Airborne Prokaryotic, Fungal and Eukaryotic Communities of an Urban Environment in the UK
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Assessment of Environmental Factors
2.3. Environmental Scanning Electronic Microscopy
2.4. DNA Extraction, PCR, and Sequencing
2.5. Sequence Analysis
2.6. Quantitative PCR
2.7. Statistical Analysis
3. Results
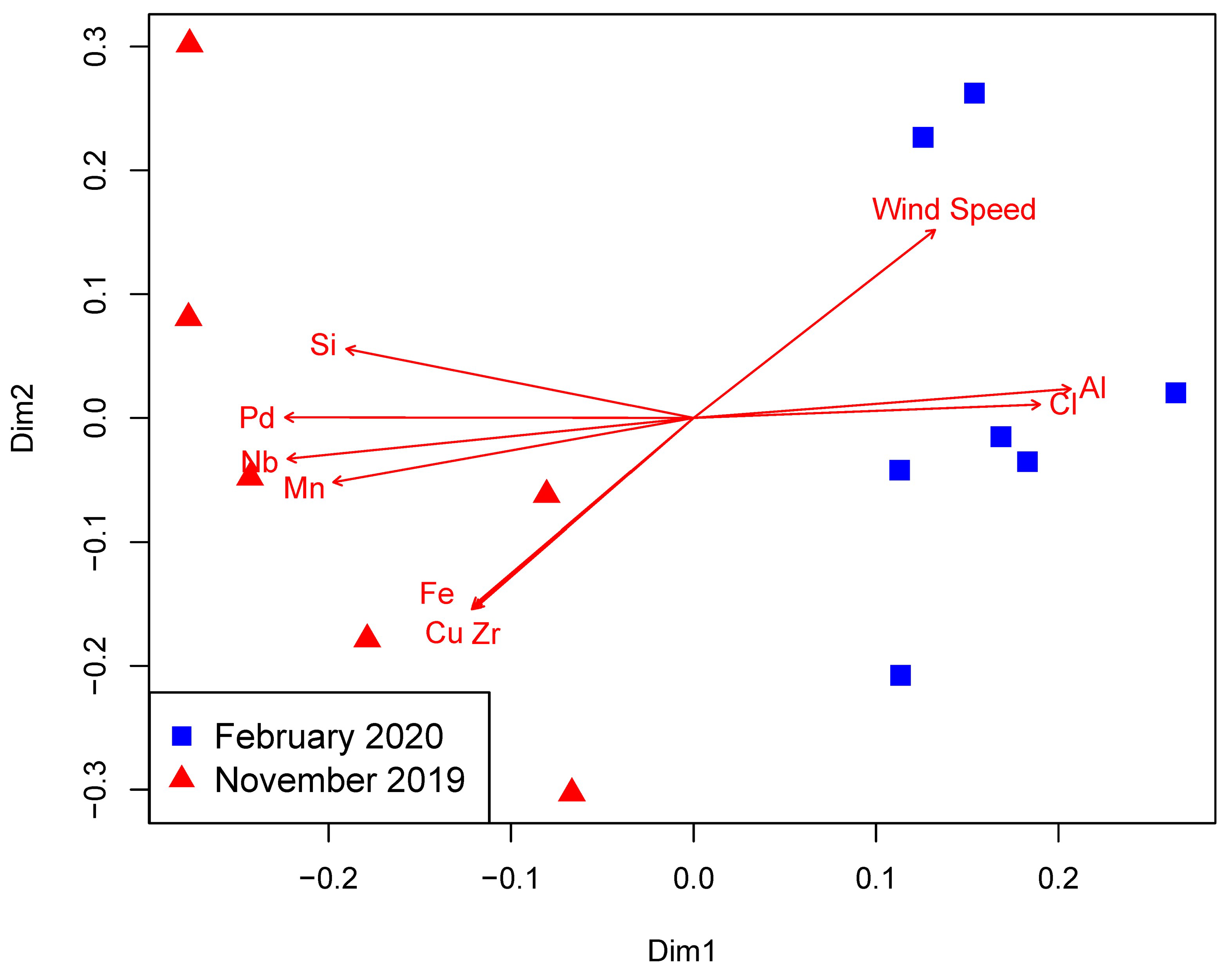
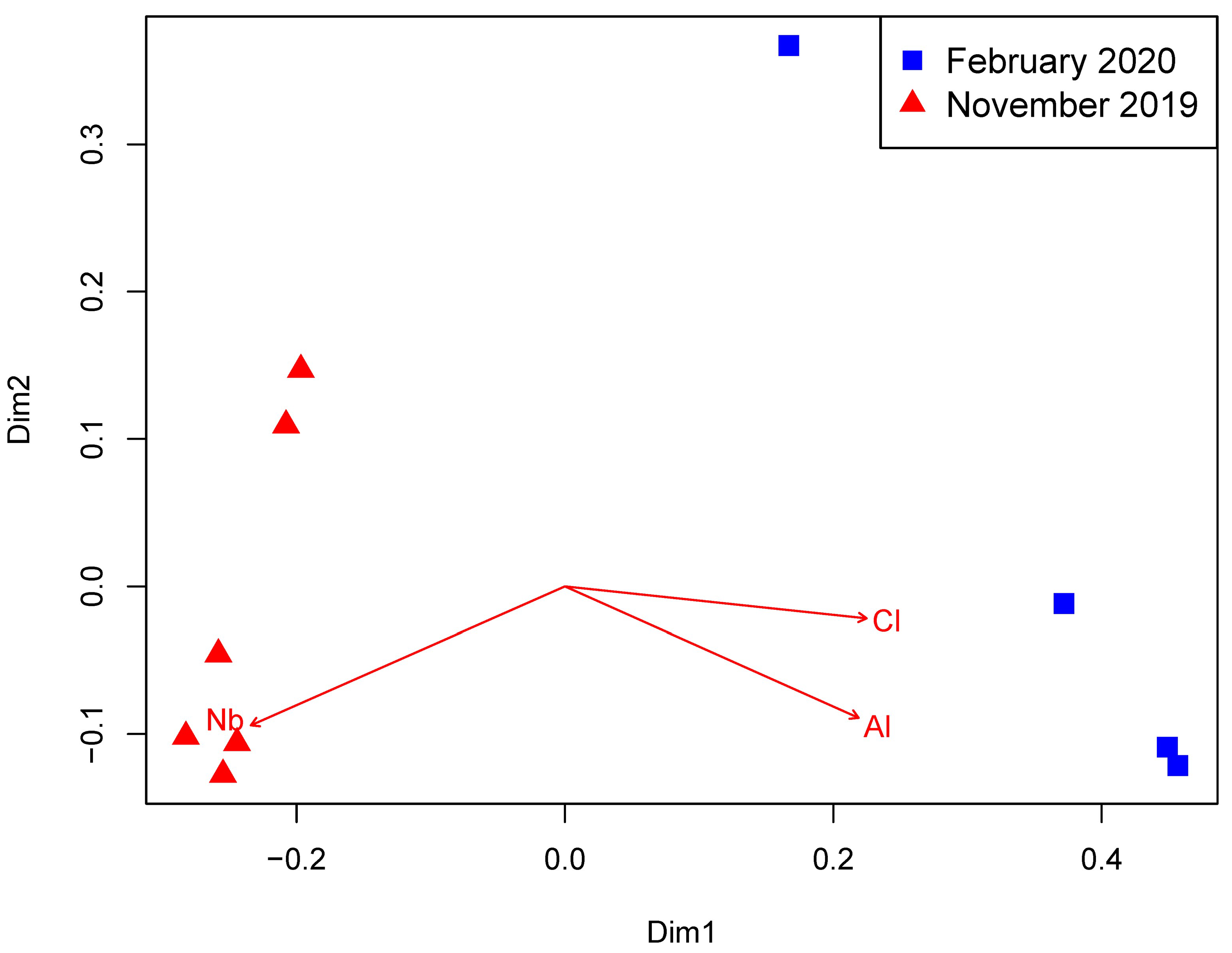
3.1. Environmental Parameters
3.2. Prokaryotic Community Structure and Diversity
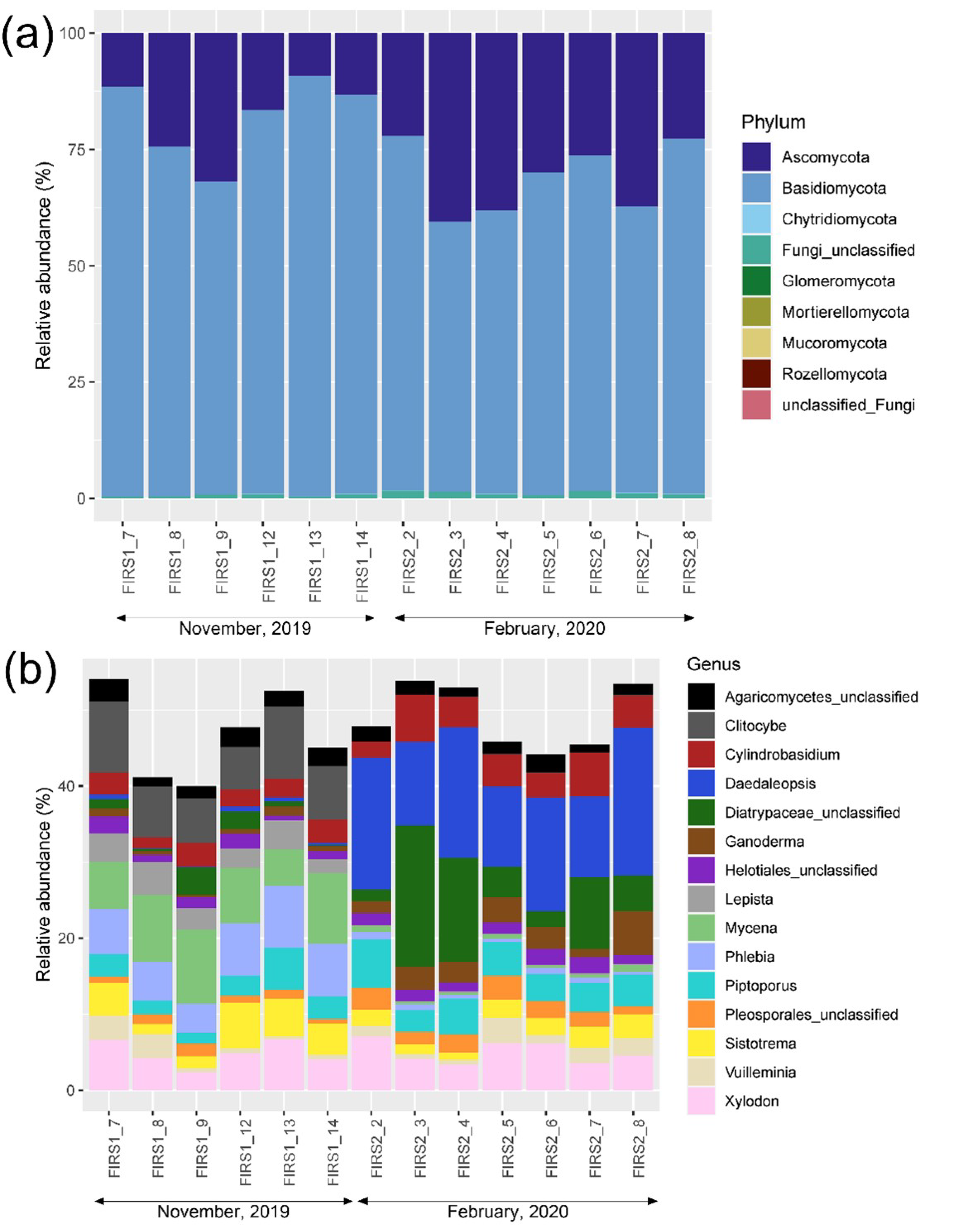
3.3. Fungal Community Structure and Diversity
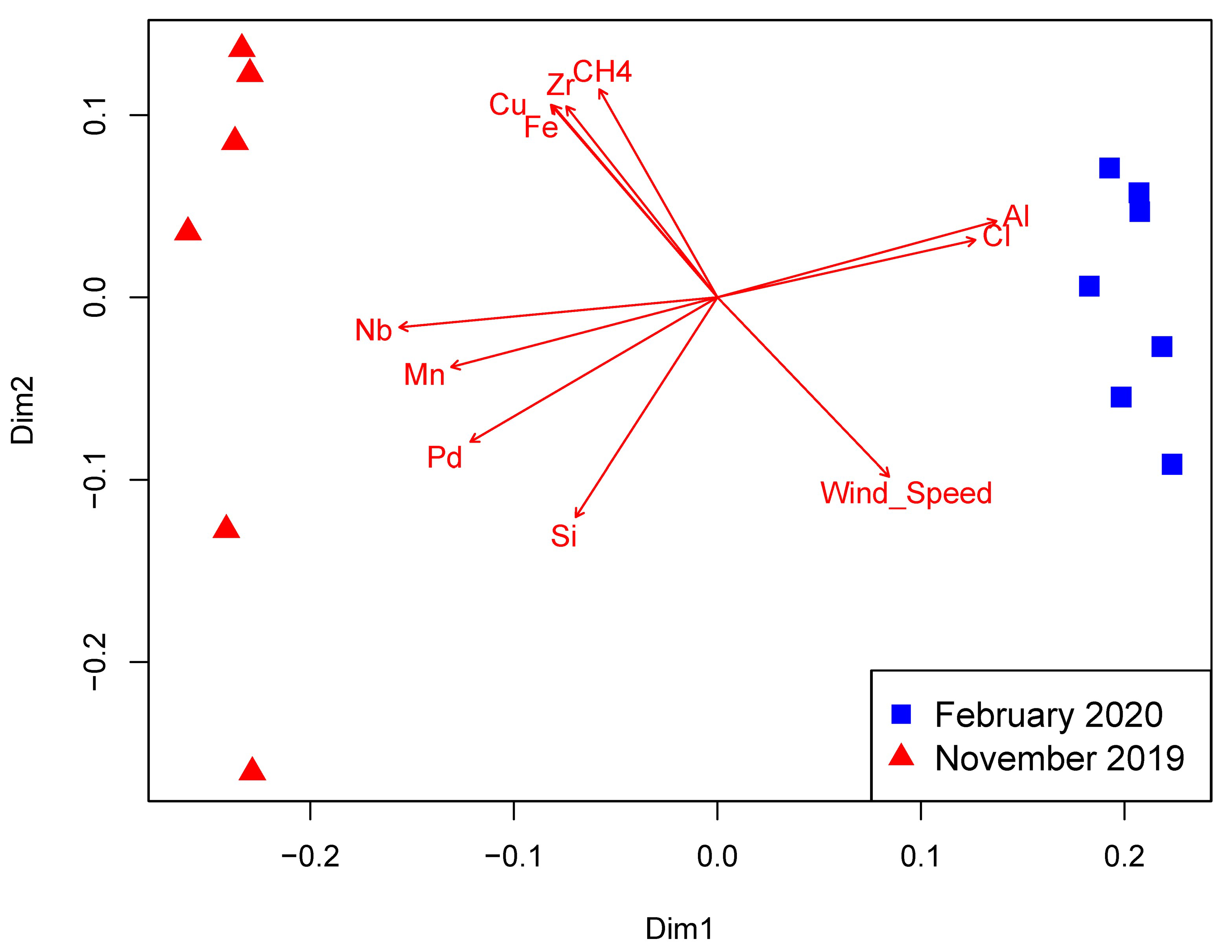
3.4. Eukaryotic Community Structure and Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef] [Green Version]
- Wittmaack, K.; Wehnes, H.; Heinzmann, U.; Agerer, R. An overview on bioaerosols viewed by scanning electron microscopy. Sci. Total Environ. 2005, 346, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Crouzy, B.; Stella, M.; Konzelmann, T.; Calpini, B.; Clot, B. All-Optical automatic pollen identification: Towards an operational system. Atmos. Environ. 2016, 140, 202–212. [Google Scholar] [CrossRef]
- Šaulienė, I.; Šukienė, L.; Daunys, G.; Valiulis, G.; Vaitkevičius, L.; Matavulj, P.; Brdar, S.; Panic, M.; Sikoparija, B.; Clot, B.; et al. Automatic pollen recognition with the Rapid-E particle counter: The first-level procedure, experience and next steps. Atmos. Meas. Tech. 2019, 12, 3435–3452. [Google Scholar] [CrossRef] [Green Version]
- Crawford, I.; Topping, D.; Gallagher, M.; Forde, E.; Lloyd, J.R.; Foot, V.; Stopford, C.; Kaye, P. Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers. Atmosphere 2020, 11, 1039. [Google Scholar] [CrossRef]
- Huffman, J.A.; Perring, A.E.; Savage, N.J.; Clot, B.; Crouzy, B.; Tummon, F.; Shoshanim, O.; Damit, B.; Schneider, J.; Sivaprakasam, V.; et al. Real-Time sensing of bioaerosols: Review and current perspectives. Aerosol Sci. Technol. 2020, 54, 465–495. [Google Scholar] [CrossRef] [Green Version]
- Amoo, A.E.; Babalola, O.O.; Stewart, F.J. Microbial Diversity of Temperate Pine and Native Forest Soils Profiled by 16S rRNA Gene Amplicon Sequencing. Microbiol. Resour. Announc. 2021, 10, e00298-21. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, Y.; Hu, Y.; Wang, Z.; Lu, X. Soil Bacterial Communities and Diversity in Alpine Grasslands on the Tibetan Plateau Based on 16S rRNA Gene Sequencing. Front. Ecol. Evol. 2021, 9, 630722. [Google Scholar] [CrossRef]
- Semenov, M.V. Metabarcoding and Metagenomics in Soil Ecology Research: Achievements, Challenges, and Prospects. Biol. Bull. Rev. 2021, 11, 40–53. [Google Scholar] [CrossRef]
- Pearman, J.K.; Thomson-Laing, G.; Howarth, J.D.; Vandergoes, M.J.; Thompson, L.; Rees, A.; Wood, S.A. Investigating variability in microbial community composition in replicate environmental DNA samples down lake sediment cores. PLoS ONE 2021, 16, e0250783. [Google Scholar] [CrossRef]
- Ghate, S.D.; Shastry, R.P.; Arun, A.B.; Rekha, P.D. Unraveling the bacterial community composition across aquatic sediments in the Southwestern coast of India by employing high-throughput 16S rRNA gene sequencing. Reg. Stud. Mar. Sci. 2021, 46, 101890. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, J.; Nawaz, M.Z.; Mahboob, S.; Al-Ghanim, K.A.; Khan, I.A.; Lu, Z.; Chen, T. Seasonal succession and spatial distribution of bacterial community structure in a eutrophic freshwater Lake, Lake Taihu. Sci. Total Environ. 2019, 669, 29–40. [Google Scholar] [CrossRef]
- Cavaco, M.A.; St. Louis, V.L.; Engel, K.; St. Pierre, K.A.; Schiff, S.L.; Stibal, M.; Neufeld, J.D. Freshwater microbial community diversity in a rapidly changing High Arctic watershed. FEMS Microbiol. Ecol. 2019, 95, fiz161. [Google Scholar] [CrossRef]
- Malki, K.; Rosario, K.; Sawaya, N.A.; Székely, A.J.; Tisza, M.J.; Breitbart, M.; Moran, M.A. Prokaryotic and Viral Community Composition of Freshwater Springs in Florida, USA. mBio 2020, 11, e00436-20. [Google Scholar] [CrossRef] [Green Version]
- Ul-Hasan, S.; Bowers, R.M.; Figueroa-Montiel, A.; Licea-Navarro, A.F.; Beman, J.M.; Woyke, T.; Nobile, C.J. Community ecology across bacteria, archaea and microbial eukaryotes in the sediment and seawater of coastal Puerto Nuevo, Baja California. PLoS ONE 2019, 14, e0212355. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, Y.; Wang, J.; Luo, T.; Zhang, R.; Sun, J.; Zheng, Q.; Jiao, N. Seasonal dynamics of bacterial communities in the surface seawater around subtropical Xiamen Island, China, as determined by 16S rRNA gene profiling. Mar. Pollut. Bull. 2019, 142, 135–144. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Yuan, T.; Zhang, H.; Feng, Q.; Wu, X.; Zhang, Y.; McCarthy, A.J.; Sekar, R. Changes in Fungal Community Structure in Freshwater Canals across a Gradient of Urbanization. Water 2020, 12, 1917. [Google Scholar] [CrossRef]
- Sommermann, L.; Geistlinger, J.; Wibberg, D.; Deubel, A.; Zwanzig, J.; Babin, D.; Schlüter, A.; Schellenberg, I. Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-Amplicon sequencing. PLoS ONE 2018, 13, e0195345. [Google Scholar] [CrossRef]
- Banerji, A.; Bagley, M.; Elk, M.; Pilgrim, E.; Martinson, J.; Santo Domingo, J. Spatial and temporal dynamics of a freshwater eukaryotic plankton community revealed via 18S rRNA gene metabarcoding. Hydrobiologia 2018, 818, 71–86. [Google Scholar] [CrossRef]
- Müller, C.A.; de Mattos Pereira, L.; Lopes, C.; Cares, J.; dos Anjos Borges, L.G.; Giongo, A.; Graeff-Teixeira, C.; Loureiro Morassutti, A. Meiofaunal diversity in the Atlantic Forest soil: A quest for nematodes in a native reserve using eukaryotic metabarcoding analysis. For. Ecol. Manag. 2019, 453, 117591. [Google Scholar] [CrossRef]
- Tanaka, D.; Sato, K.; Goto, M.; Fujiyoshi, S.; Maruyama, F.; Takato, S.; Shimada, T.; Sakatoku, A.; Aoki, K.; Nakamura, S. Airborne Microbial Communities at High-Altitude and Suburban Sites in Toyama, Japan Suggest a New Perspective for Bioprospecting. Front. Bioeng. Biotechnol. 2019, 7, 12. [Google Scholar] [CrossRef]
- Stewart, J.D.; Shakya, K.M.; Bilinski, T.; Wilson, J.W.; Ravi, S.; Choi, C.S. Variation of near surface atmosphere microbial communities at an urban and a suburban site in Philadelphia, PA, USA. Sci. Total Environ. 2020, 724, 138353. [Google Scholar] [CrossRef]
- Smith, D.J.; Ravichandar, J.D.; Jain, S.; Griffin, D.W.; Yu, H.; Tan, Q.; Thissen, J.; Lusby, T.; Nicoll, P.; Shedler, S.; et al. Airborne Bacteria in Earth’s Lower Stratosphere Resemble Taxa Detected in the Troposphere: Results From a New NASA Aircraft Bioaerosol Collector (ABC). Front. Microbiol. 2018, 9, 1752. [Google Scholar] [CrossRef]
- Kraaijeveld, K.; de Weger, L.A.; Ventayol García, M.; Buermans, H.; Frank, J.; Hiemstra, P.S.; den Dunnen, J.T. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 2015, 15, 8–16. [Google Scholar] [CrossRef]
- Banchi, E.; Ametrano, C.G.; Stanković, D.; Verardo, P.; Moretti, O.; Gabrielli, F.; Lazzarin, S.; Borney, M.F.; Tassan, F.; Tretiach, M.; et al. DNA metabarcoding uncovers fungal diversity of mixed airborne samples in Italy. PLoS ONE 2018, 13, e0194489. [Google Scholar] [CrossRef]
- Xie, W.; Li, Y.; Bai, W.; Hou, J.; Ma, T.; Zeng, X.; Zhang, L.; An, T. The source and transport of bioaerosols in the air: A review. Front. Environ. Sci. Eng. 2020, 15, 44. [Google Scholar] [CrossRef]
- Smith, D.J.; Timonen, H.J.; Jaffe, D.A.; Griffin, D.W.; Birmele, M.N.; Perry, K.D.; Ward, P.D.; Roberts, M.S. Intercontinental Dispersal of Bacteria and Archaea by Transpacific Winds. Appl. Environ. Microbiol. 2013, 79, 1134–1139. [Google Scholar] [CrossRef] [Green Version]
- Mu, F.; Li, Y.; Lu, R.; Qi, Y.; Xie, W.; Bai, W. Source identification of airborne bacteria in the mountainous area and the urban areas. Atmos. Res. 2020, 231, 104676. [Google Scholar] [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [Green Version]
- Taleghani, M. Air Pollution within Different Urban Forms in Manchester, UK. Climate 2022, 10, 26. [Google Scholar] [CrossRef]
- Gollakota, A.R.K.; Gautam, S.; Santosh, M.; Sudan, H.A.; Gandhi, R.; Sam Jebadurai, V.; Shu, C.-M. Bioaerosols: Characterization, pathways, sampling strategies, and challenges to geo-environment and health. Gondwana Res. 2021, 99, 178–203. [Google Scholar] [CrossRef]
- Ravindra, K.; Wauters, E.; Van Grieken, R. Variation in particulate PAHs levels and their relation with the transboundary movement of the air masses. Sci. Total Environ. 2008, 396, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Wu, Z.; Poulain, L.; van Pinxteren, M.; Merkel, M.; Assmann, D.; Herrmann, H.; Wiedensohler, A. Source apportionment of the organic aerosol over the Atlantic Ocean from 53° N to 53° S: Significant contributions from marine emissions and long-range transport. Atmos. Chem. Phys. 2018, 18, 18043–18062. [Google Scholar] [CrossRef] [Green Version]
- Barker, P.A.; Allen, G.; Flynn, M.; Riddick, S.; Pitt, J.R. Measurement of Recreational N2o Emissions from an Urban Environment in Manchester, UK. 2022. Available online: https://ssrn.com/abstract=4071514 (accessed on 2 July 2022).
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-End assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Westcott, S.L.; Schloss, P.D.; McMahon, K.; Watson, M.; Pollard, K. OptiClust, an Improved Method for Assigning Amplicon-Based Sequence Data to Operational Taxonomic Units. Msphere 2017, 2, e00073-17. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 47, D23–D28. [Google Scholar] [CrossRef] [Green Version]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Clarke, K.; Gorley, R. PRIMER: Getting started with v6.; PRIMER-E Ltd.: Plymouth, UK, 2005; Volume 931, p. 932. [Google Scholar]
- Stein, A.F.; Draxler, R.R.; Rolph, G.D.; Stunder, B.J.B.; Cohen, M.D.; Ngan, F. NOAA’s HYSPLIT Atmospheric Transport and Dispersion Modeling System. Bull. Am. Meteorol. Soc. 2015, 96, 2059–2077. [Google Scholar] [CrossRef]
- Lai, C.C.; Cheng, A.; Liu, W.L.; Tan, C.K.; Huang, Y.T.; Chung, K.P.; Lee, M.R.; Hsueh, P.R. Infections caused by unusual Methylobacterium species. J. Clin. Microbiol. 2011, 49, 3329–3331. [Google Scholar] [CrossRef] [Green Version]
- Szwetkowski, K.J.; Falkinham, J.O. Methylobacterium spp. as Emerging Opportunistic Premise Plumbing Pathogens. Pathogens 2020, 9, 149. [Google Scholar] [CrossRef] [Green Version]
- Krzyściak, W.; Pluskwa, K.K.; Jurczak, A.; Kościelniak, D. The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1361–1376. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.A.; Das, A.; Ton-That, H. Adhesion by pathogenic corynebacteria. Adv. Exp. Med. Biol. 2011, 715, 91–103. [Google Scholar] [CrossRef]
- Idris, R.; Kuffner, M.; Bodrossy, L.; Puschenreiter, M.; Monchy, S.; Wenzel, W.W.; Sessitsch, A. Characterization of Ni-tolerant methylobacteria associated with the hyperaccumulating plant Thlaspi goesingense and description of Methylobacterium goesingense sp. nov. Syst. Appl. Microbiol. 2006, 29, 634–644. [Google Scholar] [CrossRef]
- Photolo, M.M.; Sitole, L.; Mavumengwana, V.; Tlou, M.G. Genomic and Physiological Investigation of Heavy Metal Resistance from Plant Endophytic Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum. Int. J. Environ. Res. Public Health 2021, 18, 997. [Google Scholar] [CrossRef]
- Rinke, C.; Rubino, F.; Messer, L.F.; Youssef, N.; Parks, D.H.; Chuvochina, M.; Brown, M.; Jeffries, T.; Tyson, G.W.; Seymour, J.R.; et al. A phylogenomic and ecological analysis of the globally abundant Marine Group II archaea (Ca. Poseidoniales ord. nov.). ISME J. 2019, 13, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Dupont, C.L.; Rusch, D.B.; Yooseph, S.; Lombardo, M.-J.; Richter, R.A.; Valas, R.; Novotny, M.; Yee-Greenbaum, J.; Selengut, J.D.; Haft, D.H.; et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012, 6, 1186–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma Ghimire, P.; Joshi, D.R.; Tripathee, L.; Chen, P.; Sajjad, W.; Kang, S. Seasonal taxonomic composition of microbial communal shaping the bioaerosols milieu of the urban city of Lanzhou. Arch. Microbiol. 2022, 204, 222. [Google Scholar] [CrossRef] [PubMed]
- Núñez, A.; Amo de Paz, G.; Rastrojo, A.; Ferencova, Z.; Gutiérrez-Bustillo, A.M.; Alcamí, A.; Moreno, D.A.; Guantes, R. Temporal patterns of variability for prokaryotic and eukaryotic diversity in the urban air of Madrid (Spain). Atmos. Environ. 2019, 217, 116972. [Google Scholar] [CrossRef]
- Li, H.; Zhou, X.-Y.; Yang, X.-R.; Zhu, Y.-G.; Hong, Y.-W.; Su, J.-Q. Spatial and seasonal variation of the airborne microbiome in a rapidly developing city of China. Sci. Total Environ. 2019, 665, 61–68. [Google Scholar] [CrossRef]
- Tanaka, D.; Fujiyoshi, S.; Maruyama, F.; Goto, M.; Koyama, S.; Kanatani, J.-i.; Isobe, J.; Watahiki, M.; Sakatoku, A.; Kagaya, S.; et al. Size resolved characteristics of urban and suburban bacterial bioaerosols in Japan as assessed by 16S rRNA amplicon sequencing. Sci. Rep. 2020, 10, 12406. [Google Scholar] [CrossRef]
- Woo, C.; An, C.; Xu, S.; Yi, S.-M.; Yamamoto, N. Taxonomic diversity of fungi deposited from the atmosphere. ISME J. 2018, 12, 2051–2060. [Google Scholar] [CrossRef] [Green Version]
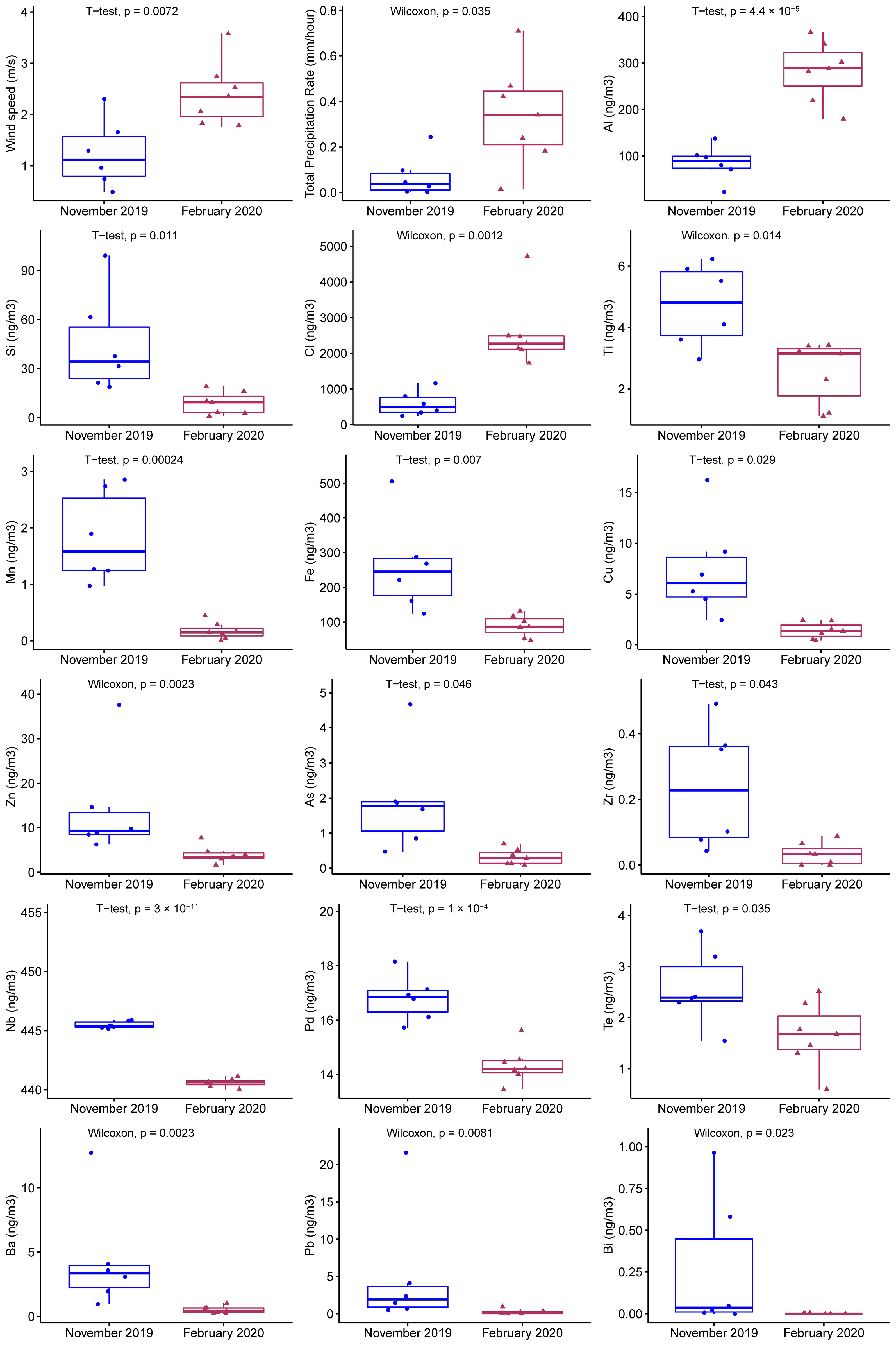
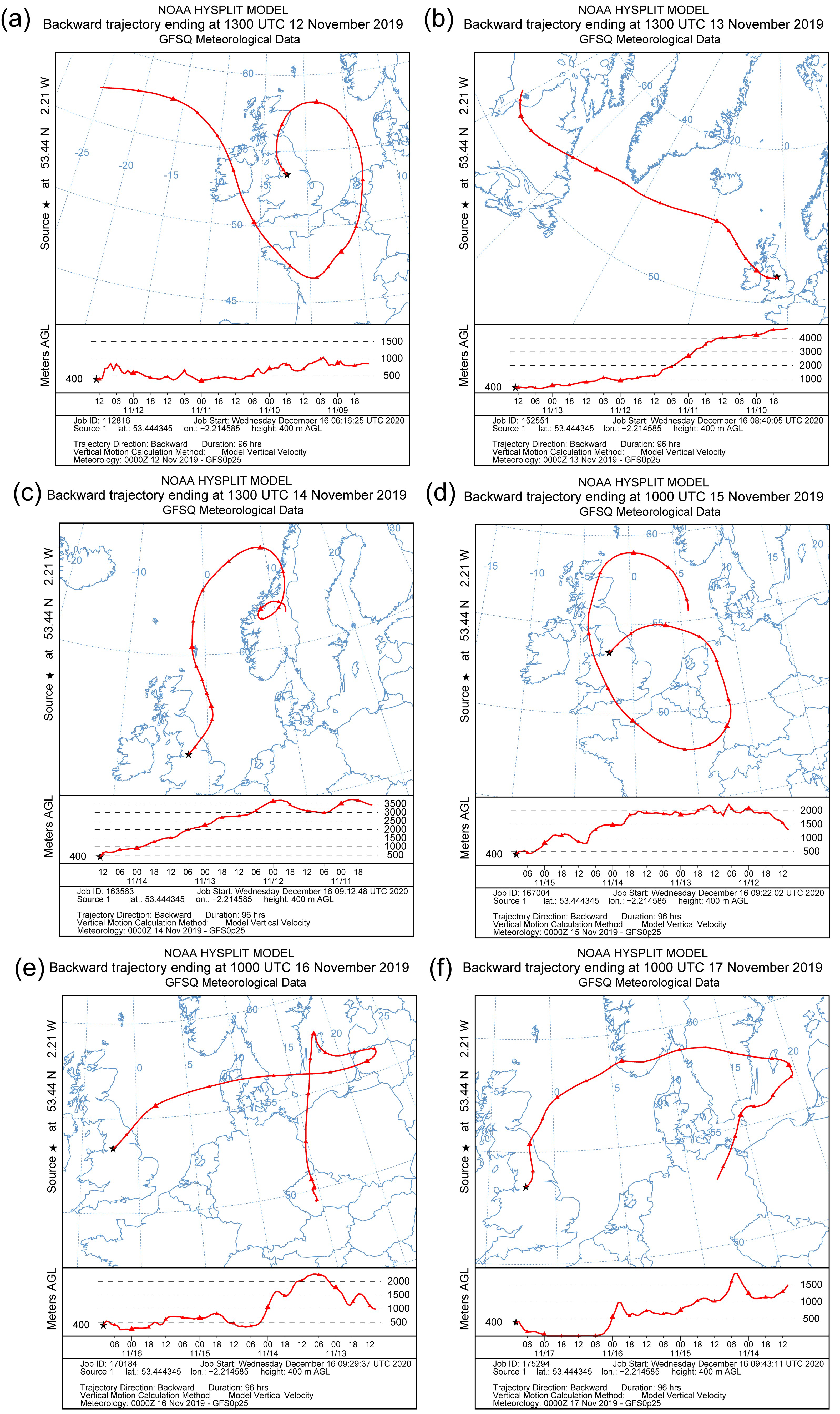
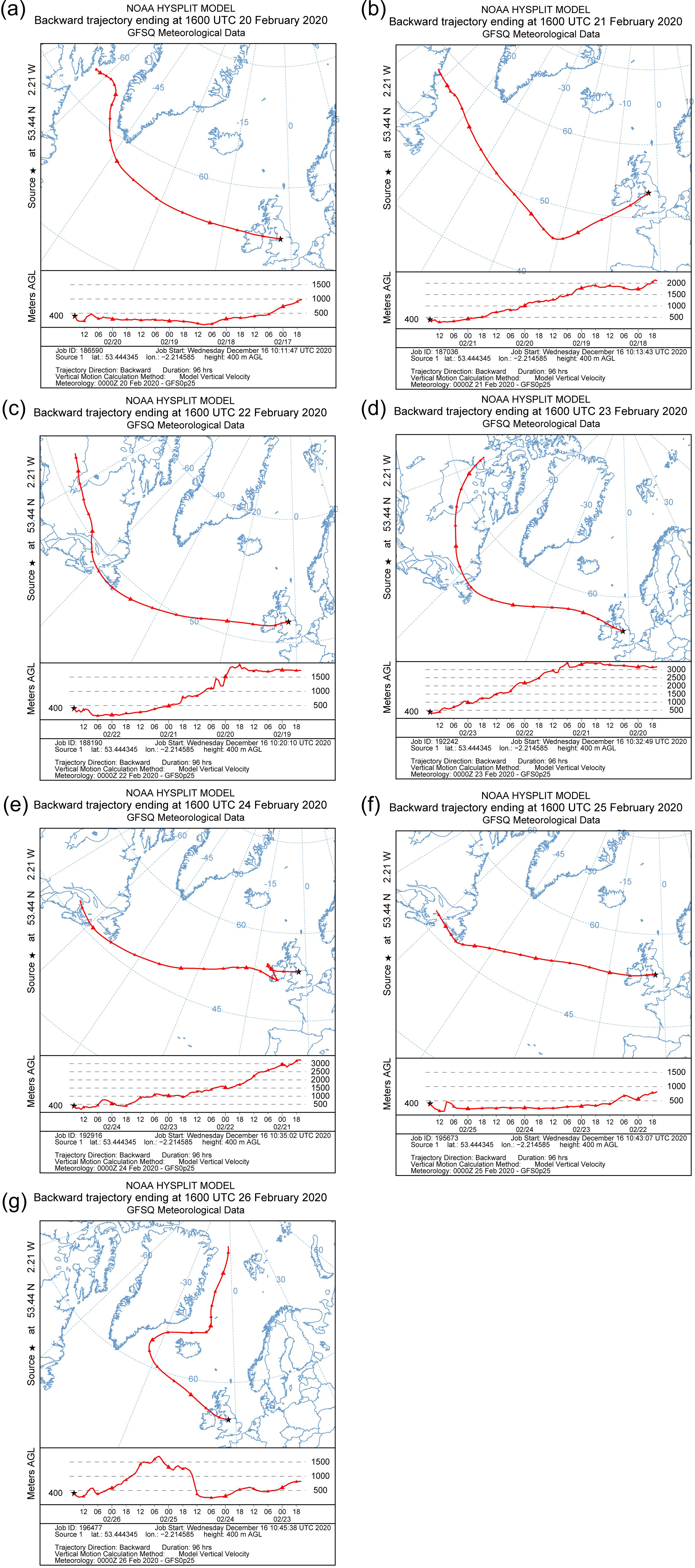
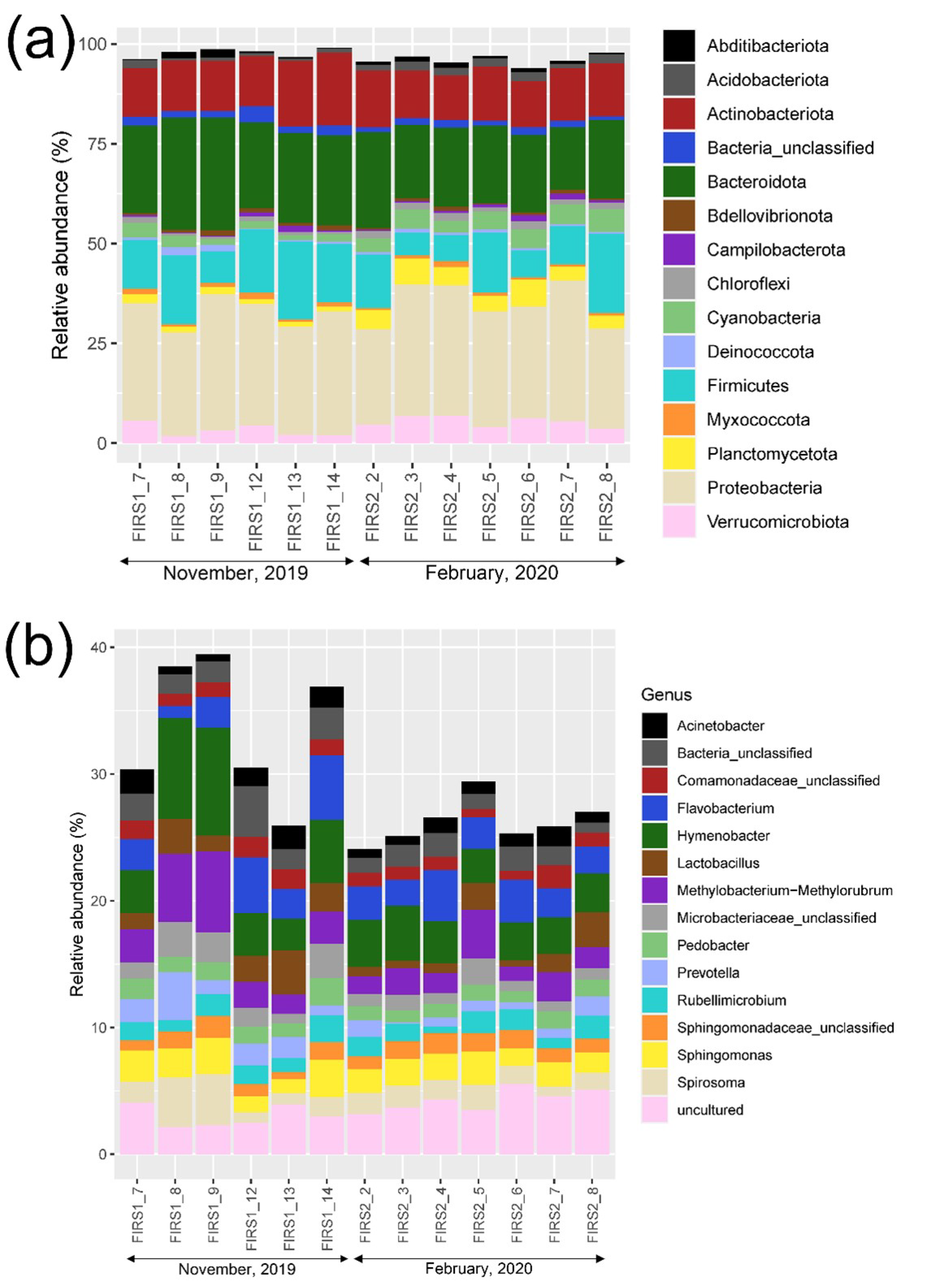

| Sample ID | Start Time | End Time |
|---|---|---|
| FIRS1_7 | 12 November 2019 13:16 | 13 November 2019 13:15 |
| FIRS1_8 | 13 November 2019 13:16 | 14 November 2019 13:15 |
| FIRS1_9 | 14 November 2019 13:16 | 15 November 2019 10:40 |
| FIRS1_12 | 15 November 2019 10:40 | 16 November 2019 10:40 |
| FIRS1_13 | 16 November 2019 10:40 | 17 November 2019 10:40 |
| FIRS1_14 | 17 November 2019 10:40 | 18 November 2019 10:20 |
| FIRS2_2 | 20 February 2020 16:25 | 21 February 2020 16:25 |
| FIRS2_3 | 21 February 2020 16:25 | 22 February 2020 16:25 |
| FIRS2_4 | 22 February 2020 16:25 | 23 February 2020 16:25 |
| FIRS2_5 | 23 February 2020 16:25 | 24 February 2020 16:25 |
| FIRS2_6 | 24 February 2020 16:25 | 25 February 2020 16:25 |
| FIRS2_7 | 25 February 2020 16:25 | 26 February 2020 16:25 |
| FIRS2_8 | 26 February 2020 16:25 | 27 February 2020 16:25 |
| OTU ID | p Value | Nov_2019 Average (%) | Feb_2020 Average (%) | Taxonomy Based on the Silva Database | BLAST against NCBI nt Database | ||
|---|---|---|---|---|---|---|---|
| Taxonomy | Similarity (%) | E-Value | |||||
| Otu000020 | 0.045 | 1.136 | 0.187 | Methylobacterium-Methylorubrum | Methylobacterium bullatum, Methylobacterium marchantiae | 100 | 6 × 10−120 |
| Otu000014 | 0.017 | 0.762 | 0.470 | Rubellimicrobium | Rubellimicrobium aerolatum | 100 | 9 × 10−112 |
| Otu000019 | 0.032 | 0.648 | 0.433 | Pedobacter | Pedobacter miscanthi, Pedobacter helvus, and etc. | 100 | 2 × 10−125 |
| Otu000048 | 0.016 | 0.654 | 0.046 | Bacteria_unclassified | Calycina alstrupii | 88.29 | 9 × 10−73 |
| Otu000047 | 0.012 | 0.406 | 0.201 | Streptococcus | Streptococcus gallolyticus, Streptococcus pasteurianus, and etc. | 100 | 8 × 10−125 |
| Otu000054 | 0.018 | 0.604 | 0.021 | Moraxellaceae_ge | Agitococcus lubricus | 97.62 | 3 × 10−117 |
| Otu000058 | 0.020 | 0.367 | 0.161 | Lactobacillus | Lactobacillus johnsonii, Lactobacillus paragasseri, and etc. | 100 | 2 × 10−125 |
| Otu000053 | 0.037 | 0.097 | 0.345 | Rickettsiella | Diplorickettsia massiliensis 20B | 98.02 | 3 × 10−118 |
| Otu000099 | 0.036 | 0.331 | 0.076 | Prevotella | Prevotella hominis | 99.6 | 1 × 10−123 |
| Otu000089 | 0.018 | 0.371 | 0.039 | Spirosoma | Spirosoma oryzae | 96.83 | 2 × 10−114 |
| Otu000102 | 0.003 | 0.283 | 0.103 | Aureimonas | Aureimonas glaciei | 100 | 2 × 10−125 |
| Otu000122 | 0.036 | 0.296 | 0.062 | Pseudomonas | Paucimonas lemoignei, Pseudomonas versuta, and etc. | 100 | 2 × 10−125 |
| Otu000090 | 0.005 | 0.051 | 0.270 | uncultured | Roseimicrobium gellanilyticum | 87.3 | 6 × 10−82 |
| Otu000120 | 0.023 | 0.315 | 0.041 | Staphylococcaceae_unclassified | Mammaliicoccus fleurettii, Mammaliicoccus sciuri, and etc. | 100 | 2 × 10−125 |
| Otu000108 | 0.030 | 0.212 | 0.115 | Dyadobacter | Dyadobacter frigoris, Dyadobacter hamtensis | 99.6 | 1 × 10−123 |
| Otu000129 | 0.041 | 0.238 | 0.090 | Chryseobacterium | Chryseobacterium solani, Epilithonimonas ginsengisoli, and etc. | 100 | 2 × 10−125 |
| Otu000111 | 0.038 | 0.206 | 0.115 | Corynebacterium | Corynebacterium freneyi, Corynebacterium xerosis | 100 | 7 × 10−126 |
| Otu000091 | 0.037 | 0.063 | 0.215 | Pseudarcobacter | Arcobacter suis, Arcobacter caeni | 100 | 2 × 10−125 |
| Otu000087 | 0.002 | 0.000 | 0.251 | Marine_Group_II_ge | Methanobrevibacter cuticularis | 79.45 | 3 × 10−54 |
| Otu000148 | 0.013 | 0.174 | 0.080 | Spirosoma | Spirosoma pomorum | 96.83 | 2 × 10−114 |
| Otu000115 | 0.011 | 0.023 | 0.208 | SAR86_clade_ge | Pseudomonas nabeulensis | 89.33 | 8 × 10−87 |
| Otu000143 | 0.003 | 0.014 | 0.194 | Corynebacteriales_unclassified | Rhodococcus aerolatus | 100 | 2 × 10−125 |
| Otu000202 | 0.010 | 0.201 | 0.032 | Comamonadaceae_unclassified | Xylophilus rhododendri, Ramlibacter rhizophilus, and etc. | 100 | 2 × 10−125 |
| Otu000141 | 0.001 | 0.033 | 0.173 | Sphingomonas | Sphingomonas flava | 99.6 | 1 × 10−123 |
| Otu000147 | 0.033 | 0.037 | 0.155 | Marinimicrobia__ge | Acinetobacter piscicola, Acinetobacter marinus | 80.57 | 2 × 10−57 |
| Otu000162 | 0.043 | 0.024 | 0.164 | Scytonema_UTEX_2349 | Hassallia antarctica | 99.6 | 1 × 10−123 |
| Otu000246 | 0.025 | 0.169 | 0.035 | 1174-901-12 | Lichenihabitans psoromatis, Beijerinckia mobilis | 95.63 | 4 × 10−110 |
| Otu000183 | 0.004 | 0.003 | 0.164 | Crocinitomicaceae_unclassified | Wandonia haliotis | 95.63 | 4 × 10−110 |
| Otu000230 | 0.016 | 0.005 | 0.155 | Cyanobacteriia_unclassified | Lobosphaera incisa | 87.7 | 5 × 10−83 |
| Otu000206 | 0.035 | 0.023 | 0.136 | Calothrix_PCC-6303 | Macrochaete lichenoides | 99.21 | 1 × 10−122 |
| OTU ID | p Value | Nov_2019 Average (%) | Feb_2020 Average (%) | Taxonomy Based on the UNITE Database | BLAST against NCBI nt Database | ||
|---|---|---|---|---|---|---|---|
| Taxonomy | Similarity (%) | E-Value | |||||
| Otu000003 | 0.000 | 0.405 | 14.467 | Daedaleopsis_unclassified | Daedaleopsis confragosa, Lenzites betulinus, and etc. | 100 | 0 |
| Otu000004 | 0.000 | 6.017 | 0.522 | Phlebia_unclassified | Phlebia radiata | 100 | 0 |
| Otu000006 | 0.002 | 6.225 | 0.001 | Clitocybe_nebularis | Leucopaxillus tricolor, Lepista nebularis | 100 | 0 |
| Otu000009 | 0.018 | 2.468 | 4.198 | Cylindrobasidium_evolvens | Polyporus gayanus | 99.567 | 0 |
| Otu000010 | 0.003 | 4.192 | 0.006 | Mycena_metata | Mycena arcangeliana | 98.966 | 0 |
| Otu000013 | 0.029 | 2.621 | 0.600 | Sistotrema_oblongisporum | Clavulina cristata | 83.252 | 2.91 × 10−95 |
| Otu000016 | 0.001 | 2.451 | 0.036 | Lepista_nuda | Lepista nuda | 99.208 | 0 |
| Otu000017 | 0.006 | 0.609 | 2.896 | Ganoderma_australe | Ganoderma australe | 99.728 | 0 |
| Otu000022 | 0.001 | 1.635 | 0.000 | Infundibulicybe_geotropa | Ampulloclitocybe clavipes | 94.01 | 8.94 × 10−160 |
| Otu000023 | 0.017 | 1.503 | 0.061 | Peniophora_unclassified | Peniophora piceae | 96.961 | 2.34 × 10−170 |
| Otu000026 | 0.001 | 1.510 | 0.000 | Paralepista_flaccida | Paralepista gilva | 99.73 | 0 |
| Otu000029 | 0.007 | 1.164 | 0.325 | Radulomyces_molaris | Cuphophyllus colemannianus | 90.517 | 1.63 × 10−77 |
| Otu000031 | 0.035 | 0.487 | 0.268 | Coprinellus_micaceus | Coprinellus micaceus, Coprinus rufopruinatus | 100 | 0 |
| Otu000034 | 0.001 | 0.364 | 1.184 | Heterobasidion_unclassified | Podoscypha multizonata, Podoscypha involuta | 100 | 0 |
| Otu000035 | 0.001 | 0.764 | 0.101 | Hypholoma_fasciculare | Hypholoma fasciculare | 100 | 0 |
| Otu000037 | 0.034 | 1.030 | 0.088 | Trechispora_byssinella | Trechispora byssinella | 99.189 | 0 |
| Otu000040 | 0.033 | 0.440 | 1.002 | Antrodia_xantha | Amyloporia xantha, Antrodia xantha | 100 | 0 |
| Otu000042 | 0.001 | 0.200 | 1.328 | Polyporaceae_unclassified | Trametes gibbosa | 100 | 0 |
| Otu000043 | 0.001 | 0.058 | 1.506 | Diatrypaceae_unclassified | Eutypa lata | 100 | 9.68 × 10−169 |
| Otu000044 | 0.041 | 0.514 | 0.776 | Russulales_unclassified | Peniophora incarnata | 100 | 0 |
| Otu000045 | 0.000 | 0.990 | 0.034 | Clitocybe_unclassified | Clitocybe vibecina | 99.733 | 0 |
| Otu000046 | 0.017 | 0.454 | 0.856 | Xenasmatella_unclassified | Phlebiella borealis | 98.864 | 1.74 × 10−176 |
| Otu000048 | 0.006 | 0.069 | 1.288 | Xylariales_unclassified | Eutypa lata | 100 | 2.70 × 10−169 |
| Otu000049 | 0.012 | 0.442 | 0.716 | Hyphodontia_pallidula | Hyphodontia pallidula | 99.446 | 0 |
| Otu000050 | 0.001 | 0.193 | 1.048 | Pleosporales_unclassified | Phaeosphaeria caricicola | 94.375 | 7.65 × 10−135 |
| Otu000051 | 0.001 | 0.886 | 0.000 | Rhodocollybia_butyracea | Rhodocollybia butyracea | 99.542 | 0 |
| Otu000052 | 0.002 | 0.270 | 0.885 | Resinicium_bicolor | Resinicium bicolor | 100 | 0 |
| Otu000054 | 0.003 | 0.202 | 0.665 | Flammulina_velutipes | Flammulina velutipes | 100 | 0 |
| Otu000056 | 0.001 | 0.587 | 0.105 | Pleurotus_ostreatus | Pleurotus sapidus, Pleurotus ostreatus, and etc. | 100 | 0 |
| Otu000058 | 0.027 | 0.606 | 0.042 | Hyaloscyphaceae_unclassified | Lachnum virgineum | 93.631 | 3.52 × 10−128 |
| OTU ID | p Value | Nov_2019 Average (%) | Feb_2020 Average (%) | Taxonomy Based on the Silva Database | BLAST against NCBI nt Database | ||
|---|---|---|---|---|---|---|---|
| Taxonomy | Similarity (%) | E-Value | |||||
| Otu00011 | 0.006 | 0.000 | 56.006 | Embryophyta_unclassified | Taxus wallichiana | 100 | 3.06 × 10−49 |
| Otu00001 | 0.000 | 17.691 | 0.270 | Agaricales_unclassified | Lepista sordida, Lepista saeva, etc. | 100 | 1.09 × 10−48 |
| Otu00002 | 0.014 | 6.563 | 0.323 | Polyporales_unclassified | Fomitopsis pinicola, Antrodia albida, etc. | 100 | 1.09 × 10−48 |
| Otu00003 | 0.013 | 5.611 | 0.749 | Basidiomycota_unclassified | Sistotrema brinkmannii, Sistotrema oblongisporum | 100 | 1.09 × 10−48 |
| Otu00004 | 0.008 | 4.137 | 1.057 | Hyphodontia | Hyphodontia rimosissima | 95.413 | 2.37 × 10−40 |
| Otu00029 | 0.038 | 0.708 | 5.812 | Chlorophyta_ph_unclassified | Trebouxia impressa | 100 | 1.09 × 10−48 |
| Otu00006 | 0.002 | 3.249 | 0.092 | Agaricomycetes_unclassified | Phlebia radiata | 100 | 1.09 × 10−48 |
| Otu00008 | 0.011 | 2.879 | 0.377 | Russulales_ge | Peniophora nuda | 98.165 | 1.82 × 10−46 |
| Otu00012 | 0.000 | 2.193 | 0.038 | Baeospora | Baeospora myosura | 100 | 1.09 × 10−48 |
| Otu00015 | 0.014 | 2.013 | 0.275 | Agaricomycetes_unclassified | Rogersella griseliniae | 95.413 | 2.37 × 10−40 |
| Otu00016 | 0.011 | 2.077 | 0.000 | Magnoliophyta_ge | Parietaria judaica | 100 | 8.61 × 10−50 |
| Otu00017 | 0.023 | 1.714 | 0.367 | Eukaryota_unclassified | Sterigmatomyces halophilu | 90.991 | 5.16 × 10−32 |
| Otu00136 | 0.006 | 0.000 | 2.890 | Eukaryota_unclassified | Taxus wallichiana | 99.09 | 1.00 × 10−45 |
| Otu00024 | 0.013 | 1.218 | 0.005 | Basidiomycota_unclassified | Chamaeota sinica | 93.578 | 5.12 × 10−37 |
| Otu00028 | 0.038 | 0.942 | 0.199 | Eukaryota_unclassified | Hyphodontia crustosa | 92.661 | 2.38 × 10−35 |
| Otu00035 | 0.025 | 0.744 | 0.248 | Agaricomycetes_unclassified | Burgoa anomala, Sistotrema octosporum, etc. | 100 | 1.09 × 10−48 |
| Otu00031 | 0.002 | 0.726 | 0.162 | Agaricomycetes_unclassified | Rogersella griseliniae | 91.818 | 1.11 × 10−33 |
| Otu00032 | 0.013 | 0.802 | 0.005 | Agaricales_unclassified | Mycena galericulata | 98.165 | 2.35 × 10−45 |
| Otu00258 | 0.006 | 0.000 | 1.068 | Embryophyta_unclassified | Taxus wallichiana | 99.09 | 1.00 × 10−45 |
| Otu00039 | 0.000 | 0.665 | 0.022 | Agaricomycetes_unclassified | Mycena galericulata | 95.413 | 2.37 × 10−40 |
| Otu00041 | 0.042 | 0.550 | 0.102 | Hyphodontia | Hyphodontia nespori | 100 | 1.09 × 10−48 |
| Otu00036 | 0.013 | 0.600 | 0.005 | Sporidiobolaceae_unclassified | Sporobolomyces carnicolor, Sporobolomyces patagonicus, etc. | 100 | 1.09 × 10−48 |
| Otu00040 | 0.024 | 0.571 | 0.027 | Eukaryota_unclassified | Tulasnella violea | 97.222 | 3.88 × 10−43 |
| Otu00049 | 0.011 | 0.528 | 0.075 | Trechispora | Trechispora alnicola | 93.578 | 5.12 × 10−37 |
| Otu00043 | 0.011 | 0.536 | 0.000 | Agaricales_unclassified | Chrysomphalina grossula | 96.33 | 5.08 × 10−42 |
| Otu00057 | 0.011 | 0.503 | 0.000 | Pleosporales_unclassified | Cochliobolus kusanoi, Epicoccum nigrum | 100 | 1.09 × 10−48 |
| Otu00051 | 0.008 | 0.453 | 0.070 | Agaricales_unclassified | Chondrostereum purpureum | 100 | 8.45 × 10−45 |
| Otu00063 | 0.022 | 0.439 | 0.049 | Sordariomycetes_unclassified | Lopadostoma polynesium, Monographella lycopodina, etc. | 100 | 1.09 × 10−48 |
| Otu00054 | 0.039 | 0.385 | 0.124 | Eukaryota_unclassified | Jaculispora submersa | 98.165 | 2.35 × 10−45 |
| Otu00060 | 0.023 | 0.403 | 0.049 | Eukaryota_unclassified | Repetobasidium conicum | 92.661 | 2.38 × 10−35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Marsden, N.; Lloyd, J.R.; Robinson, C.H.; Boothman, C.; Crawford, I.; Gallagher, M.; Coe, H.; Allen, G.; Flynn, M. Airborne Prokaryotic, Fungal and Eukaryotic Communities of an Urban Environment in the UK. Atmosphere 2022, 13, 1212. https://doi.org/10.3390/atmos13081212
Song H, Marsden N, Lloyd JR, Robinson CH, Boothman C, Crawford I, Gallagher M, Coe H, Allen G, Flynn M. Airborne Prokaryotic, Fungal and Eukaryotic Communities of an Urban Environment in the UK. Atmosphere. 2022; 13(8):1212. https://doi.org/10.3390/atmos13081212
Chicago/Turabian StyleSong, Hokyung, Nicholas Marsden, Jonathan R. Lloyd, Clare H. Robinson, Christopher Boothman, Ian Crawford, Martin Gallagher, Hugh Coe, Grant Allen, and Michael Flynn. 2022. "Airborne Prokaryotic, Fungal and Eukaryotic Communities of an Urban Environment in the UK" Atmosphere 13, no. 8: 1212. https://doi.org/10.3390/atmos13081212
APA StyleSong, H., Marsden, N., Lloyd, J. R., Robinson, C. H., Boothman, C., Crawford, I., Gallagher, M., Coe, H., Allen, G., & Flynn, M. (2022). Airborne Prokaryotic, Fungal and Eukaryotic Communities of an Urban Environment in the UK. Atmosphere, 13(8), 1212. https://doi.org/10.3390/atmos13081212










