Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research
Abstract
1. Introduction
2. The Formation Mechanisms of Polyploid Plants
3. Polyploidy-Driven Adaptation and Domestication in Wild and Cultivated Plants
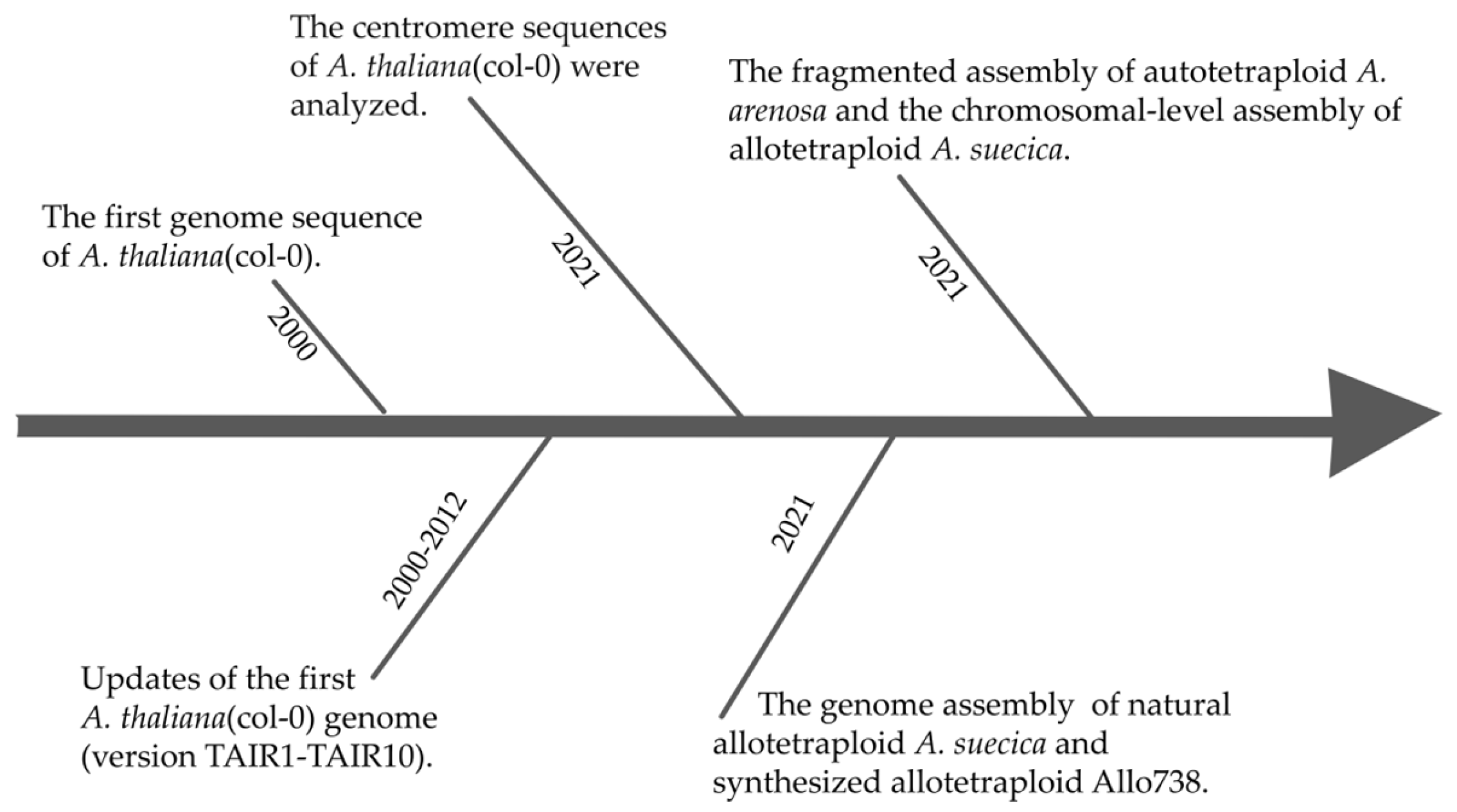
4. Advances in Genome Sequencing Technologies: Lessons from the Arabidopsis Genome
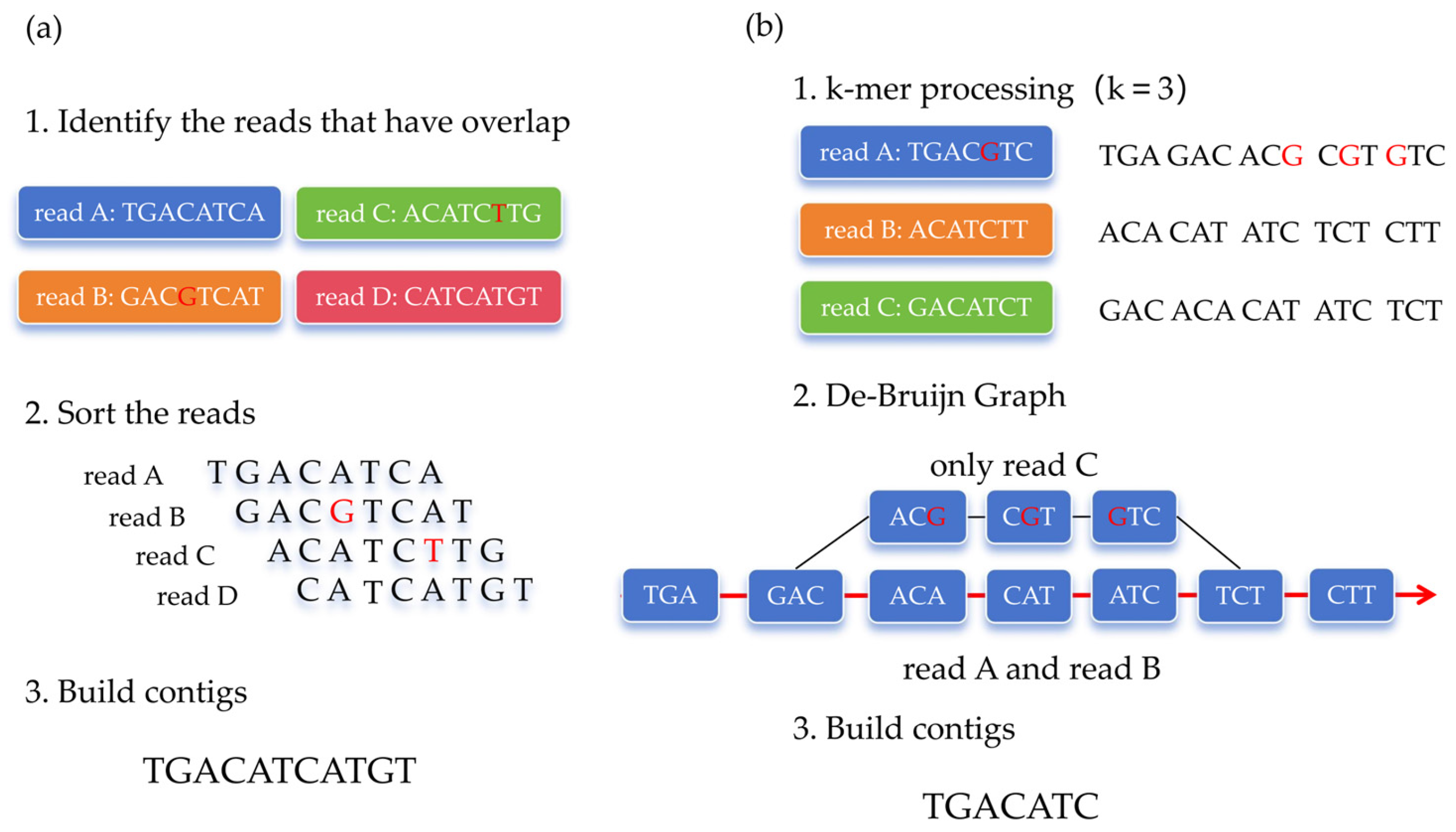
5. Advancements in Genome (Contig) Assembly Algorithms
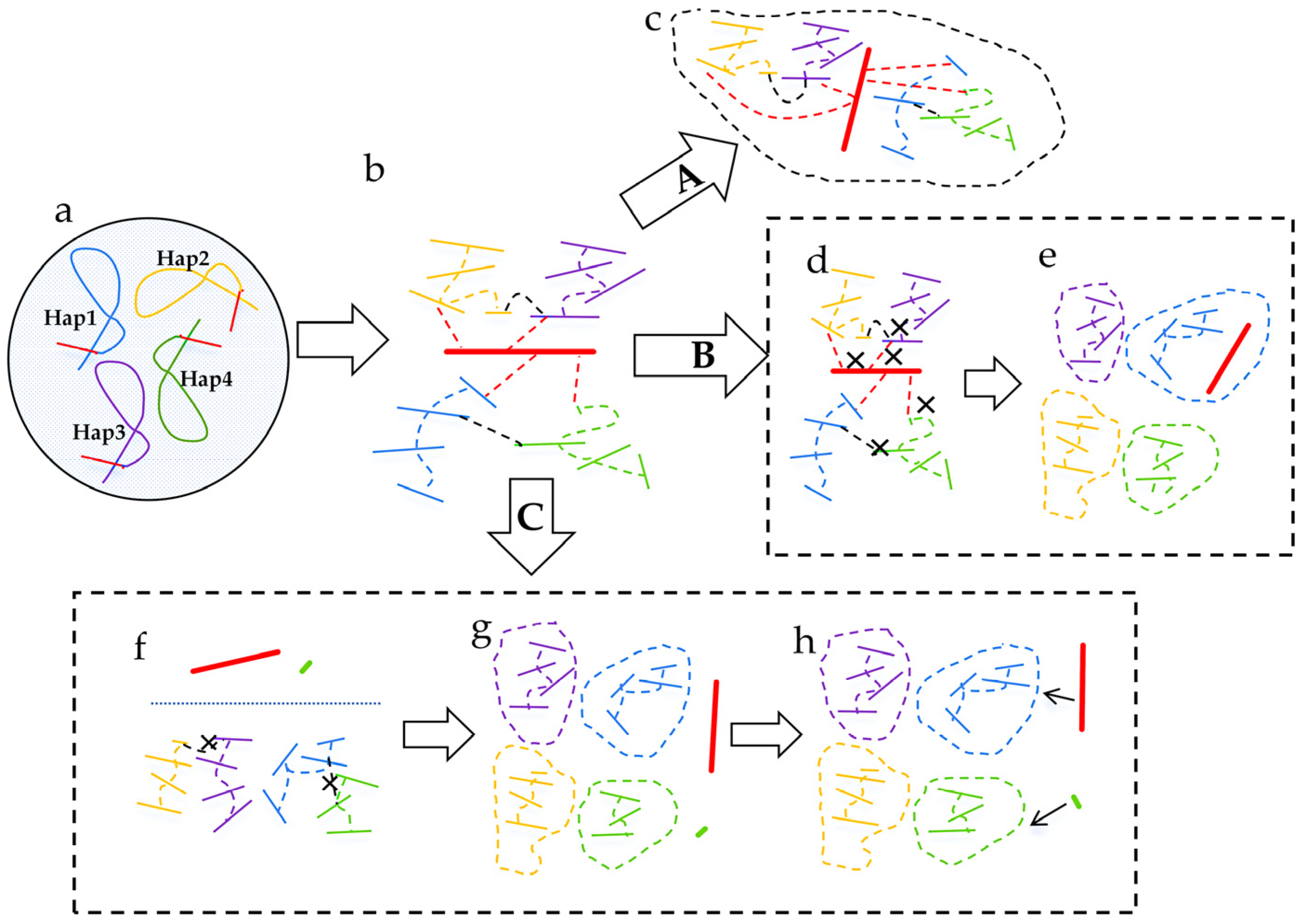
6. Current Challenges and Strategies in Polyploid Plant Genome Assembly
7. Genomic Assembly Studies of Polyploid Plants
8. Future Development and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, H. Disentangling a polyploid genome. Nat. Plants 2017, 3, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Visger, C.J.; Soltis, P.S. The polyploidy revolution then… and now: Stebbins revisited. Am. J. Bot. 2014, 101, 1057–1078. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Diallo, A.M.; Nielsen, L.R.; Kjær, E.D.; Petersen, K.K.; Ræbild, A. Polyploidy can confer superiority to West African Acacia senegal (L.) Willd. trees. Front. Plant Sci. 2016, 7, 821. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Wang, L.; Cao, S.; Wang, P.; Lu, K.; Song, Q.; Zhao, F.-J.; Chen, Z.J. DNA hypomethylation in tetraploid rice potentiates stress-responsive gene expression for salt tolerance. Proc. Natl. Acad. Sci. USA 2021, 118, e2023981118. [Google Scholar] [CrossRef]
- Levy, A.A.; Feldman, M. Evolution and origin of bread wheat. Plant Cell 2022, 34, 2549–2567. [Google Scholar] [CrossRef]
- Peng, R.; Xu, Y.; Tian, S.; Unver, T.; Liu, Z.; Zhou, Z.; Cai, X.; Wang, K.; Wei, Y.; Liu, Y. Evolutionary divergence of duplicated genomes in newly described allotetraploid cottons. Proc. Natl. Acad. Sci. USA 2022, 119, e2208496119. [Google Scholar] [CrossRef]
- Sun, H.; Jiao, W.-B.; Krause, K.; Campoy, J.A.; Goel, M.; Folz-Donahue, K.; Kukat, C.; Huettel, B.; Schneeberger, K. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 2022, 54, 342–348. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Asri, M.; Lucas, J.; Koren, S.; Li, H. Scalable telomere-to-telomere assembly for diploid and polyploid genomes with double graph. Nat. Methods 2024, 21, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Magnuson-Ford, K.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Recently formed polyploid plants diversify at lower rates. Science 2011, 333, 1257. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef]
- Kreiner, J.M.; Kron, P.; Husband, B.C. Evolutionary dynamics of unreduced gametes. Trends Genet. 2017, 33, 583–593. [Google Scholar] [CrossRef]
- Mason, A.S.; Pires, J.C. Unreduced gametes: Meiotic mishap or evolutionary mechanism? Trends Genet. 2015, 31, 5–10. [Google Scholar] [CrossRef]
- Hagerup, O. The spontaneous formation of haploid, polyploid, and aneuploid embryos in some orchids. Kongel Dan. Vidensk. Selsk. Biol. Meddelelser. 1947, 20, 1. [Google Scholar]
- Levin, D.A. Polyploidy and novelty in flowering plants. Am. Nat. 1983, 122, 1–25. [Google Scholar] [CrossRef]
- Stebbins, G.L., Jr. Types of polyploids: Their classification and significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar]
- Zhuang, Y.; Wang, X.; Li, X.; Hu, J.; Fan, L.; Landis, J.B.; Cannon, S.B.; Grimwood, J.; Schmutz, J.; Jackson, S.A. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat. Plants 2022, 8, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Mohammadin, S.; Edger, P.P. Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Maere, S.; Van De Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef]
- Wu, S.; Han, B.; Jiao, Y. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef]
- Doyle, J.J.; Coate, J.E. Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 2019, 180, 1–52. [Google Scholar] [CrossRef]
- Prost-Boxoen, L.; Bafort, Q.; Van de Vloet, A.; Almeida-Silva, F.; Paing, Y.T.; Casteleyn, G.; D’hondt, S.; De Clerck, O.; Van de Peer, Y. Asymmetric genome merging leads to gene expression novelty through nucleo-cytoplasmic disruptions and transcriptomic shock in Chlamydomonas triploids. New Phytol. 2025, 245, 869–884. [Google Scholar] [CrossRef]
- Burns, R.; Mandáková, T.; Gunis, J.; Soto-Jiménez, L.M.; Liu, C.; Lysak, M.A.; Novikova, P.Y.; Nordborg, M. Gradual evolution of allopolyploidy in Arabidopsis suecica. Nat. Ecol. Evol. 2021, 5, 1367–1381. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Pires, J.C.; Kovarik, A.; Tate, J.A.; Mavrodiev, E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. 2004, 82, 485–501. [Google Scholar] [CrossRef][Green Version]
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Wang, Y.; Brown, L.H.; Adams, T.M.; Cheung, Y.W.; Li, J.; Young, V.; Todd, D.T.; Armstrong, M.R.; Neugebauer, K.; Kaur, A. SMRT–AgRenSeq-d in potato (Solanum tuberosum) as a method to identify candidates for the nematode resistance Gpa5. Hortic. Res. 2023, 10, uhad211. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, C.; Liu, K.; Leng, Z.; Wang, Y.; Meng, W.; Li, D.; Zhang, C.; Ma, J. Altered reactive oxygen species scavenging and hormonal signaling in tetraploid rice are associated with blast resistance. Plant Physiol. 2025, 197, kiae547. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Xie-Kui, C.; Ao, C.-Q.; Zhang, Q.; Chen, L.-T.; Liu, J.-Q. Diploid and tetraploid distribution of Allium przewalskianum Regel. (Liliaceae) in the Qinghai-Tibetan Plateau and adjacent regions. Caryologia 2008, 61, 192–200. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Hon, T.; Mars, K.; Young, G.; Tsai, Y.-C.; Karalius, J.W.; Landolin, J.M.; Maurer, N.; Kudrna, D.; Hardigan, M.A.; Steiner, C.C. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 2020, 7, 399. [Google Scholar] [CrossRef]
- Naish, M.; Alonge, M.; Wlodzimierz, P.; Tock, A.J.; Abramson, B.W.; Schmücker, A.; Mandáková, T.; Jamge, B.; Lambing, C.; Kuo, P. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 2021, 374, eabi7489. [Google Scholar] [CrossRef]
- Jiang, X.; Song, Q.; Ye, W.; Chen, Z. Concerted genomic and epigenomic changes accompany stabilization of Arabidopsis allopolyploids. Nat. Ecol. Evol. 2021, 5, 1382–1393. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Reinert, K.; Myers, E.W. The greedy path-merging algorithm for contig scaffolding. J. ACM 2002, 49, 603–615. [Google Scholar] [CrossRef]
- Cherukuri, Y.; Janga, S.C. Benchmarking of de novo assembly algorithms for Nanopore data reveals optimal performance of OLC approaches. BMC Genom. 2016, 17, 95–105. [Google Scholar] [CrossRef][Green Version]
- Compeau, P.E.; Pevzner, P.A.; Tesler, G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011, 29, 987–991. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Sun, Z.; Hu, B.; Ayoola, A.O.; Liang, F.; Li, J.; Sandoval, J.R.; Cooper, D.N.; Ye, K. NextDenovo: An efficient error correction and accurate assembly tool for noisy long reads. Genome Biol. 2024, 25, 107. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, J.; Jiang, M.; Lei, W.; Zhang, X.; Tang, H. Sequencing and assembly of polyploid genomes. Polyploidy Methods Protoc. 2023, 2545, 429–458. [Google Scholar]
- Yuan, Y.; Chung, C.Y.-L.; Chan, T.-F. Advances in optical mapping for genomic research. Comput. Struct. Biotechnol. J. 2020, 18, 2051–2062. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 2019, preprint. [Google Scholar] [CrossRef]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Ghurye, J.; Pop, M.; Koren, S.; Bickhart, D.; Chin, C.-S. Scaffolding of long read assemblies using long range contact information. BMC Genom. 2017, 18, 527. [Google Scholar] [CrossRef]
- Zhou, C.; McCarthy, S.A.; Durbin, R. YaHS: Yet another Hi-C scaffolding tool. Bioinformatics 2023, 39, btac808. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Zhao, Q.; Ming, R.; Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 2019, 5, 833–845. [Google Scholar] [CrossRef]
- Zhang, Q.; Qi, Y.; Pan, H.; Tang, H.; Wang, G.; Hua, X.; Wang, Y.; Lin, L.; Li, Z.; Li, Y. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 2022, 54, 885–896. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, Y.; Hua, X.; Wang, Y.; Wang, B.; Qi, Y.; Huang, Y.; Yu, Z.; Gao, R.; Zhang, Y. The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum. Nat. Genet. 2025, 57, 242–253. [Google Scholar] [CrossRef]
- Zeng, X.; Yi, Z.; Zhang, X.; Du, Y.; Li, Y.; Zhou, Z. Chromosome-level scaffolding of haplotype-resolved assemblies using Hi-C data without reference genomes. Nat. Plants 2024, 10, 1184–1200. [Google Scholar] [CrossRef]
- Jiang, Z.; Peng, Z.; Wei, Z.; Sun, J.; Luo, Y.; Bie, L.; Zhang, G.; Wang, Y. A deep learning-based method enables the automatic and accurate assembly of chromosome-level genomes. Nucleic Acids Res. 2024, 52, e92. [Google Scholar] [CrossRef]
- Jia, K.H.; Wang, Z.X.; Wang, L.; Li, G.Y.; Zhang, W.; Wang, X.L.; Xu, F.J.; Jiao, S.Q.; Zhou, S.S.; Liu, H. SubPhaser: A robust allopolyploid subgenome phasing method based on subgenome-specific k-mers. New Phytol. 2022, 235, 801–809. [Google Scholar] [CrossRef]
- Li, X.; Yu, S.; Cheng, Z.; Chang, X.; Yun, Y.; Jiang, M.; Chen, X.; Wen, X.; Li, H.; Zhu, W. Origin and evolution of the triploid cultivated banana genome. Nat. Genet. 2024, 56, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Li, C.; Li, G.; Wang, P.; Peng, Z.; Cheng, L.; Li, H.; Zhang, Z.; Li, Y.; Huang, W. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 2022, 15, 1211–1226. [Google Scholar] [CrossRef]
- Lu, X.-M.; Yu, X.-F.; Li, G.-Q.; Qu, M.-H.; Wang, H.; Liu, C.; Man, Y.-P.; Jiang, X.-H.; Li, M.-Z.; Wang, J. Genome assembly of autotetraploid Actinidia arguta highlights adaptive evolution and enables dissection of important economic traits. Plant Commun. 2024, 5, 100856. [Google Scholar] [CrossRef]
- Yang, Z.; He, F.; Mai, Y.; Fan, S.; An, Y.; Li, K.; Wu, F.; Tang, M.; Yu, H.; Liu, J.; et al. A near-complete assembly of the Houttuynia cordata genome provides insights into the regulatory mechanism of flavonoid biosynthesis in Yuxingcao. Plant Commun. 2024, 5, 101075. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, Y.; Gong, W.; Zhao, G.; Xiao, S.; Lin, H.; Li, Y.; Liao, Z.; Zhang, S.; Hu, G. The tetraploid Camellia oleifera genome provides insights into evolution, agronomic traits, and genetic architecture of oil Camellia plants. Cell Rep. 2024, 43, 114902. [Google Scholar] [CrossRef]
- Shen, F.; Xu, S.; Shen, Q.; Bi, C.; Lysak, M.A. The allotetraploid horseradish genome provides insights into subgenome diversification and formation of critical traits. Nat. Commun. 2023, 14, 4102. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Liu, Y.; Wu, S.; Sun, H.; Wu, J.; Li, Y.; Zheng, Y.; Ren, H.; Yang, Y. Haplotype-resolved genome assembly and resequencing provide insights into the origin and breeding of modern rose. Nat. Plants 2024, 10, 1659–1671. [Google Scholar] [CrossRef]
- He, Q.; Li, W.; Miao, Y.; Wang, Y.; Liu, N.; Liu, J.; Li, T.; Xiao, Y.; Zhang, H.; Wang, Y. The near-complete genome assembly of hexaploid wild oat reveals its genome evolution and divergence with cultivated oats. Nat. Plants 2024, 10, 2062–2078. [Google Scholar] [CrossRef]
- Consortium, I.W.G.S.; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Miao, L.; Tan, K.; Guo, W.; Xin, B.; Appels, R.; Jia, J.; Lai, J.; Lu, F.; Ni, Z. Near-complete assembly and comprehensive annotation of the wheat Chinese Spring genome. Mol. Plant 2025, 18, 892–907. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Su, J.; Wang, H.; Zhang, Z.; Zhang, X.; Van de Peer, Y.; Chen, F.; Fang, W.; Guan, Z.; Zhang, F. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat. Commun. 2023, 14, 2021. [Google Scholar] [CrossRef]
- Jin, X.; Du, H.; Zhu, C.; Wan, H.; Liu, F.; Ruan, J.; Mower, J.P.; Zhu, A. Haplotype-resolved genomes of wild octoploid progenitors illuminate genomic diversifications from wild relatives to cultivated strawberry. Nat. Plants 2023, 9, 1252–1266. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Guo, C.; Zhao, L.; Mao, L.; Hu, X.-Z.; Yang, Y.-Z.; Qian, K.-C.; Ma, P.-F.; Guo, Z.-H.; Li, D.-Z. Haplotype-resolved nonaploid genome provides insights into in vitro flowering in bamboos. Hortic. Res. 2024, 11, uhae250. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.; Wai, C.M. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Liston, A.; Wei, N.; Tennessen, J.A.; Li, J.; Dong, M.; Ashman, T.-L. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 2–4. [Google Scholar] [CrossRef]
- Feng, C.; Wang, J.; Harris, A.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol. Biol. Evol. 2021, 38, 478–485. [Google Scholar] [CrossRef]
- Huang, G.; Bao, Z.; Feng, L.; Zhai, J.; Wendel, J.F.; Cao, X.; Zhu, Y. A telomere-to-telomere cotton genome assembly reveals centromere evolution and a Mutator transposon-linked module regulating embryo development. Nat. Genet. 2024, 56, 1953–1963. [Google Scholar] [CrossRef]
- Secomandi, S.; Gallo, G.R.; Rossi, R.; Rodríguez Fernandes, C.; Jarvis, E.D.; Bonisoli-Alquati, A.; Gianfranceschi, L.; Formenti, G. Pangenome graphs and their applications in biodiversity genomics. Nat. Genet. 2025, 57, 13–26. [Google Scholar] [CrossRef]
- Jiao, C.; Xie, X.; Hao, C.; Chen, L.; Xie, Y.; Garg, V.; Zhao, L.; Wang, Z.; Zhang, Y.; Li, T. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 2024, 637, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Z.; You, C.; Qi, Z.; You, J.; Grover, C.E.; Long, Y.; Huang, X.; Lu, S.; Wang, Y. Convergence and divergence of diploid and tetraploid cotton genomes. Nat. Genet. 2024, 56, 2562–2573. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Li, X.; He, H.; Yuan, Q.; Song, Y.; Wei, Z.; Lin, H.; Hu, M.; Zhao, F.; Zhang, C. A super pan-genomic landscape of rice. Cell Res. 2022, 32, 878–896. [Google Scholar] [CrossRef]
- Guo, L.; Wang, X.; Ayhan, D.H.; Rhaman, M.S.; Yan, M.; Jiang, J.; Wang, D.; Zheng, W.; Mei, J.; Ji, W. Super pangenome of Vitis empowers identification of downy mildew resistance genes for grapevine improvement. Nat. Genet. 2025, 57, 741–753. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, M.; Tan, J.; Huang, M.; Chu, X.; Li, Y.; Han, X.; Fang, T.; Tian, Y.; Jarret, R. Telomere-to-telomere Citrullus super-pangenome provides direction for watermelon breeding. Nat. Genet. 2024, 56, 1750–1761. [Google Scholar] [CrossRef]
| Tools | Step in Assembly | Method Breakthrough | Publishing Date | Reference |
|---|---|---|---|---|
| Canu | Contig assembly | MinHash Alignment Process solved the problem of low alignment efficiency due to high error rate in long-read assembly | 2017 | Koren, et al. [45] |
| SubPhaser | Subgenome partitioning | Partitioning polyploid subgenomes based on k-mer frequency statistics | 2022 | Jia, et al. [60] |
| HiFiasm | Contig Assembly | Combining the string graph with the phased assembly graph enables haplotype-resolved genome assembly | 2021 | Cheng, et al. [47] |
| ALLHiC | Scaffolding | An allele table was constructed from the genome of a closely related species to assist in the scaffolding process | 2019 | Zhang et al. [55] |
| HapHiC | Scaffolding | Chromosome anchoring of polyploid genomes can be achieved without relying on reference genomes | 2024 | Zeng et al. [58] |
| Chromosome Ploidy | Common Name | Species | Assembly Size | Publishing Date | Reference |
|---|---|---|---|---|---|
| Autotriploid | Banana | Musa acuminata cv. Cavendish | 1.48 Gb | 2024 | Li et al. [61] |
| M. acuminata cv. Gros Michel | 1.33 Gb | ||||
| Autotetraploid | Alfalfa | M. sativa | 2.738 Gb | 2020 | Chen et al. [62] |
| Wild sugarcane | Saccharum spontaneum | 2.761 Gb | 2022 | Zhang et al. [56] | |
| Potato | Solanum tuberosum C88 | 3,16 Gb | 2022 | Bao et al. [63] | |
| Hardy kiwifruit | Actinidia arguta | 2.61 Gb | 2024 | Lu et al. [64] | |
| Fish mint | Houttuynia cordata | 2.24 Gb | 2024 | Yang et al. [65] | |
| Oil tea tree | Camellia oleifera | 11.06 Gb | 2024 | Zhang et al. [66] | |
| Allotetraploid | Horseradish | Armracia rusticana | 610.05 Mb | 2023 | Shen et al. [67] |
| China rose | Rosa chinensis | 2.51 Gb | 2024 | Zhang et al. [68] | |
| Autohexaploid | Wild oat | Avena sterilis | 10.99 Gb | 2024 | He et al. [69] |
| Allohexaploid | Wheat | Triticum aestivum Chinese Spring v1.0 | 14.5 Gb | 2018 | IWGSC et al. [70] |
| T. aestivum Chinese Spring v2.1 | 14.41 Gb | 2021 | Zhu et al. [71] | ||
| T. aestivum Chinese Spring | 14.446 Gb | 2025 | Wang et al. [72] | ||
| Garden Mum | Chrysanthemum morifolium | 8.15 Gb | 2023 | Song et al. [73] | |
| Allooctoploid | Strawberry | Fragaria chiloensis | 1.64 Gb | 2023 | Jin et al. [74] |
| F. virginiana | 1.54 Gb | ||||
| Allononaploid | Bamboo | Bambusa odashimae | 3.36 Gb | 2024 | Wang et al. [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Shi, T. Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research. Genes 2025, 16, 636. https://doi.org/10.3390/genes16060636
Zhao Z, Shi T. Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research. Genes. 2025; 16(6):636. https://doi.org/10.3390/genes16060636
Chicago/Turabian StyleZhao, Zhenning, and Tao Shi. 2025. "Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research" Genes 16, no. 6: 636. https://doi.org/10.3390/genes16060636
APA StyleZhao, Z., & Shi, T. (2025). Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research. Genes, 16(6), 636. https://doi.org/10.3390/genes16060636








