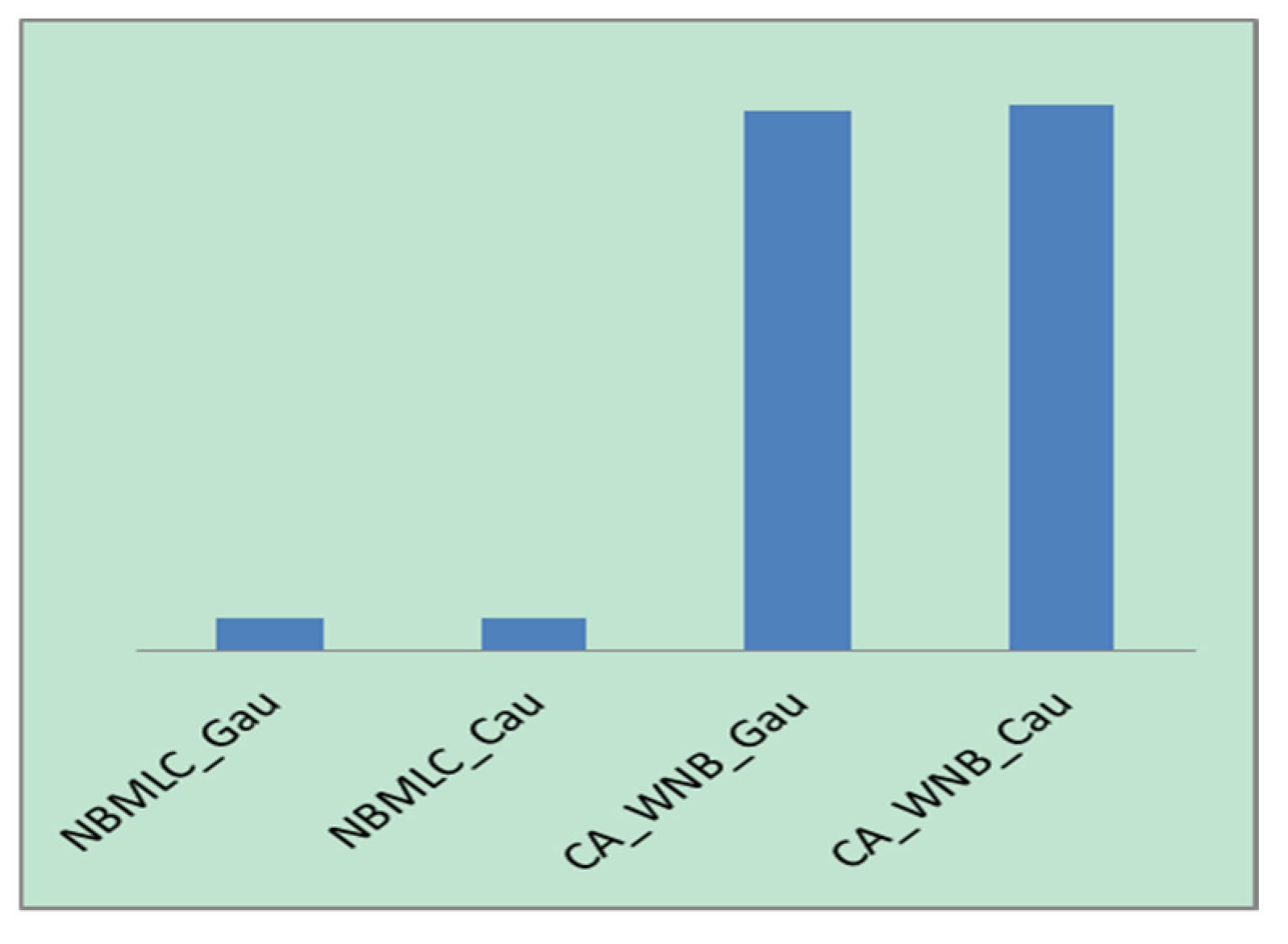
- Based on Figure 11, the CA-WNB algorithm consumes a significantly longer computation time than the NBMLC algorithm.
- (2)
- If one omits computational cost, the CA-WNB algorithm is evidently superior to the simple NBMLC algorithm in terms of classification performance. The percentage improvement in classification accuracy with the CA-WNB algorithm was most pronounced in the “emotions” dataset, followed by the “scene”, “yeast”, and CAL500 datasets. An analysis of the characteristics of these datasets revealed that the use of stratified sampling to disrupt the “emotions” and “scene” datasets did not significantly affect their classification accuracies. Meanwhile, the effects of this operation on the CAL500 and “yeast” datasets are more pronounced. This illustrates that the “emotions” and “scene” datasets have relatively uniform distributions of data, whereas the CAL500 and “yeast” datasets have highly nonuniform data distributions. If the training dataset is strongly representative of the dataset, the classification efficacy of the algorithm will be substantially improved by the training process. Otherwise, the improvement in classification efficacy will not be very significant. Based on this observation, weighted naïve Bayes can be used to optimize classification efficacy if the distribution of the dataset is relatively uniform and if the time requirements of the experiment are not particularly stringent.
- (3)
- A comparison of Table 10 and Table 11 reveals that the improvement in the average classification accuracy yielded by Prediction Method 2 (6.79% and 7.43%) is always higher than that yielded by Prediction Method 1 (6.54% and 7.20%) regardless of whether Gaussian or Cauchy fitting is used. Furthermore, the improvement in average classification accuracy is always higher with the Gaussian fitting (7.20% and 7.43%) than with the Cauchy fitting (6.54% and 6.79%). A comparison of the results with Cauchy fitting and Gaussian fitting reveals that the results of the CA-WNB algorithm with Cauchy fitting varied substantially. Moreover, the weights obtained by the CA also exert a higher impact on the Cauchy fitting. Therefore, the results with Cauchy fitting are unstable to a certain degree. Nonetheless, the average classification accuracy of the CA-WNB algorithm with Cauchy fitting is superior to that of the CA-WNB algorithm with Gaussian fitting.
- (4)
- Table 10 and Table 11 reveal that the highest average classification accuracies for the four experimental datasets were obtained by individuals with Ranks 10–20, according to Prediction Method 2. Conversely, the worst classification accuracies were obtained by individuals with Ranks 20–30. It is established that the weights that have the best fit with training set instances may not be the optimal weights for classifying test set instances. After a certain number of evolutions in the population, excessive fitting may have occurred because the fitting curves were adjusted too finely, thereby resulting in less-than-ideal classification accuracies for the test set’s instances. Consequently, the fitting curves produced by the weights of individuals with Ranks 10–20 were better for classifying the instances of the test set.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsoumakas, G.; Katakis, I.; Vlahavas, I. Mining multi-label data. In Data Mining and Knowledge Discovery Handbook; Springer: Boston, MA, USA, 2010; pp. 667–685. [Google Scholar]
- Streich, A.P.; Buhmann, J.M. Classification of multi-labeled data: A generative approach. In Machine Learning and Knowledge Discovery in Databases; Springer: Berlin/Heidelberg, Germany, 2008; pp. 390–405. [Google Scholar]
- Kazawa, H.; Izumitani, T.; Taira, H.; Maeda, E. Maximal margin labeling for multi-topic text categorization. In Advances in Neural Information Processing Systems; MIT Press: Vancouver, BC, Canada, 2004; pp. 649–656. [Google Scholar]
- Snoek, C.G.; Worring, M.; Van Gemert, J.C.; Geusebroek, J.M.; Smeulders, A.W. The challenge problem for automated detection of 101 semantic concepts in multimedia. In Proceedings of the 14th annual ACM International Conference on Multimedia, Santa Barbara, CA, USA, 23–27 October 2006; ACM: New York, NY, USA, 2006; pp. 421–430. [Google Scholar]
- Vens, C.; Struyf, J.; Schietgat, L.; Džeroski, S.; Blockeel, H. Decision trees for hierarchical multi-label classification. Mach. Learn. 2008, 73, 185–214. [Google Scholar] [CrossRef]
- Boutell, M.R.; Luo, J.; Shen, X.; Brown, C.M. Learning multi-label scene classification. Pattern Recognit. 2004, 37, 1757–1771. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, K.; Yang, Y. Multi-Label Classification with Weighted Classifier Selection and Stacked Ensemble. Inf. Sci. 2020. [Google Scholar] [CrossRef]
- Qian, W.; Xiong, C.; Wang, Y. A ranking-based feature selection for multi-label classification with fuzzy relative discernibility. Appl. Soft Comput. 2021, 102, 106995. [Google Scholar] [CrossRef]
- Yao, Y.; Li, Y.; Ye, Y.; Li, X. MLCE: A Multi-Label Crotch Ensemble Method for Multi-Label Classification. Int. J. Pattern Recognit. Artif. Intell. 2020. [Google Scholar] [CrossRef]
- Yang, B.; Tong, K.; Zhao, X.; Pang, S.; Chen, J. Multilabel Classification Using Low-Rank Decomposition. Discret. Dyn. Nat. Soc. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Kumar, A.; Abhishek, K.; Kumar Singh, A.; Nerurkar, P.; Chandane, M.; Bhirud, S.; Busnel, Y. Multilabel classification of remote sensed satellite imagery. Trans. Emerg. Telecommun. Technol. 2020, 4, 118–133. [Google Scholar] [CrossRef]
- Huang, S.J.; Li, G.X.; Huang, W.Y.; Li, S.Y. Incremental Multi-Label Learning with Active Queries. J. Comput. Sci. Technol. 2020, 35, 234–246. [Google Scholar] [CrossRef]
- Zhang, M.L.; Peña, J.M.; Robles, V. Feature selection for multi-label naive Bayes classification. Inf. Sci. 2009, 179, 3218–3229. [Google Scholar] [CrossRef]
- De Carvalho, A.C.; Freitas, A.A. A tutorial on multi-label classification techniques. Found. Comput. Intell. 2009, 5, 177–195. [Google Scholar]
- Spyromitros, E.; Tsoumakas, G.; Vlahavas, I. An empirical study of lazy multilabel classification algorithms. In Artificial Intelligence: Theories, Models and Applications; Springer: Berlin/Heidelberg, Germany, 2008; pp. 401–406. [Google Scholar]
- Rousu, J.; Saunders, C.; Szedmak, S.; Shawe-Taylor, J. Kernel-based learning of hierarchical multilabel classification models. J. Mach. Learn. Res. 2006, 7, 1601–1626. [Google Scholar]
- Yang, Y.; Chute, C.G. An example-based mapping method for text categorization and retrieval. ACM Trans. Inf. Syst. (TOIS) 1994, 12, 252–277. [Google Scholar] [CrossRef]
- Grodzicki, R.; Mańdziuk, J.; Wang, L. Improved multilabel classification with neural networks. Parallel Probl. Solving Nat. Ppsn X 2008, 5199, 409–416. [Google Scholar]
- Gonçalves, E.C.; Freitas, A.A.; Plastino, A. A Survey of Genetic Algorithms for Multi-Label Classification. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, 29 January 2018; pp. 1–8. [Google Scholar]
- McCallum, A.; Nigam, K. A comparison of event models for naive bayes text classification. AAAI-98 Workshop Learn. Text Categ. 1998, 752, 41–48. [Google Scholar]
- Gao, S.; Wu, W.; Lee, C.H.; Chua, T.S. A MFoM learning approach to robust multiclass multi-label text categorization. In Proceedings of the Twenty-First International Conference on Machine Learning; ACM: New York, NY, USA, 2004; pp. 329–336. [Google Scholar]
- Ghamrawi, N.; McCallum, A. Collective multi-label classification. In Proceedings of the 14th ACM International Conference on Information and Knowledge Management; ACM: New York, NY, USA, 2005; pp. 195–200. [Google Scholar]
- Zhang, M.L.; Zhou, Z.H. ML-KNN: A lazy learning approach to multi-label learning. Pattern Recognit. 2007, 40, 2038–2048. [Google Scholar] [CrossRef]
- Xu, X.S.; Jiang, Y.; Peng, L.; Xue, X.; Zhou, Z.H. Ensemble approach based on conditional random field for multi-label image and video annotation. In Proceedings of the 19th ACM International Conference on Multimedia; ACM: New York, NY, USA, 2011; pp. 1377–1380. [Google Scholar]
- Qu, G.; Zhang, H.; Hartrick, C.T. Multi-label classification with Bayes’ theorem. In Proceedings of the 2011 4th International Conference on Biomedical Engineering and Informatics (BMEI), Shanghai, China, 15–17 October 2011; pp. 2281–2285. [Google Scholar]
- Wu, J.; Cai, Z. Attribute weighting via differential evolution algorithm for attribute weighted naive bayes (wnb). J. Comput. Inf. Syst. 2011, 7, 1672–1679. [Google Scholar]
- Wu, J.; Cai, Z. A naive Bayes probability estimation model based on self-adaptive differential evolution. J. Intell. Inf. Syst. 2014, 42, 671–694. [Google Scholar] [CrossRef]
- Sucar, L.E.; Bielza, C.; Morales, E.F.; Hernandez-Leal, P.; Zaragoza, J.H.; Larrañaga, P. Multi-label classification with Bayesian network-based chain classifiers. Pattern Recognit. Lett. 2014, 41, 14–22. [Google Scholar] [CrossRef]
- Reyes, O.; Morell, C.; Ventura, S. Evolutionary feature weighting to improve the performance of multi-label lazy algorithms. Integr. Comput. Aided Eng. 2014, 21, 339–354. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.W. Memetic feature selection algorithm for multi-label classification. Inf. Sci. 2015, 293, 80–96. [Google Scholar] [CrossRef]
- Yan, X.; Wu, Q.; Sheng, V.S. A Double Weighted Naive Bayes with Niching Cultural Algorithm for Multi-Label Classification. Int. J. Pattern Recognit. Artif. Intell. 2016, 30, 1–23. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, H.; Yan, X. Multi-label classification algorithm research based on swarm intelligence. Clust. Comput. 2016, 19, 2075–2085. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, D.W.; Sun, X.Y.; Guo, Y.N. A PSO-based multi-objective multi-label feature selection method in classification. Sci. Rep. 2017, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, H.; Yan, X.; Liu, X. MapReduce-based adaptive random forest algorithm for multi-label classification. Neural Comput. Appl. 2019, 31, 8239–8252. [Google Scholar] [CrossRef]
- Moyano, J.M.; Gibaja, E.L.; Cios, K.J.; Ventura, S. An evolutionary approach to build ensembles of multi-label classifiers. Inf. Fusion 2019, 50, 168–180. [Google Scholar] [CrossRef]
- Guo, Y.N.; Zhang, P.; Cheng, J.; Wang, C.; Gong, D. Interval Multi-objective Quantum-inspired Cultural Algorithms. Neural Comput. Appl. 2018, 30, 709–722. [Google Scholar] [CrossRef]
- Yan, X.; Zhu, Z.; Hu, C.; Gong, W.; Wu, Q. Spark-based intelligent parameter inversion method for prestack seismic data. Neural Comput. Appl. 2019, 31, 4577–4593. [Google Scholar] [CrossRef]
- Wu, B.; Qian, C.; Ni, W.; Fan, S. The improvement of glowworm swarm optimization for continuous optimization problems. Expert Syst. Appl. 2012, 39, 6335–6342. [Google Scholar] [CrossRef]
- Lu, C.; Gao, L.; Li, X.; Zheng, J.; Gong, W. A multi-objective approach to welding shop scheduling for makespan, noise pollution and energy consumption. J. Clean. Prod. 2018, 196, 773–787. [Google Scholar] [CrossRef]
- Wu, Q.; Zhu, Z.; Yan, X.; Gong, W. An improved particle swarm optimization algorithm for AVO elastic parameter inversion problem. Concurr. Comput. Pract. Exp. 2019, 31, 1–16. [Google Scholar] [CrossRef]
- Yu, P.; Yan, X. Stock price prediction based on deep neural network. Neural Comput. Appl. 2020, 32, 1609–1628. [Google Scholar] [CrossRef]
- Gong, W.; Cai, Z. Parameter extraction of solar cell models using repaired adaptive differential evolution. Solar Energy 2013, 94, 209–220. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Zhou, A.; Tang, K. An estimation of distribution algorhim for mixed-variable Newsvendor problems. IEEE Trans. Evol. Comput. 2020, 24, 479–493. [Google Scholar]
- Wang, G.G. Improving Metaheuristic Algorithms with Information Feedback Models. IEEE Trans. Cybern. 2017, 99, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Li, P.; Tang, K.; Gao, L.; Wang, L. Clonal Selection Based Intelligent Parameter Inversion Algorithm for Prestack Seismic Data. Inf. Sci. 2020, 517, 86–99. [Google Scholar] [CrossRef]
- Yan, X.; Yang, K.; Hu, C.; Gong, W. Pollution source positioning in a water supply network based on expensive optimization. Desalination Water Treat. 2018, 110, 308–318. [Google Scholar] [CrossRef]
- Wang, R.; Zhou, Z.; Ishibuchi, H.; Liao, T.; Zhang, T. Localized weighted sum method for many-objective optimization. IEEE Trans. Evol. Comput. 2018, 22, 3–18. [Google Scholar] [CrossRef]
- Lu, C.; Gao, L.; Yi, J. Grey wolf optimizer with cellular topological structure. Expert Syst. Appl. 2018, 107, 89–114. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Zhou, A. A particle swarm optimization algorithm for mixed-variable optimization problems. Swarm Evol. Comput. 2021, 60, 100808. [Google Scholar] [CrossRef]
- Yan, X.; Zhao, J. Multimodal optimization problem in contamination source determination of water supply networks. Swarm Evol. Comput. 2019, 47, 66–71. [Google Scholar] [CrossRef]
- Yan, X.; Hu, C.; Sheng, V.S. Data-driven pollution source location algorithm in water quality monitoring sensor networks. Int. J. Bio-Inspir Compu. 2020, 15, 171–180. [Google Scholar] [CrossRef]
- Hu, C.; Dai, L.; Yan, X.; Gong, W.; Liu, X.; Wang, L. Modified NSGA-III for Sensor Placement in Water Distribution System. Inf. Sci. 2020, 509, 488–500. [Google Scholar] [CrossRef]
- Wang, R.; Li, G.; Ming, M.; Wu, G.; Wang, L. An efficient multi-objective model and algorithm for sizing a stand-alone hybrid renewable energy system. Energy 2017, 141, 2288–2299. [Google Scholar] [CrossRef]
- Li, S.; Gong, W.; Yan, X.; Hu, C.; Bai, D.; Wang, L. Parameter estimation of photovoltaic models with memetic adaptive differential evolution. Solar Energy 2019, 190, 465–474. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, M.; Wu, Q. Big-Data-Driven Pre-Stack Seismic Intelligent Inversion. Inf. Sci. 2021, 549, 34–52. [Google Scholar] [CrossRef]
- Wang, F.; Li, Y.; Liao, F.; Yan, H. An ensemble learning based prediction strtegy for dynamic multi-objective optimization. Appl. Soft Comput. 2020, 96, 106592. [Google Scholar] [CrossRef]
- Yan, X.; Li, T.; Hu, C. Real-time localization of pollution source for urban water supply network in emergencies. Clust. Comput. 2019, 22, 5941–5954. [Google Scholar] [CrossRef]
- Reynolds, R.G. Cultural algorithms: Theory and applications. In New Ideas in Optimization; McGraw-Hill Ltd.: Berkshire, UK, 1999; pp. 367–378. [Google Scholar]
- Reynolds, R.G.; Zhu, S. Knowledge-based function optimization using fuzzy cultural algorithms with evolutionary programming. IEEE Trans. Syst. Man Cybern. Part B 2001, 31, 1–18. [Google Scholar] [CrossRef]
- Zhang, H.; Sheng, S. Learning weighted naïve Bayes with accurate ranking. In Proceedings of the 4th IEEE International Conference on Data Mining, Brighton, UK, 1–4 November 2004; pp. 567–570. [Google Scholar]
- Xie, T.; Liu, R.; Wei, Z. Improvement of the Fast Clustering Algorithm Improved by K-Means in the Big Data. Appl. Math. Nonlinear Sci. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Yan, X.; Li, W.; Wu, Q.; Sheng, V.S. A Double Weighted Naive Bayes for Multi-label Classification. In International Symposium on Computational Intelligence and Intelligent Systems; Springer: Singapore, 2015; pp. 382–389. [Google Scholar]



| 1 | 0 | 1 | ||||
| 2 | 1 | 0 | ||||
| 3 | 0 | 0 | ||||
| 4 | 1 | 1 |
| Data Set | Definition Domain | No. of Samples in Sample Set | No. of Attributes | No. of Class Labels | ||
|---|---|---|---|---|---|---|
| Training Set | Test Set | Numerical Type | Noun Type | |||
| yeast | Biology | 1500 | 917 | 103 | 0 | 14 |
| scene | Image | 1211 | 1196 | 294 | 0 | 6 |
| emotions | Music | 391 | 202 | 72 | 0 | 6 |
| CAL500 | Music | 351 | 151 | 68 | 0 | 174 |
| Data Set | Gaussian | Cauchy | Disperse_10 | ||||||
| MAX | MIN | AVE | MAX | MIN | AVE | MAX | MIN | AVE | |
| CAL500 | 0.8732 | 0.8574 | 0.8622 | 0.8662 | 0.8588 | 0.8635 | 0.8838 | 0.8620 | 0.8740 |
| emotions | 0.6976 | 0.6798 | 0.6884 | 0.7022 | 0.6798 | 0.6892 | 0.8361 | 0.7968 | 0.8168 |
| scene | 0.8239 | 0.8195 | 0.8212 | 0.8241 | 0.8188 | 0.8151 | 0.8707 | 0.8552 | 0.8614 |
| yeast | 0.7749 | 0.7636 | 0.7688 | 0.7720 | 0.7603 | 0.7673 | 0.8023 | 0.7799 | 0.7958 |
| Data Set | Gau-Cau | Dis-Gau | Dis-Cau |
|---|---|---|---|
| CAL500 | −0.0013 | 0.0118 | 0.0105 |
| emotions | −0.0008 | 0.1284 | 0.1276 |
| scene | 0.0061 | 0.0402 | 0.0463 |
| yeast | 0.0015 | 0.027 | 0.0285 |
| Parameter | Population Size | Maximum Number of Iterations | Initial Acceptance Ratio |
|---|---|---|---|
| Value | 100 | 200 | 0.2 |
| Data Set | Algorithm | Gaussian | Cauchy | ||||
|---|---|---|---|---|---|---|---|
| MAX | MIN | AVE | MAX | MIN | AVE | ||
| CAL_500 | NBMLC | 0.8732 | 0.8574 | 0.8622 | 0.8721 | 0.8554 | 0.8635 |
| CA-WNB-P1 | 0.8893 | 0.8750 | 0.8813 | 0.8871 | 0.8737 | 0.8800 | |
| CA-WNB-P2 | 0.8897 | 0.8751 | 0.8825 | 0.8890 | 0.8744 | 0.8811 | |
| emotions | NBMLC | 0.6976 | 0.6798 | 0.6884 | 0.6976 | 0.6787 | 0.6892 |
| CA-WNB-P1 | 0.8059 | 0.7853 | 0.7938 | 0.8215 | 0.7850 | 0.8040 | |
| CA-WNB-P2 | 0.8115 | 0.7900 | 0.7993 | 0.8215 | 0.7869 | 0.8044 | |
| scene | NBMLC | 0.8239 | 0.8195 | 0.8212 | 0.8195 | 0.8098 | 0.8151 |
| CA-WNB-P1 | 0.8693 | 0.8564 | 0.8630 | 0.8841 | 0.8652 | 0.8732 | |
| CA-WNB-P2 | 0.8714 | 0.8592 | 0.8654 | 0.8848 | 0.8615 | 0.8744 | |
| yeast | NBMLC | 0.7749 | 0.7636 | 0.7688 | 0.7739 | 0.7688 | 0.7673 |
| CA-WNB-P1 | 0.8045 | 0.7787 | 0.7901 | 0.8129 | 0.7873 | 0.7948 | |
| CA-WNB-P2 | 0.8051 | 0.7851 | 0.7933 | 0.8126 | 0.7831 | 0.7952 | |
| Data Set | Algorithm | Gaussian | Cauchy | ||||
|---|---|---|---|---|---|---|---|
| MAX | MIN | AVE | MAX | MIN | AVE | ||
| CAL_500 | NBMLC | 0.8732 | 0.8574 | 0.8622 | 0.8721 | 0.8554 | 0.8635 |
| CA-WNB-P1 | 0.8885 | 0.8697 | 0.8821 | 0.8884 | 0.8727 | 0.8801 | |
| CA-WNB-P2 | 0.8897 | 0.8716 | 0.8829 | 0.8890 | 0.8744 | 0.8811 | |
| emotions | NBMLC | 0.6976 | 0.6798 | 0.6884 | 0.6976 | 0.6787 | 0.6892 |
| CA-WNB-P1 | 0.8012 | 0.7799 | 0.7939 | 0.8224 | 0.7822 | 0.8039 | |
| CA-WNB-P2 | 0.8143 | 0.7900 | 0.8011 | 0.8271 | 0.7869 | 0.8070 | |
| scene | NBMLC | 0.8239 | 0.8195 | 0.8212 | 0.8195 | 0.8098 | 0.8151 |
| CA-WNB-P1 | 0.8698 | 0.8592 | 0.8647 | 0.8836 | 0.8638 | 0.8725 | |
| CA-WNB-P2 | 0.8714 | 0.8592 | 0.8656 | 0.8848 | 0.8626 | 0.8746 | |
| yeast | NBMLC | 0.7749 | 0.7636 | 0.7688 | 0.7739 | 0.7688 | 0.7673 |
| CA-WNB-P1 | 0.8040 | 0.7898 | 0.7953 | 0.8115 | 0.7875 | 0.7943 | |
| CA-WNB-P2 | 0.8051 | 0.7851 | 0.7938 | 0.8126 | 0.7831 | 0.7941 | |
| Data Set | Algorithm | Gaussian | Cauchy | ||||
|---|---|---|---|---|---|---|---|
| MAX | MIN | AVE | MAX | MIN | AVE | ||
| CAL_500 | NBMLC | 0.8732 | 0.8574 | 0.8622 | 0.8721 | 0.8554 | 0.8635 |
| CA-WNB-P1 | 0.8884 | 0.8690 | 0.8823 | 0.8884 | 0.8740 | 0.8801 | |
| CA-WNB-P2 | 0.8903 | 0.8717 | 0.8832 | 0.8890 | 0.8744 | 0.8812 | |
| emotions | NBMLC | 0.6976 | 0.6798 | 0.6884 | 0.6976 | 0.6787 | 0.6892 |
| CA-WNB-P1 | 0.8021 | 0.7843 | 0.7939 | 0.8231 | 0.7812 | 0.8071 | |
| CA-WNB-P2 | 0.8143 | 0.7900 | 0.8013 | 0.8271 | 0.7869 | 0.8088 | |
| scene | NBMLC | 0.8239 | 0.8195 | 0.8212 | 0.8195 | 0.8098 | 0.8151 |
| CA-WNB-P1 | 0.8714 | 0.8573 | 0.8653 | 0.8839 | 0.8620 | 0.8724 | |
| CA-WNB-P2 | 0.8714 | 0.8592 | 0.8658 | 0.8848 | 0.8626 | 0.8750 | |
| yeast | NBMLC | 0.7749 | 0.7636 | 0.7688 | 0.7739 | 0.7688 | 0.7673 |
| CA-WNB-P1 | 0.8060 | 0.7868 | 0.7955 | 0.8109 | 0.7824 | 0.7927 | |
| CA-WNB-P2 | 0.8091 | 0.7913 | 0.7972 | 0.8139 | 0.7831 | 0.7943 | |
| Data Set | Algorithm | Gaussian | Cauchy | ||||
|---|---|---|---|---|---|---|---|
| MAX | MIN | AVE | MAX | MIN | AVE | ||
| CAL_500 | NBMLC | 0.8732 | 0.8574 | 0.8622 | 0.8721 | 0.8554 | 0.8635 |
| CA-WNB-P1 | 0.8888 | 0.8695 | 0.8822 | 0.8890 | 0.8722 | 0.8793 | |
| CA-WNB-P2 | 0.8893 | 0.8716 | 0.8829 | 0.8890 | 0.8722 | 0.8800 | |
| emotions | NBMLC | 0.6976 | 0.6798 | 0.6884 | 0.6976 | 0.6787 | 0.6892 |
| CA-WNB-P1 | 0.8031 | 0.7797 | 0.7937 | 0.8187 | 0.7812 | 0.7984 | |
| CA-WNB-P2 | 0.8059 | 0.7853 | 0.7938 | 0.8215 | 0.7812 | 0.8040 | |
| scene | NBMLC | 0.8239 | 0.8195 | 0.8212 | 0.8195 | 0.8098 | 0.8151 |
| CA-WNB-P1 | 0.8702 | 0.8580 | 0.8654 | 0.8788 | 0.8594 | 0.8701 | |
| CA-WNB-P2 | 0.8693 | 0.8564 | 0.8660 | 0.8848 | 0.8626 | 0.8735 | |
| yeast | NBMLC | 0.7749 | 0.7636 | 0.7688 | 0.7739 | 0.7688 | 0.7673 |
| CA-WNB-P1 | 0.8104 | 0.7870 | 0.7951 | 0.8087 | 0.7814 | 0.7904 | |
| CA-WNB-P2 | 0.8045 | 0.7850 | 0.7921 | 0.8126 | 0.7792 | 0.7932 | |
| Data Set | Algorithm | Average Classification Accuracy | Improved Percentage | |||
|---|---|---|---|---|---|---|
| NBMLC | CA-WNB-P1 | CA-WNB-P2 | CA-WNB-P1 | CA-WNB-P2 | ||
| CAL500 | best | 0.8622 | 0.8813 | 0.8825 | 2.22% | 2.35% |
| Top 10 | 0.8821 | 0.8829 | 2.31% | 2.40% | ||
| Top 20 | 0.8823 | 0.8832 | 2.33% | 2.43% | ||
| Top 30 | 0.8822 | 0.8829 | 2.32% | 2.41% | ||
| emotions | best | 0.6884 | 0.7938 | 0.7993 | 15.32% | 16.11% |
| Top 10 | 0.7939 | 0.8011 | 15.33% | 16.38% | ||
| Top 20 | 0.7939 | 0.8013 | 15.33% | 16.41% | ||
| Top 30 | 0.7937 | 0.7938 | 15.31% | 15.32% | ||
| scene | best | 0.8212 | 0.8647 | 0.8656 | 5.30% | 5.41% |
| Top 10 | 0.8653 | 0.8658 | 5.37% | 5.43% | ||
| Top 20 | 0.8654 | 0.8660 | 5.38% | 5.46% | ||
| Top 30 | 0.8630 | 0.8654 | 5.09% | 5.39% | ||
| yeast | best | 0.7688 | 0.7901 | 0.7933 | 2.77% | 3.19% |
| Top 10 | 0.7953 | 0.7938 | 3.44% | 3.24% | ||
| Top 20 | 0.7955 | 0.7972 | 3.47% | 3.69% | ||
| Top 30 | 0.7951 | 0.7921 | 3.42% | 3.03% | ||
| Mean | 0.7851 | 0.8336 | 0.8354 | 6.54% | 6.79% | |
| Data Set | Algorithm | Average Classification Accuracy | Improved Percentage | |||
|---|---|---|---|---|---|---|
| NBMLC | CA-WNB-P1 | CA-WNB-P2 | CA-WNB-P1 | CA-WNB-P2 | ||
| CAL500 | best | 0.8635 | 0.8800 | 0.8811 | 1.91% | 2.04% |
| Top 10 | 0.8801 | 0.8811 | 1.92% | 2.04% | ||
| Top 20 | 0.8801 | 0.8812 | 1.92% | 2.04% | ||
| Top 30 | 0.8793 | 0.8800 | 1.83% | 1.91% | ||
| emotions | best | 0.6892 | 0.8040 | 0.8044 | 16.65% | 16.71% |
| Top 10 | 0.8039 | 0.8070 | 16.63% | 17.08% | ||
| Top 20 | 0.8071 | 0.8088 | 17.09% | 17.34% | ||
| Top 30 | 0.7984 | 0.8040 | 15.84% | 16.65% | ||
| scene | best | 0.8151 | 0.8732 | 0.8744 | 7.13% | 7.28% |
| Top 10 | 0.8725 | 0.8746 | 7.04% | 7.30% | ||
| Top 20 | 0.8724 | 0.8750 | 7.04% | 7.35% | ||
| Top 30 | 0.8701 | 0.8735 | 6.75% | 7.17% | ||
| yeast | best | 0.7673 | 0.7948 | 0.7952 | 3.59% | 3.64% |
| Top 10 | 0.7943 | 0.7941 | 3.53% | 3.49% | ||
| Top 20 | 0.7927 | 0.7943 | 3.32% | 3.52% | ||
| Top 30 | 0.7904 | 0.7932 | 3.01% | 3.38% | ||
| Mean | 0.7838 | 0.8371 | 0.8389 | 7.20% | 7.43% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Wu, B.; Hu, C.; Yan, X. Evolutionary Multilabel Classification Algorithm Based on Cultural Algorithm. Symmetry 2021, 13, 322. https://doi.org/10.3390/sym13020322
Wu Q, Wu B, Hu C, Yan X. Evolutionary Multilabel Classification Algorithm Based on Cultural Algorithm. Symmetry. 2021; 13(2):322. https://doi.org/10.3390/sym13020322
Chicago/Turabian StyleWu, Qinghua, Bin Wu, Chengyu Hu, and Xuesong Yan. 2021. "Evolutionary Multilabel Classification Algorithm Based on Cultural Algorithm" Symmetry 13, no. 2: 322. https://doi.org/10.3390/sym13020322
APA StyleWu, Q., Wu, B., Hu, C., & Yan, X. (2021). Evolutionary Multilabel Classification Algorithm Based on Cultural Algorithm. Symmetry, 13(2), 322. https://doi.org/10.3390/sym13020322





