Abstract
Advances in proteome research have opened the gateway to understanding numerous metabolic pathways and fundamental mechanisms involved in abiotic stress tolerance. In the present study, the antioxidant capacity of four tomato genotypes i.e., Kashi Amrit, Kashi Anupam, EC-317-6-1, and WIR-4360 was determined under drought stress to ascertain the scavenging potential for reactive oxygen species (ROS). A significant increase in the superoxide dismutase (SOD), Ascorbate peroxidase (APX), and catalase (CAT) activities in all the four genotypes under drought stress was observed, which seemed to be associated with a protective role against ROS (p < 0.001). Based on the antioxidant enzyme activities, a proteomic approach was applied to study differential protein expression in two selected genotypes from different species i.e., EC-317-6-1 (Solanum pimpinellifolium) and Kashi Amrit (Solanum lycopersicum) grown under irrigated, drought, and re-watering conditions. To reveal the protein network regulated under these conditions, two-dimensional gel electrophoresis was employed to identify and quantify the number of proteins in drought-sensitive (Kashi Amrit) and tolerant (EC-317-6-1) genotypes. Matrix-assisted laser desorption/ionization-time of flight analysis (MALDI-TOF) revealed a total of 453 spots after fine-tuning factors i.e., smoothness, saliency, and minimum area that responded to drought. Out of 453 total spots, 93 spots were identified in Kashi Amrit and 154 in EC-317-6-1 under irrigated conditions, whereas 4 spots were identified in Kashi Amrit and 77 spots in EC-317-6-1 under drought conditions. Furthermore, differentially expressed proteins were distinguished according to the fold change of their expression. Information provided in this report will be useful for the selection of proteins or genes in analyzing or improving drought tolerance in tomato cultivars. These findings may assist in the construction of a complete proteome database encompassing various divergent species which could be a valuable source for the improvement of crops under drought-stress conditions in the future.
1. Introduction
Tomato is a well-studied species belonging to the family Solanaceae, primarily because of its short generation time, rudimentary diploid hereditary qualities, outstanding genetic transformation methodology, and an immensely well-characterized genetic resource [1]. In 2015, Tomato was ranked 7th globally in terms of production which was approximately 1.64 billion tons on an area of nearly 4.8 million hectares [2]. In India, tomato is cultivated on an 813 thousand hectares area with 21.2 million metric tons production [3]. Its consumption, as well as production, is increasing because of its antioxidant and anti-cancerous properties [4]. Tomato has been a model plant for molecular studies aiming at improving fruit quality and stress tolerance to different abiotic stress factors [5,6].
Abiotic stresses caused by different environmental factors could negatively influence crop growth and development. Crop plants respond to different abiotic stresses via various modifications at morphological, cellular, physiological, biochemical, and molecular levels. Among abiotic stress factors, drought is one of the major limiting factors for tomato production that adversely affects its performance and threatens its productivity. Tomato has been found to show limited tolerance against high temperature and drought stress [7] and therefore is extremely sensitive to drought stress [8]. During drought stress, many physiological and biochemical processes are disturbed [9]. An understanding of the multitude of processes through which plants respond to drought stress has been taken up as a challenge to improve drought tolerance in crops [10]. Adverse abiotic factors like extreme temperatures, salinity, or drought cause significant biochemical and physiological changes. Drought stress causes the excessive generation of reactive oxygen species (ROS), which results in oxidative damage to biomolecules and ultimately leads to cell death [11,12]. Accumulation of reactive oxygen species may be increased through different ways. For example, reduction of CO2 fixation owing to closure of stomata results in reduced NADP+ regeneration via the Calvin cycle, which in integration with changes in photosystem activities and photosynthetic transport capacity, causes higher electron leakage to O2 and consequent increase in ROS production by chloroplast Mehler reaction, in contrast to unstressed plants. Increased ROS levels result in lipid peroxidation, protein oxidation, enzymatic activity inhibition, oxidative damage to RNA and DNA and finally cell death [13]. The ROS scavenging machinery consisting of enzymatic and non-enzymatic antioxidants plays a pivotal role in stress tolerance [14]. Various studies have reported that environmental stresses induce an increase in the number of antioxidant phytochemicals and osmolytes against oxidative stress [15,16,17]. Maintenance of elevated levels of antioxidants to detoxify the ROS is causally related to enhanced tolerance to abiotic stresses [14,18]. To cope with environmental stresses, plants have developed strategies constituting of a complex array of enzymatic (catalase, CAT; superoxide dismutase, SOD; glutathione reductase, GR; ascorbate peroxidase, APX; monodehydroascorbate reductase, MDHAR: dehydroascorbate reductase, DHAR; glutathione peroxidase, GPX; and glutathione-S- transferase, GST) and non-enzymatic (ascorbic acid, ASH; glutathione, GSH; alkaloids, phenolic compounds, non-protein amino acids, and α-tocopherols) antioxidative defense mechanisms that can avoid cell damage by timely scavenging of harmful ROS, thereby, regulating intra-cellular environment [14].
Proteome plays an important part in plant stress response both in the form of structural proteins and proteins that regulate plant epigenome, transcriptome, and metabolome. This observation comes to light because proteins are directly involved in the enhancement of stress tolerance, being closer to phenotype than transcripts. The role of proteomics in studying the complex and dynamic plant proteomes depends on protein identification and its modifications under environmental stresses to develop improved crop varieties [19]. In the recent past, the evolution of high-throughput proteomics has enabled the research community to study plant proteome responses to various stress factors in an exceedingly advanced approach. Abiotic stresses induce profound changes on plant proteomes including relative abundance of protein, post-transcriptional and post-translational modifications, protein–protein interactions, and ultimately biological functions of proteins aimed at metabolic adjustments to changing environment and an improvement to plant-stress tolerance [20].
Drought tolerance involves a myriad of physio-biochemical processes, the understanding of which is critical for developing cultivars having improved resistance to drought stress, thus making crops withstand dry spells in regions where the availability of water is a critical factor. Furthermore, this will help in the identification of the new and emerging sources for improved drought-stress resistance. In the recent study, we have evaluated the four genotypes viz., Kashi Amrit, Kashi Anupam, EC-317-6-1, and WIR-4360 for biochemical as well as physiological studies under drought stress [21]. The performance of tomato genotypes used in the study showed significant differences in all studied traits, suggesting that they could be considered when selecting for drought tolerance. EC-317-6-171 and WIR-4360 had good yield performance under deficit irrigation treatment. Moreover, results indicate that biochemical and physiological parameters are more useful for the screening of drought-tolerant tomato genotypes. The present investigation is designed to understand the proteomic behavior of two different tomato genotypes differing in antioxidant potential and drought-stress tolerance through 2D gel electrophoresis and mass spectrometry. The proteomic insight may help in understanding the behavior of contrasting tomato genotypes under drought stress by spotting the proteins, which may play an important role in drought tolerance in the agroclimatic zone of Jammu and Kashmir.
2. Results
2.1. Antioxidant Enzyme Activities of Tomato Genotypes
To examine the detoxification of the oxidative damage, three important enzyme activities and their native PAGE profiles were evaluated. The four tomato genotypes showed differential antioxidant potential measured in terms of SOD, CAT, and APX activity under drought stress. In comparison to irrigated conditions (control), all the four tomato genotypes showed a significant increase in the activities of SOD, CAT, and APX during drought conditions, which reflects their ROS homeostasis potential. EC-317-6-1 genotype showed a substantial increase in all the three antioxidant enzymes viz. SOD (641.40%), CAT (159.60%), and APX (83.48%), followed by WIR-4360 under water stress compared to plants under irrigated conditions (p < 0.001). A minimum increase in SOD activity was found in Kashi Anupam followed by Kashi Amritas compared to irrigated conditions. Furthermore, a minimum increase in CAT and APX activities was found in Kashi Amrit followed by Kashi Anupam in drought as compared to irrigated conditions (Figure 1). Native PAGE was further used to check antioxidant enzyme activities, showing a similar trend to SOD, CAT, and APX activities under irrigated, drought, and re-watering conditions (Figure 2). Our findings indicate a substantial increase in comparative enzyme activity and native PAGE expression of three antioxidant enzymes during drought stress in all the four tomato genotypes taken under investigation.
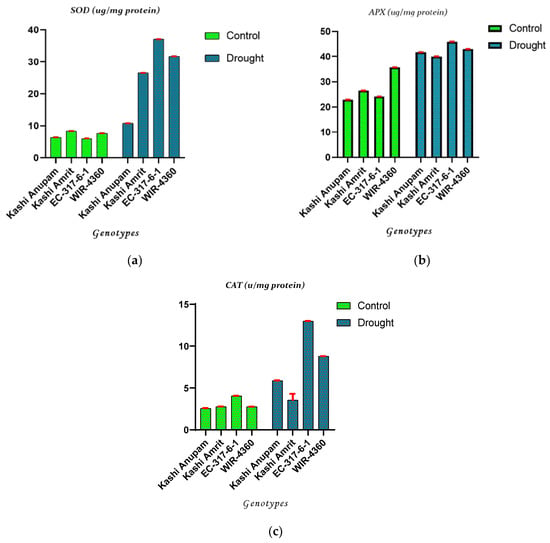
Figure 1.
Antioxidant enzyme activities (μmol/g) of SOD (a), APX (b), and CAT (c) in tomato genotypes under irrigated and drought conditions. Values are the means of three replicates with standard deviations. Different bar letters show significant differences among treatments separately.
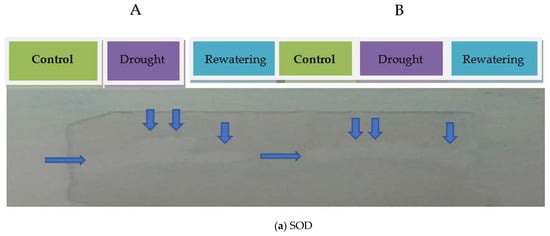
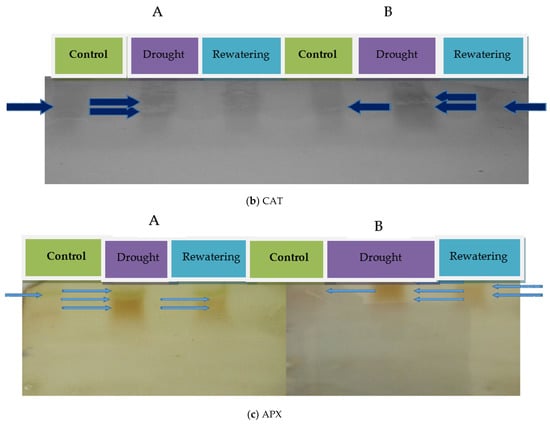
Figure 2.
Antioxidant enzyme activity of SOD (a), CAT (b) and APX (c) confirmed using native PAGE (A) Kashi Amrit and (B) EC-317-6-1.
Based on antioxidant enzyme activities, the proteome of two selected genotypes i.e., drought-tolerant (EC-317-6-1) and sensitive (Kashi Amrit) were analyzed for systematic proteomic analysis under drought conditions (Figure 3). To investigate proteins in tomato leaves under drought and re-watering conditions (Figure 4), 2D gel electrophoresis followed by mass spectrometry was used. All the gels were then examined for differential spot identification. As a result of fine-tuning the parameters such as smoothness, saliency, and minimum area, 453 spots were identified, and out of them, 93 were identified in Kashi Amrit and 154 in EC-317-6-1 in the control sample while 4 were identified in Kashi Amrit in drought condition. In re-watering conditions, 125 spots were identified in Kashi Amrit and 77 spots in EC-317-6-1 (Figure 5). Also, differential expression of proteins was observed based on fold change of their expression.
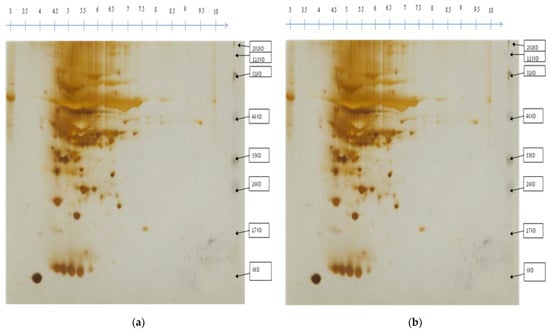
Figure 3.
Representative gel image following two-dimensional PAGE analysis of leaf protein extracts of Kashi Amrit (a) and EC-317-6-1 (b). Protein spots circled are the proteins present in the controlled condition.
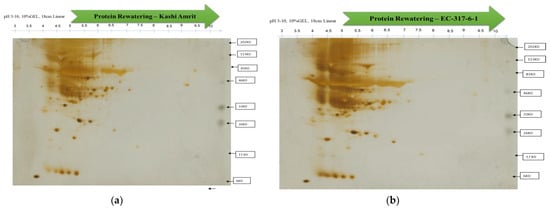
Figure 4.
2-D gel electrophoresis of proteins isolated from Kashi Amrit (a) and EC-317-6-1 (b) under rewatering conditions.

Figure 5.
2-D gel electrophoresis of proteins isolated from Kashi Amrit (a) and EC-317-6-1 (b) under drought conditions.
2.2. Identification and Functional Classification of Drought-Responsive Tomato Leaf Proteins
Further differentially expressed proteins in tomato genotypes were excised from 2-D gels and evaluated with matrix-assisted laser desorption/ionization-time of flight analysis (MALDI-TOF MS). Proteins identified by MS had functional annotations in the universal protein data bank. These findings specify that most of the proteins identified in drought had a substantial correlation to the defense response, oxidative stress, detoxification, protein synthesis, energy, and carbon metabolism, and mitochondrial small heat shock proteins (HSPs). These outcomes provide a summary of the proteins in tomato genotypes and unique insight for acclimatization under drought stress. Using gene ontology (www.geneontology.com) [22], 15 identified proteins from tomato leaves were grouped into 10 different functional clusters (Figure 6).
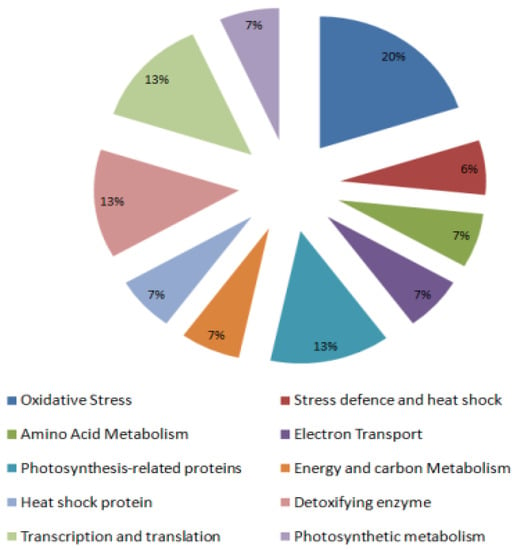
Figure 6.
Categorization of functional proteins into 10 different functional clusters.
2.2.1. Defense-Related Proteins
Stress receptive proteins function in the mitigation of various abiotic stresses. Six percent of the proteins fall under this group and are linked to both abiotic and biotic stresses. GRP-like proteins 2 are abiotic stress-related proteins and were identified in tomato leaves, and the cell wall. GRPs like protein 2 are proposed to act as an agglutinating agent for the deposition of constituents of the cell wall [23]. Besides acting as molecular chaperones in protein processing, HSPs and luminal-binding proteins express a defensive role in plants.
2.2.2. Oxidative Stress-Related Proteins
The presence of ~20% proteins viz. GPX, APX, and glycine-rich proteins correlate that these proteins provide defense against oxidative stress. The ROS scavenging proteins include GPX, peroxidase, and APX. Various investigations of ascorbate metabolism also support our results [24].
2.2.3. Protein Synthesis and Processing
Seven percent of proteins were found to be associated with protein synthesis and processing out of the total known proteins such as chloroplast elongation factor and polypeptide-associated complex subunit.
2.2.4. Photosynthesis-Related Proteins and Photosynthetic Metabolism
Seven percent of identified proteins viz. rubisco activase and rubisco small subunit were noted to be associated with photosynthesis-related proteins. Earlier research also described photosynthesis-associated proteins in spinach and 33 kDa precursor protein as oxygen-evolving complex proteins [25].
2.2.5. Electron Transport
Four percent of the total proteins that were associated with electron transport include putative ferredoxin NADP reductase. Ferredoxin (flavodoxin)-NADP(H) reductases are universal flavoenzymes associated with providing NADPH or ferredoxin, flavodoxin, adrenodoxin to redox-based metabolic pathways in plastids and mitochondria.
2.2.6. Energy and Carbon Metabolism
Only 7% of proteins were identified as energy and carbon metabolism associated proteins under drought stress that included pyruvate dehydrogenase complex (PDC).
2.2.7. Detoxifying Enzyme
Thirteen percent of proteins among the total identified proteins viz. ferritin- and osmotin-like proteins play a vital role in detoxification. Ferritin stores iron in a soluble, non-toxic, easily available form while the iron is taken up in the ferrous form and stored as ferric hydroxides when oxidized. It has been reported that osmotin or osmotin-like proteins are accumulated in response to diverse stresses in plants.
2.2.8. Mitochondrial Small Heat Shock Protein
Seven percent of the total identified proteins constitute mitochondrial small heat shock proteins. The heat shock response involves the temporal modification of metabolic behavior in the cell characterized by the disruption of normal protein synthesis and a sturdy expression of HSPs. MT-sHSPs plays a most important part in the heat tolerance of plant mitochondria.
3. Discussion
In the present investigation, an attempt was made to identify the response of susceptible and drought-tolerant tomato genotypes under water deficit conditions by determining antioxidant activity and using proteomics approaches for the identification of drought-responsive proteins. A significant phenomenon of drought stress in crop plants is the excess biosynthesis of various ROS such as superoxide anion, hydrogen peroxide, and hydroxyl radicals particularly in mitochondriaand chloroplast in cells [26,27] causing cellular damage by triggering off a chain reaction. Results revealed that the antioxidant activities (SOD, CAT, and APX) were up-regulated under severe drought stress in all four genotypes (p < 0.001). Oxidative damage is a main cytotoxic effect of ROS damage. SOD plays a significant role by facilitating the dismutation of superoxide radicals into H2O2 [28,29]. Similarly, CAT and APX play a major role in the conversion of toxic H2O2 in H2O [30,31]. APX is known to engage with glucose via the pentose phosphate shunt and reducing power (NADPH) to yield the reduced form of GSH from its oxidized form (GSSG). We found that the activities of APX and CAT were up-regulated markedly under drought stress. As summarized in results, drought-sensitive species also activate their antioxidative system. Though, results point out that not only the magnitude of activation could be decisive, but also the activated enzymes. For example, the activation of the APX and CAT is stronger in species tolerant to drought in contrast to their sensitive counterparts. Thus, our results propose that antioxidant machinery may provide a stratagem to engineer plant tolerance to drought stress, which has been also reported by various previous investigations [32,33,34].
The proteomic analysis of tomato plants has identified various proteins (drought-responsive) that are involved primarily in oxidative stress response, regulation of redox status, protein synthesis, and processing, photosynthesis, and energy production. In our study, mitochondrial heat shock proteins, GRP-like protein 2, and a luminal binding protein (33 kDa precursor protein) were identified. Their upregulation suggests that they play a key role in preventing denatured protein aggregation and in facilitating protein refolding under stress conditions [35,36]. Proteomic studies highlighted specific changes in components involved in transcription/translation machinery and/or in structural elements regulating cytoplasm hydration [37]. A proteomic study of drought-stressed leaves of S. lycopersicum identified several differentially accumulated proteins, with a majority in the down-regulated fraction in both drought-tolerant genotypes. These belonged to categories related to cellular metabolic activities and protein translation [38].
In a recent study, drought-induced changes were studied in Capsicum annum L. The proteome analysis of apoplast showed that there was an increase in peroxidase and phenylalanine ammonia-lyase activities with a concomitant reduction in CAT activity on the other side, and LC-MS analysis showed that there was differential expression of proteins under drought-induced stress conditions. Fourty-three stress-related protein species viz. defensins, polygalacturonase inhibitor proteins, SOD, and peroxidases were found to be up-regulated and 20 unique protein species were recognized in drought-treated plant proteome [39]. Verma and co-workers performed a genome-wide analysis of abiotic-stress responsive SOD gene family in B. juncea and B. rapa and found 29 and 18 SOD genes, respectively. Using the available FPKM analysis SRA data, 14 and 10 abiotic stress-responsive SOD genes were identified in B. rapa and B. juncea that could help in engineering the plant abiotic stress resistance. This experiment was the first investigation to explain the SOD gene family through genome-wide analysis in B. rapa and B. juncea and will further assist in conducting future research in cloning and functional validation of SOD genes for crop improvement in Brassica crop [40]. Various proteins involved in photosynthesis, redox regulation, oxidative defense system, and chaperone have been characterized in sugar beet leaves under water deficit conditions [41]. In Glycine max seedlings, the root proteome was studied under drought stress. In their study of underwater deficit, a total of 45 proteins were observed, two of them were new proteins, five proteins were found to be up-regulated and 21 were down-regulated. However, after re-watering conditions, the concentration of proteins was like the control levels [42]. An analogous trend was detected in the present investigation. In T. aestivum plants, a total of 36 protein spots were detected, in which 12 proteins were up-regulated and 24 down-regulated under drought-stress conditions [43]. Using 2-DE, mass spectrometry, and MALDI-TOF, rice proteomics was investigated in a study under water deficit and 18 proteins were detected, out of which 12 were up-regulated and 12 down-regulated [44]. In barley plants, drought-induced effects in leaf proteome were investigated by a group of researchers and a substantial decrement in yield and biomass of plants was observed, but the photosynthetic potential was found to remain unchanged [45]. The experimental analysis also strongly advocated that drought-stress response induces changes at the proteomics level, thus enabling plants to modulate specific adaptive mechanisms, which could differ in genotypes of drought-tolerant and drought-sensitive plants, and similar results were also obtained by Yu and coworkers [46]. The results here clearly revealed that drought-induced impairment in photosynthesis coupled with an inhibited synthesis of proteins in tomato plants, which are in line with previous studies [1,47]. These differential expression patterns of proteins in the tolerant and sensitive tomato genotypes and the changes observed in drought-responsive proteins that were positively regulated only in the tolerant genotype, could have the ability to be used as molecular markers to screen and select drought-tolerant lines. Hence, this study reveals that a comprehensive proteome database should be constructed which will cover all the proteomic information of the different species of tomato, which could provide a valuable source for future research.
4. Materials and Methodology
4.1. Tomato Genotypes and Drought Stress Treatment
Four tomato genotypes i.e., Kashi Amrit, Kashi Anupam, EC-317-6-1 and WIR-4360 were selected in the present study based on the combined investigation of their morphological, physiological and biochemical parameters, for investigating the antioxidant enzyme activities under drought stress and to determine their antioxidant potential. The authentic seeds of tomatoes were procured from IIVR, Varanasi, Uttar Pradesh, India. Healthy seeds were sown in pots under controlled environmental conditions (temperature: 25 °C ± 2 and relative humidity: 55% ± 5) at the School of Biotechnology, Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Jammu and Kashmir, India. The experiment was built up of 26 pots with 6 pots per genotype in two complete sets (3 control and 3 stressed pots for each genotype) that were arranged according to a completely randomized design. The plants were then subjected to drought stress at the growth stage (45 days) till the temporary wilting point (Figure 7), and irrigated plants were grown under normal conditions for the same time. All plants were irrigated to field capacity (FC) until the pre-flowering stage, four weeks after transplanting, after which water restriction treatment was imposed. Half of the seedlings of each genotype (3 plants) were regularly watered as control plants and the remaining half were submitted to drying and re-watering cycles. Control plants were irrigated daily in the evening, with an amount of water to replenish FC. In contrast, drought-stressed plants (ds) received no water for 10 days as soon as the stomatal conductance reached values near zero and plants started wilting. Recovery of plants was carried out by re-watering of pots to FC for 7 days. The second cycle of drought stress was imposed by withholding irrigation for 6 days followed by a final re-watering. Irrigation was given manually using a measuring cylinder. Careful attention was taken for the homogenous application of irrigation water in the pots throughout the whole growing period of the plants.

Figure 7.
In vivo phenotyping of early detection of drought in tomato genotypes Kashi Amrit (a) and EC-317-6-1 (b) under normal, drought and rewatering conditions.
4.2. Estimation of Drought Tolerance Linked Biomarkers
4.2.1. Protein Extraction and Estimation
For antioxidant enzyme assays, frozen leaves were ground to a fine powder with liquid nitrogen and extracted with ice-cold 0.1 M Tris-HCl buffer (pH 7.5) and 0.1% β-mercaptoethanol (3:1 buffer volume/FW). The homogenate was centrifuged at 10,000× g for 20 min, at 4 °C, and the supernatant was used for enzyme activity and protein determinations. Preparations for enzyme extraction and enzyme assay were carried out at 4 °C. Estimation of proteins was done by Bradford’s method. The standard curve was prepared with the bovine serum albumin standard solution and the absorbance was read at 480 nm.
4.2.2. Native Polyacrylamide Gel Electrophoresis (Native PAGE) and Activity Staining
Plant extracts containing equal amounts of protein were subjected to discontinuous PAGE under non-denaturing and non-reducing conditions, as described by Laemmli [48]. Native PAGE of SOD, CAT and APX was performed on a 10% resolving gel and 5% stacking gel at 140 V and 4 °C.
4.3. Antioxidant Isoenzyme Profiling
4.3.1. Superoxide Dismutase
After electrophoretic separation, the activity staining for SOD Isozyme was performed as reported by Beauchamp and Fridovich [47]. Gels were incubated in 2.5 M NBT for 20 min followed by incubation in 50 mM K-phosphate buffer (pH 7.8), containing 28 mM riboflavin and 28 mM tetramethylethylenediamine (TEMED) in darkness for another 20 min, and then exposed to a light box until the SOD activity bands became visible. The enzymes appeared as colorless bands. The isoenzyme was identified and characterized by selective inhibition with KCN or H2O2. The gel was incubated for 20 min in 50 mM K-phosphate buffer, pH 7.8, containing either 3 mM KCN or 5 mM H2O2 before staining for activity. Cu/Zn SODs were inhibited by KCN and H2O2; Fe SODs were resistant to KCN but were inactivated by H2O2; Mn SODs were resistant to both inhibitors.
4.3.2. Ascorbate Peroxidase
After electrophoretic separation, the activity staining for APX isozyme was performed by equilibration with 50 mM phosphate buffer (pH−7.5) containing 2.0 mM ascorbate for 30 min. The gel was incubated in 2.0 mM H2O2 for 30 min. in the dark. Gel was washed with Buffer (pH−7.5). The reaction was continued for 10 min and stopped by a wash with water.
4.3.3. Catalase
For the presence of CAT, the gel was washed with ddH2O and 50 mL solution containing potassium ferric cyanide (250 mg) and FeCl2 (250 mg) was added to visualize yellowish bands.
4.4. Protein Expression and Identification
4.4.1. Protein Sample Preparation for 2DE
One gram sample of fresh leaf from each genotype was ground in liquid N2 with the help of pre-chilled mortar and pestle and 1 mL phosphate buffer (pH 6.8) was added in Eppendorf tubes and mixed vigorously. The tubes were centrifuged at 14,000 rpm for 15 min at 4 °C, and the supernatant was collected in a fresh tube. An equal volume of TCAAEB (Trichloroacetic acid/acetone extraction buffer) was added and stored at −20 °C for 2 h. The suspension was again centrifuged at 10,000 rpm for 20 min at 4 °C. The pellet obtained was washed with a wash buffer 4 times and once with pure acetone. Finally, the pellet was dissolved in lysis buffer.
4.4.2. D Gel Electrophoresis
Loading of protein samples (250 µg of protein in 250 µL of rehydration buffer) containing IPG buffer (0.5%, pH 3–10) was done in a re-swelling tray. Immobilized linear pH gradient strips (pH 3–10, 13 cm, GE Healthcare, UK) were rehydrated overnight. IEF was performed using an Ettan IPGPhor3 Isoelectric Focusing System (GE Healthcare, Chalfont Saint Giles, UK). After carrying IEF, the strips were incubated for 15 min each in SDS equilibration buffer (6 M urea, 75 mM Tris HCl, pH 8.8, glycerol 29.3%, SDS 2%, BPB 0.002%) first supplemented with DTT (10 mg mL−1) and then replaced with equilibration buffer containing IAA (25 mg mL−1). The equilibrated strips were then placed on a 12.5 percent Tris-glycine SDS- PAGE (18 × 16 cm)19 and were sealed with a sealing solution (Laemmli buffer, agarose (0.5%), and bromophenol blue (0.002%). 2D gel electrophoresis was carried out using the Hoefer SE600 Ruby electrophoresis unit (GE Healthcare, Chalfont Saint Giles, UK) at 30 mA gel−1 at 25 °C [49]. The gels were stained overnight with CBB solution [50] and then destained with a solution containing 40% NaCl solution overnight.
4.4.3. Analysis of 2D Gels for Differential Spots Identification
All gels stained with 2D Coomassie Brilliant Blue (CBB stain) were scanned with high-resolution GE Image Scanner III. Gel images (Mel format) were analyzed using Image Master 2-D Platinum V.7.0 software (GE Healthcare, Chalfont Saint Giles, UK). Protein spots were detected by fine-tuning smoothness, saliency, and minimum area to enhance optimal clarity. To avoid erroneous interpretation, spots that were detected near the edges of the gel were deleted manually. 2D gel match sets were grouped into classes by the task of analysis. Protein extraction data of different treatments was obtained from technical replicates.
4.4.4. In-Gel Digestion and Protein Identification
Based on antioxidant potential, the proteome of two tomato genotypes i.e., drought-tolerant (EC-317-6-1) and sensitive (Kashi Amrit) were studied for systematic proteomic analysis under drought. The 30 noticeable differentially expressed spots were selected manually and digested by trypsin for MS analysis. The excised protein spots were washed, dehydrated with acetonitrile and ammonium bicarbonate, and then reduced by 15 mM DTT at 60 °C for 1 h. Further, 100 mM isoamyl alcohol was used to alkylate gel slices in dark for 15 min followed by rehydration with ammonium bicarbonate, and then drying-up in a speed vacuum for 15 min. The dried gel slices were rehydrated using 15 μL of trypsin (Sequencing grade, Promega, Madison, WI, USA) overnight at 37 °C. The supernatant was taken for further extraction, and 20% of acetonitrile and 1% formic acid was added to the remaining gel slice. The volume was reduced to 25–50 μL by drying the final supernatant in speed vacuum and this final volume was analyzed on AB Sciex (Applied Biosciences, New York, NY, USA) MALDI-TOF MS. During protein database searches performed on a local Mascot (Matrix Science, London, UK) server, peptide tolerance of 150 ppm, fragment mass tolerance of ±0.4 Da, and peptide charge of 1+ were selected. Significant hits were accepted based on MASCOT probability analysis (p < 0.05) and finally, peptides were searched in the NCBI database, the taxonomy of green plants.
4.5. Statistical Analysis
The statistical analysis was done by using GraphPad Prism 8.0.2 (263) software. All the data were analyzed by two-way analysis of variance (ANOVA). The means comparison and p-value Tukey test-HSD were used and considered the significance level at p < 0.001.
5. Conclusions
Our research report provided a systematic proteome analysis of tomato plants grown under irrigated, drought, and re-watering conditions using drought-tolerant (EC-317-6-1 and WIR-4360) and susceptible (Kashi Amrit and Kashi Anupam) genotypes. Fifteen proteins from tomato leaves were identified that responded to the drought-induced condition, which were classified into 10 different groups based on their functional properties, and are involved in an extensive array of cellular processes including defense/stress response, photosynthesis, electron transport, protein synthesis, and various metabolic pathways. Such proteins are present in different cellular components like mitochondria, cytoplasm, and chloroplast. Our results provide evidence for differential response of the tolerant genotypes compared to susceptible ones under drought stress. The tolerant genotypes showed a relatively higher photosynthetic rate and a more robust anti-oxidative defense system in comparison to the susceptible genotypes. Thus, it can be concluded that drought-responsive proteins can be taken as potent agents to improve drought tolerance and will help in a better understanding of the molecular biology in tomatoes under drought stress. Although our data suggest the activation of specific proteins in tomatoes under drought stress, involvement and fine regulation of such proteins need to be further investigated through the development and characterization of ad hoc designed plant mutants. Further, these drought-tolerant genotypes can be used for future tomato improvement programs for the agroecological climate in Jammu and Kashmir. Moreover, with the advancement in science and introduction of high-throughput genomics and gene editing techniques such CRISPR-Cas system, the candidate protein can be well characterized for crop enhancement programs.
Author Contributions
Conceptualization, G.K.R.; methodology, G.K.R. and A.P.; software, A.P. and R.R.K.; validation, G.K.R., A.P. and P.K.R.; formal analysis, J.P.S., A.R., G.K.R. and R.R.K.; investigation, A.P., R.R.K. and G.K.R.; resources, G.K.R.; data curation, A.P., S.D. and M.I.S.; writing—original draft preparation, G.K.R., A.P., M.A.H. and M.H.u.R.; writing—review and editing, A.I.A., M.A.A.-D., A.P., S.D., U.B., G.J. and M.I.S.; visualization, R.R.K. and A.P.; supervision, G.K.R.; project administration, G.K.R. and R.R.K. All authors have read and agreed to the final version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to acknowledge the assistance/help/effort of Milan Skalicky, Muntazir Mushtaq, Marian Brestic and Ayman EL Sabagh. We would like to thank the “Deanship of Scientific Research, University of Tabuk, KSA”, under the Research Group (S-1441-0044) for supporting us.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sant’Ana, D.V.P.; Lefsrud, M. Tomato proteomics: Tomato as a model for crop proteomics. Sci. Hortic. 2018, 239, 224–233. [Google Scholar] [CrossRef]
- Gerszberg, A.; Hnatuszko-Konka, K. Tomato tolerance to abiotic stress: A review of most often engineered target sequences. Plant Growth Regul. 2016, 83, 175–198. [Google Scholar] [CrossRef] [Green Version]
- Statista. 2020. Available online: https://www.statista.com/aboutus/ (accessed on 5 August 2021).
- Solankey, S.S.; Singh, R.K.; Baranwal, D.K. Genetic Expression of Tomato for Heat and Drought Stress Tolerance: An Overview. Int. J. Veg. Sci. 2015, 21, 496–515. [Google Scholar] [CrossRef]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vikram, A.; Deshmukh, R. Advances in omics approaches for abiotic stress tolerance in tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef] [Green Version]
- Ahammed, G.J.; Li, X.; Yang, Y.; Liu, C.; Zhou, G.; Wan, H.; Cheng, Y. Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2–mediated stomatal closure. Environ. Exp. Bot. 2020, 171, 103960. [Google Scholar] [CrossRef]
- Nahar, K.; Ullah, S.M. Morphological and physiological characters of tomato (Lycopersicon esculentum Mill) cultivars under water stress. Bangladesh J. Agric. Res. 2012, 37, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Nurrudin, M.M.; Madramootoo, C.A.; Doods, G.T. Effects of water stress at different growth stages on greenhouse tomato yield and quality. Hortscience 2003, 38, 1389–1393. [Google Scholar] [CrossRef] [Green Version]
- Soltys-Kalina, D.; Plich, J.; Strzelczyk-Zyta, D.; Sliwka, J.; Marczewski, W. The effect of drought stress on the relative water content and tuber yield of a half-sib family of ‘Katahdin’-derived potato cultivars. Breed. Sci. 2016, 66, 328–331. [Google Scholar] [CrossRef] [Green Version]
- Deikman, J.; Petracek, M.; Heard, J.E. Drought tolerance through biotechnology: Improving translation from the laboratory to farmer’s fields. Curr. Opin. Biotechnol. 2012, 23, 243–250. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, R.; Fernandes, J.L.; Abdelrahman, M.; Burritt, D.J.; Tran, L.S.P. Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Crit. Rev. Biotechnol. 2020, 40, 320–340. [Google Scholar] [CrossRef]
- Tamburino, R.; Vitale, M.; Ruggiero, A.; Sassi, M.; Sannino, L.; Arena, S.; Costa, A.; Batelli, G.; Zambrano, N.; Scaloni, A.; et al. Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicon L.). BMC Plant Biol. 2017, 17, 40. [Google Scholar]
- Zaid, A.; Wani, S.H. Reactive Oxygen Species Generation, Scavenging and Signaling in Plant Defense Responses. In Bioactive Molecules in Plant Defense; Jogaiah, S., Abdelrahman, M., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Zhou, Y.H.; Xia, X.J.; Mao, W.H.; Shi, K.; Yu, J.Q. Brassinosteroid regulates secondary metabolism in tomato towards enhanced tolerance to phenanthrene. Biol. Plant 2013, 57, 154–158. [Google Scholar] [CrossRef]
- Zaid, A.; Mohammad, F.; Wani, S.H.; Siddique, K.M. Salicylic acid enhances nickel stress tolerance by up-regulating antioxidant defense and glyoxalase systems in mustard plants. Ecotoxicol. Environ. Saf. 2019, 180, 575–587. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Elkelish, A.; Soliman, M.; Elansary, H.O.; Zaid, A.; Wani, S.H. Serratia marcescens BM1 Enhances Cadmium Stress Tolerance and Phytoremediation Potential of Soybean through Modulation of Osmolytes, Leaf Gas Exchange, Antioxidant Machinery, and Stress-Responsive Genes Expression. Antioxidants 2020, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Rodriguez, E.; Rubio-Wilhelmi, M.M.; Cervilla, L.M.; Blasco, B.; Rios, J.J.; Rosales, M.A.; Romero, L.; Ruiz, J.M. Genotypic differences in some physiological parameters symptomatic for oxidative stress under moderate drought in tomato plants. Plant Sci. 2010, 178, 30–40. [Google Scholar] [CrossRef]
- Kosova, K.; Vitamvas, P.; Urban, M.O.; Prasil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Dhindsa, R.H.; Plumb-Dhindsa, P.; Thrope, T.A. Leaf senescence correlated with increased level of membrane permeability, lipid peroxidation and decreased level of SOD and CAT. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Parveen, A.; Rai, G.K.; Mushtaq, M.; Singh, M.; Rai, P.K.; Rai, S.K.; Kundoo, A.A. Deciphering the Morphological, Physiological and Biochemical Mechanism Associated with Drought Stress Tolerance in Tomato Genotypes. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 227–255. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, A.; Hotta, Y. Glycine-rich proteins: A class of novel proteins. Appl. Biochem. Biotechnol. 2005, 120, 169–174. [Google Scholar] [CrossRef]
- Chen, L.; Ding, C.; Zhao, X.; Xu, J.; Mohammad, A.A.; Wang, S.; Ding, Y. Differential regulation of proteins in rice (Oryza sativa L.) under iron deficiency. Plant Cell Rep. 2015, 34, 83–96. [Google Scholar] [CrossRef]
- Rokka, A.; Zhang, L.; Aro, E.M. Rubisco activase: An enzyme with a temperature-dependent dual function. Plant J. 2001, 25, 463–471. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hurst, R.D.; Hancock, J.T. Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 2002, 53, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Muneer, S.; Ko, C.H.; Wei, H.; Chen, Y.; Jeong, B.R. Physiological and Proteomic Investigations to Study the Response of Tomato Graft Unions under Temperature Stress. PLoS ONE 2016, 11, e0157439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics 2019, 20, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Anjum, N.A.; Sharma, P.; Gill, S.S.; Hasanuzzaman, M.; Khan, E.A.; Kachhap, K.; Sofo, A. Catalase and ascorbate peroxidase—Representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. 2016, 23, 19002–19029. [Google Scholar] [CrossRef]
- Faize, M.; Burgos, L.; Faize, L.; Piqueras, A.; Nicolas, E.; Barba-Espin, G.; Clemente-Moreno, M.J.; Alcobendas, R.; Artlip, T.; Hernandez, J.A. Involvement of cytosolic ascorbate peroxidase and Cu/Zn-superoxide dismutase for improved tolerance against drought stress. J. Exp. Bot. 2011, 62, 2599–2613. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and drought: Can we make metabolic connections from available data. J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F.; Tomato, S.R. CAMTA transcription factors SLSR1 and SLSR3L negatively regulate disease resistance response and SLSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 286. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SLNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.; Lee, S.; Kang, K.Y.; Bahk, J.D.; Choi, M.S.; Lee, I.; Renaut, J.; Lee, B. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiol. Plant 2007, 131, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Dong, K.; Ge, P.; Bian, Y.; Dong, L.; Deng, X.; Li, X.; Yan, Y. Identification of leaf proteins differentially accumulated between wheat cultivars distinct in their levels of drought tolerance. PLoS ONE 2015, 10, e0125302. [Google Scholar] [CrossRef]
- Zhou, S.; Palmer, M.; Zhou, J.; Bhatti, S.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Differential root proteome expression in tomato genotypes with contrasting drought tolerance exposed to dehydration. J. Am. Soc. Hortic. Sci. 2013, 138, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Jaswanthi, N.; Krishna, M.S.R.; Sahitya, U.L.; Suneetha, P. Apoplast proteomic analysis reveals drought stress-responsive protein datasets in chilli (Capsicum annum L.). Data Brief 2019, 25, 104041. [Google Scholar] [CrossRef]
- Rahman, M.H.U.; Ahmad, I.; Wang, X.; Wajid, A.; Nasim, W.; Hussain, M.; Ahmad, B.; Ali, Z.; Ishaque, W.; Awais, M.; et al. Multi-model projections of future climate and climate change impacts uncertainty assessment for cotton production in Pakistan. Agric. For. Meteorol. 2018, 253–254, 94–113. [Google Scholar] [CrossRef]
- Hajheidari, M.; Abdollahian-Noghabi, M.; Askari, H.; Heidari, M.; Sadeghian, S.Y.; Ober, E.S. Proteome analysis of sugar beet leaves under drought stress. Proteomics 2005, 5, 950–960. [Google Scholar] [CrossRef]
- Larrainzar, E.; Wienkoop, S.; Weckwerth, W.; Ladrera, R.; Arrese-Igor, C.; Gonzalez, E.M. Medigo truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol. 2007, 144, 1495–1507. [Google Scholar] [CrossRef] [Green Version]
- Caruso, G.; Cavaliere, C.; Foglia, P.; Gubbiotti, R.; Guarino, C.; Samperi, R. Analysis of drought-responsive proteins in wheat (Triticum aestivum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci. 2009, 177, 570–576. [Google Scholar] [CrossRef]
- Ke, Y.; Han, G.; He, H.; Li, J. Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 2009, 379, 133–138. [Google Scholar] [CrossRef]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; James, A.T.; Yang, A.; Jones, A.; Mendoza-Porras, O.; Betrix, C.A.; Ma, H.; Colgrave, M.L. A comparative proteomic study of drought-tolerant and drought-sensitive soybean seedlings under drought stress. Crop Pasture Sci. 2016, 67, 528–540. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Newsholme, S.J.; Maleeff, B.F.; Steiner, S.; Anderson, N.L.; Schwartz, L.W. Two-dimensional electrophoresis of liver proteins: Characterization of a drug-induced hepatomegaly in rats. Electrophoresis 2000, 21, 2122–2128. [Google Scholar] [CrossRef]
- Beuchamp, C.; Fridovich, I. Superoxide Dismutase: Improved assays and an assay applicable to acrylamide gel. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).