Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta
Abstract
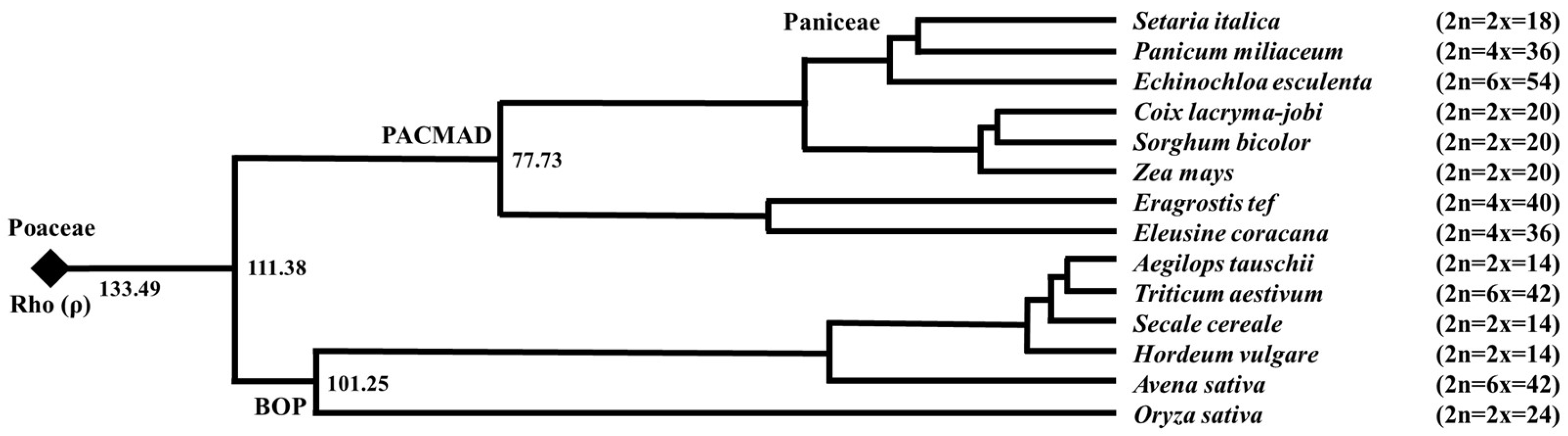
1. Introduction
2. Ploidy in Cereals
3. Ploidy and Domestication History in Genus Setaria
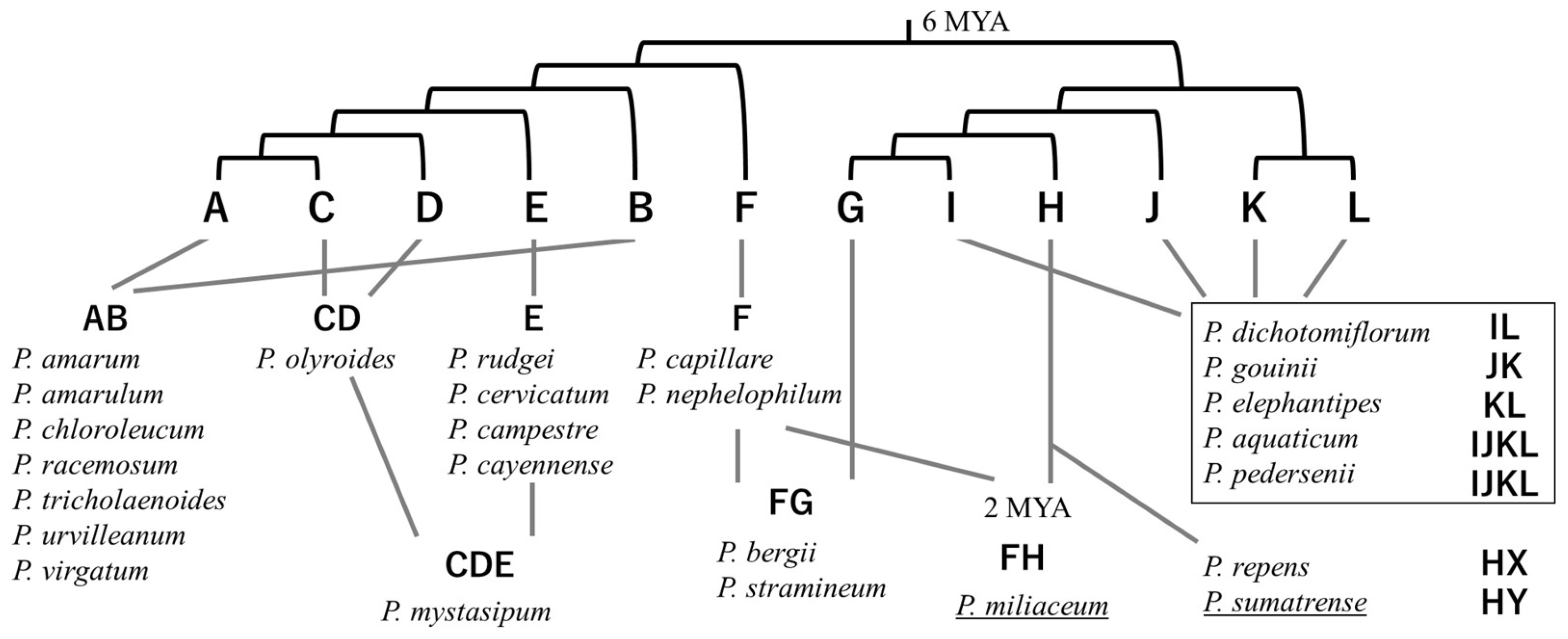
4. Ploidy and Domestication History in Genus Panicum
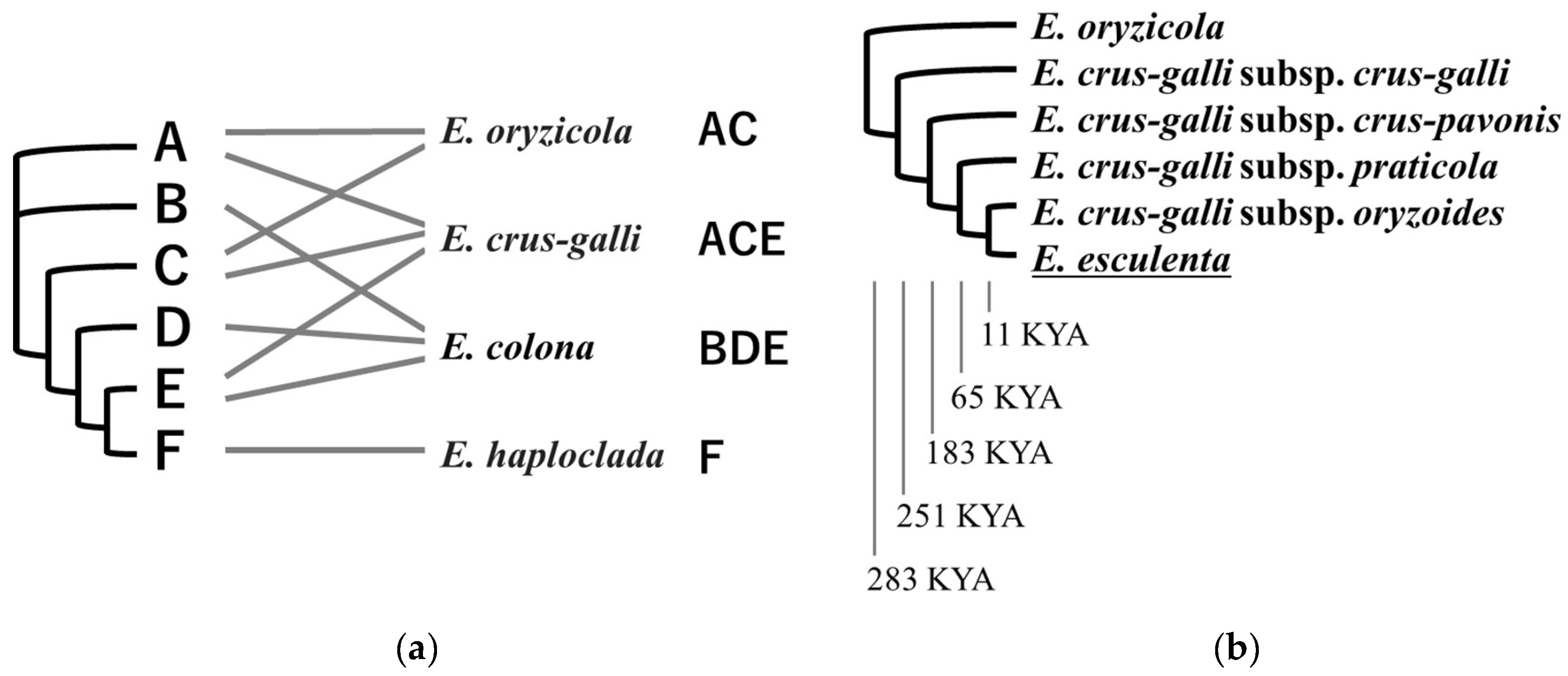
5. Ploidy and Domestication History in Genus Echinochloa
6. The Influence of Ploidy on Domestication and Selective Breeding
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Satomura, K. History of Propagation and Genetic Structures in Three Japanese Millets: Proso Millet, Foxtail Millet, and Barnyard Millet. In Phylogeographic History of Plants and Animals Coexisting with Humans in Asia; Osada, N., Suzuki, H., Kumagai, M., Endo, M., Eds.; Evolutionary Studies; Springer Nature: Singapore, 2024; pp. 107–130. ISBN 9789819768868. [Google Scholar]
- Sage, R.F. The Evolution of C4 Photosynthesis. New Phytol. 2004, 161, 341–370. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Lu, H.; Zhang, J.; Wang, C.; Huan, X. Prehistoric Evolution of the Dualistic Structure Mixed Rice and Millet Farming in China. Holocene 2017, 27, 1885–1898. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, Mechanisms, and Rates of Polyploid Formation in Flowering Plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Leitch, A.R.; Leitch, I.J. Genomic Plasticity and the Diversity of Polyploid Plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Levin, D.A. The Role of Chromosomal Change in Plant Evolution; Oxford Series in Ecology and Evolution; Oxford University Press: New York, NY, USA, 2002; ISBN 9781280834721. [Google Scholar]
- Rice, A.; Mayrose, I. The Chromosome Counts Database (CCDB). Methods Mol. Biol. 2023, 2703, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Kolář, F.; Čertner, M.; Suda, J.; Schönswetter, P.; Husband, B.C. Mixed-Ploidy Species: Progress and Opportunities in Polyploid Research. Trends Plant Sci. 2017, 22, 1041–1055. [Google Scholar] [CrossRef]
- Rice, A.; Šmarda, P.; Novosolov, M.; Drori, M.; Glick, L.; Sabath, N.; Meiri, S.; Belmaker, J.; Mayrose, I. The Global Biogeography of Polyploid Plants. Nat. Ecol. Evol. 2019, 3, 265–273. [Google Scholar] [CrossRef]
- David, K.T. Global Gradients in the Distribution of Animal Polyploids. Proc. Natl. Acad. Sci. USA 2022, 119, e2214070119. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The Evolutionary Significance of Polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Whitton, J. Polyploid Incidence and Evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Willis, J.H. Plant Speciation. Science 2007, 317, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Bartolić, P.; Morgan, E.J.; Padilla-García, N.; Kolář, F. Ploidy as a Leaky Reproductive Barrier: Mechanisms, Rates and Evolutionary Significance of Interploidy Gene Flow. Ann. Bot. 2024, 134, 537–550. [Google Scholar] [CrossRef]
- Wendel, J.F. Genome Evolution in Polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [PubMed]
- Renny-Byfield, S.; Wendel, J.F. Doubling down on Genomes: Polyploidy and Crop Plants. Am. J. Bot. 2014, 101, 1711–1725. [Google Scholar] [CrossRef] [PubMed]
- Touchell, D.H.; Palmer, I.E.; Ranney, T.G. In Vitro Ploidy Manipulation for Crop Improvement. Front. Plant Sci. 2020, 11, 722. [Google Scholar] [CrossRef]
- Akagi, T.; Jung, K.; Masuda, K.; Shimizu, K.K. Polyploidy before and after Domestication of Crop Species. Curr. Opin. Plant Biol. 2022, 69, 102255. [Google Scholar] [CrossRef]
- Abrouk, M.; Ahmed, H.I.; Cubry, P.; Šimoníková, D.; Cauet, S.; Pailles, Y.; Bettgenhaeuser, J.; Gapa, L.; Scarcelli, N.; Couderc, M.; et al. Fonio Millet Genome Unlocks African Orphan Crop Diversity for Agriculture in a Changing Climate. Nat. Commun. 2020, 11, 4488. [Google Scholar] [CrossRef]
- Kihara, H. Discovery of the DD-Analyser, One of the Ancestors of Triticum vulgare. Agric. Hortic. 1944, 19, 889–890. [Google Scholar]
- Ozkan, H.; Brandolini, A.; Schäfer-Pregl, R.; Salamini, F. AFLP Analysis of a Collection of Tetraploid Wheats Indicates the Origin of Emmer and Hard Wheat Domestication in Southeast Turkey. Mol. Biol. Evol. 2002, 19, 1797–1801. [Google Scholar] [CrossRef]
- Ozkan, H.; Brandolini, A.; Pozzi, C.; Effgen, S.; Wunder, J.; Salamini, F. A Reconsideration of the Domestication Geography of Tetraploid Wheats. Züchter Genet. Breed. Res. 2005, 110, 1052–1060. [Google Scholar] [CrossRef]
- Luo, M.-C.; Yang, Z.-L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvorak, J. The Structure of Wild and Domesticated Emmer Wheat Populations, Gene Flow between Them, and the Site of Emmer Domestication. Züchter Genet. Breed. Res. 2007, 114, 947–959. [Google Scholar] [CrossRef]
- Li, H.W.; Li, C.H.; Pao, W.K. Cytological and Genetical Studies of the Interspecific Cross of the Cultivated Foxtail Millet, Setaria Italica (L.) Beauv., and the Green Foxtail Millet, S. Viridis L. Agron. J. 1945, 37, 32–54. [Google Scholar] [CrossRef]
- Hilu, K.W. Evidence from RAPD Markers in the Evolution of Echinochloa Millets (Poaceae). Osterr. Bot. Z. 1994, 189, 247–257. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference Genome Sequence of the Model Plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef]
- Mamidi, S.; Healey, A.; Huang, P.; Grimwood, J.; Jenkins, J.; Barry, K.; Sreedasyam, A.; Shu, S.; Lovell, J.T.; Feldman, M.; et al. A Genome Resource for Green Millet Setaria viridis Enables Discovery of Agronomically Valuable Loci. Nat. Biotechnol. 2020, 38, 1203–1210. [Google Scholar] [CrossRef]
- Shi, J.; Ma, X.; Zhang, J.; Zhou, Y.; Liu, M.; Huang, L.; Sun, S.; Zhang, X.; Gao, X.; Zhan, W.; et al. Chromosome Conformation Capture Resolved near Complete Genome Assembly of Broomcorn Millet. Nat. Commun. 2019, 10, 464. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W.; et al. The Genome of Broomcorn Millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Chen, C.; Chen, L.; Li, M.; Qin, H.; Tian, X.; Hou, S.; Yang, X.; Jian, J.; et al. A Complete Reference Genome of Broomcorn Millet. Sci. Data 2024, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa Crus-Galli Genome Analysis Provides Insight into Its Adaptation and Invasiveness as a Weed. Nat. Commun. 2017, 8, 1031. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the Genome Sequence of the Flowering Plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling Angiosperm Genome Evolution by Phylogenetic Analysis of Chromosomal Duplication Events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral Polyploidy in Seed Plants and Angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One Thousand Plant Transcriptomes and the Phylogenomics of Green Plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with Double Genomes Might Have Had a Better Chance to Survive the Cretaceous-Tertiary Extinction Event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 Plant Genomes Supports a Wave of Successful Genome Duplications in Association with the Cretaceous-Paleogene Boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Demography of Duplicate Genes. J. Struct. Funct. Genom. 2003, 3, 35–44. [Google Scholar] [CrossRef]
- Paterson, A.H.; Chapman, B.A.; Kissinger, J.C.; Bowers, J.E.; Feltus, F.A.; Estill, J.C. Many Gene and Domain Families Have Convergent Fates Following Independent Whole-Genome Duplication Events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006, 22, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, C.A.; Davies, B. Gene Duplication and the Evolution of Plant MADS-Box Transcription Factors. J. Genet. Genom. 2012, 39, 157–165. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin, Germany, 1970; ISBN 9783540052258. [Google Scholar]
- Ruprecht, C.; Lohaus, R.; Vanneste, K.; Mutwil, M.; Nikoloski, Z.; Van de Peer, Y.; Persson, S. Revisiting Ancestral Polyploidy in Plants. Sci. Adv. 2017, 3, e1603195. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient Polyploidization Predating Divergence of the Cereals, and Its Consequences for Comparative Genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Paterson, A.H. Angiosperm Genome Comparisons Reveal Early Polyploidy in the Monocot Lineage. Proc. Natl. Acad. Sci. USA 2010, 107, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated Syntenic and Phylogenomic Analyses Reveal an Ancient Genome Duplication in Monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E.; et al. The Pineapple Genome and the Evolution of CAM Photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef]
- Devos, K.M. Grass Genome Organization and Evolution. Curr. Opin. Plant Biol. 2010, 13, 139–145. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, W.; Zhang, L.; Li, D.-Z.; Qi, J.; Ma, H. Phylogenomic Profiles of Whole-Genome Duplications in Poaceae and Landscape of Differential Duplicate Retention and Losses among Major Poaceae Lineages. Nat. Commun. 2024, 15, 3305. [Google Scholar] [CrossRef]
- Zhou, X.; Jellen, E.N.; Murphy, J.P. Progenitor Germplasm of Domisticated Hexaploid Oat. Crop Sci. 1999, 39, 1208–1214. [Google Scholar] [CrossRef]
- VanBuren, R.; Man Wai, C.; Wang, X.; Pardo, J.; Yocca, A.E.; Wang, H.; Chaluvadi, S.R.; Han, G.; Bryant, D.; Edger, P.P.; et al. Exceptional Subgenome Stability and Functional Divergence in the Allotetraploid Ethiopian Cereal Teff. Nat. Commun. 2020, 11, 884. [Google Scholar] [CrossRef] [PubMed]
- Devos, K.M.; Qi, P.; Bahri, B.A.; Gimode, D.M.; Jenike, K.; Manthi, S.J.; Lule, D.; Lux, T.; Martinez-Bello, L.; Pendergast, T.H., 4th; et al. Genome Analyses Reveal Population Structure and a Purple Stigma Color Gene Candidate in Finger Millet. Nat. Commun. 2023, 14, 3694. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhang, J.; Liu, K.-B.; Wu, N.; Li, Y.; Zhou, K.; Ye, M.; Zhang, T.; Zhang, H.; Yang, X.; et al. Earliest Domestication of Common Millet (Panicum Miliaceum) in East Asia Extended to 10,000 Years Ago. Proc. Natl. Acad. Sci. USA 2009, 106, 7367–7372. [Google Scholar] [CrossRef]
- Stevens, C.J.; Fuller, D.Q. The Spread of Agriculture in Eastern Asia. Lang. Dyn. Change 2017, 7, 152–186. [Google Scholar] [CrossRef]
- Barton, L.; Newsome, S.D.; Chen, F.-H.; Wang, H.; Guilderson, T.P.; Bettinger, R.L. Agricultural Origins and the Isotopic Identity of Domestication in Northern China. Proc. Natl. Acad. Sci. USA 2009, 106, 5523–5528. [Google Scholar] [CrossRef]
- Stevens, C.J.; Murphy, C.; Roberts, R.; Lucas, L.; Silva, F.; Fuller, D.Q. Between China and South Asia: A Middle Asian Corridor of Crop Dispersal and Agricultural Innovation in the Bronze Age. Holocene 2016, 26, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Robbeets, M.; Bouckaert, R.; Conte, M.; Savelyev, A.; Li, T.; An, D.-I.; Shinoda, K.-I.; Cui, Y.; Kawashima, T.; Kim, G.; et al. Triangulation Supports Agricultural Spread of the Transeurasian Languages. Nature 2021, 599, 616–621. [Google Scholar] [CrossRef]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Alam, O.; Li, H.; Zhang, H.; Xing, L.; et al. A Graph-Based Genome and Pan-Genome Variation of the Model Plant Setaria. Nat. Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef]
- Kihara, H.; Kishimoto, E. Bastarde Zwischen Setaria italica Und Setaria Viridis. Bot. Mag. Tokyo 1942, 20, 63–67. [Google Scholar]
- Doust, A.N.; Kellogg, E.A. Effect of Genotype and Environment on Branching in Weedy Green Millet (Setaria viridis) and Domesticated Foxtail Millet (Setaria italica) (Poaceae). Mol. Ecol. 2006, 15, 1335–1349. [Google Scholar] [CrossRef]
- Wang, C.; Chen, J.; Zhi, H.; Yang, L.; Li, W.; Wang, Y.; Li, H.; Zhao, B.; Chen, M.; Diao, X. Population Genetics of Foxtail Millet and Its Wild Ancestor. BMC Genet. 2010, 11, 90. [Google Scholar] [CrossRef]
- Zhu, Q.; Zheng, X.; Luo, J.; Gaut, B.S.; Ge, S. Multilocus Analysis of Nucleotide Variation of Oryza Sativa and Its Wild Relatives: Severe Bottleneck during Domestication of Rice. Mol. Biol. Evol. 2007, 24, 875–888. [Google Scholar] [CrossRef]
- Ashikari, M.; Matsuoka, M. Identification, Isolation and Pyramiding of Quantitative Trait Loci for Rice Breeding. Trends Plant Sci. 2006, 11, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Till-Bottraud, I.; Reboud, X.; Brabant, P.; Lefranc, M.; Rherissi, B.; Vedel, F.; Darmency, H. Outcrossing and Hybridization in Wild and Cultivated Foxtail Millets: Consequences for the Release of Transgenic Crops. Züchter Genet. Breed. Res. 1992, 83, 940–946. [Google Scholar] [CrossRef]
- Rice, A.; Glick, L.; Abadi, S.; Einhorn, M.; Kopelman, N.M.; Salman-Minkov, A.; Mayzel, J.; Chay, O.; Mayrose, I. The Chromosome Counts Database (CCDB)—A Community Resource of Plant Chromosome Numbers. New Phytol. 2015, 206, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Orr-Weaver, T.L. When Bigger Is Better: The Role of Polyploidy in Organogenesis. Trends Genet. 2015, 31, 307–315. [Google Scholar] [CrossRef]
- Pinto, S.C.; Stojilković, B.; Zhang, X.; Sablowski, R. Plant Cell Size: Links to Cell Cycle, Differentiation and Ploidy. Curr. Opin. Plant Biol. 2024, 78, 102527. [Google Scholar] [CrossRef]
- Li, Y.; Jia, J.; Wang, Y.; Wu, S. Intraspecific and Interspecific Variation in Setaria Revealed by RAPD Analysis. Genet. Resour. Crop Evol. 1998, 45, 279–285. [Google Scholar] [CrossRef]
- Benabdelmouna, A.; Abirached-Darmency, M.; Darmency, H. Phylogenetic and Genomic Relationships in Setaria italica and Its Close Relatives Based on the Molecular Diversity and Chromosomal Organization of 5S and 18S-5.8S-25S RDNA Genes. Züchter Genet. Breed. Res. 2001, 103, 668–677. [Google Scholar] [CrossRef]
- Benabdelmouna, A.; Shi, Y.; Abirached-Darmency, M.; Darmency, H. Genomic in Situ Hybridization (GISH) Discriminates between the A and the B Genomes in Diploid and Tetraploid Setaria Species. Genome 2001, 44, 685–690. [Google Scholar] [CrossRef]
- Wang, Y.; Zhi, H.; Li, W.; Li, H.; Wang, Y.; Huang, Z.; Diao, X. A Novel Genome of C and the First Autotetraploid Species in the Setaria Genus Identified by Genomic in Situ Hybridization. Genet. Resour. Crop Evol. 2009, 56, 843–850. [Google Scholar] [CrossRef]
- Li, W.; Zhi, H.; Wang, Y.-F.; Li, H.-Q.; Diao, X.-M. Assessment of Genetic Relationship of Foxtail Millet with Its Wild Ancestor and Close Relatives by ISSR Markers. J. Integr. Agric. 2012, 11, 556–566. [Google Scholar] [CrossRef]
- Zhao, M.; Zhi, H.; Doust, A.N.; Li, W.; Wang, Y.; Li, H.; Jia, G.; Wang, Y.; Zhang, N.; Diao, X. Novel Genomes and Genome Constitutions Identified by GISH and 5S RDNA and Knotted1 Genomic Sequences in the Genus Setaria. BMC Genom. 2013, 14, 244. [Google Scholar] [CrossRef]
- Gould, F.W. Chromosome Numbers of Texas Grasses. Can. J. Bot. 1968, 46, 1315–1325. [Google Scholar] [CrossRef]
- Reeder, J.R. Notes on Mexican Grasses IX. Miscellaneous Chromosome Numbers-3. Brittonia 1971, 23, 105–117. [Google Scholar] [CrossRef]
- Li, S.-X.; Wang, Z.-H.; Malhi, S.S.; Li, S.-Q.; Gao, Y.-J.; Tian, X.-H. Chapter 7 Nutrient and Water Management Effects on Crop Production, and Nutrient and Water Use Efficiency in Dryland Areas of China. In Advances in Agronomy; Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2009; pp. 223–265. ISBN 9780123748188. [Google Scholar]
- Li, C.; Liu, M.; Sun, F.; Zhao, X.; He, M.; Li, T.; Lu, P.; Xu, Y. Genetic Divergence and Population Structure in Weedy and Cultivated Broomcorn Millets (Panicum miliaceum L.) Revealed by Specific-Locus Amplified Fragment Sequencing (SLAF-Seq). Front. Plant Sci. 2021, 12, 688444. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Liu, M.; Guo, W.; Wang, Y.; He, Q.; Chen, W.; Liao, Y.; Zhang, W.; Gao, Y.; et al. Pangenome Analysis Reveals Genomic Variations Associated with Domestication Traits in Broomcorn Millet. Nat. Genet. 2023, 55, 2243–2254. [Google Scholar] [CrossRef]
- de Wet, J.M.J.; Brink, D.E.; Rao, K.E.P.; Mengesha, M.H. Diversity in Kodo Millet, Paspalum scrobiculatum. Econ. Bot. 1983, 37, 159–163. [Google Scholar] [CrossRef]
- Weber, S.; Kashyap, A. The Vanishing Millets of the Indus Civilization. Archaeol. Anthropol. Sci. 2016, 8, 9–15. [Google Scholar] [CrossRef]
- Hiremath, S.C.; Patil, G.N.V.; Salimath, S.S. Genome Homology and Origin of Panicum Sumatrense (Gramineae). Cytologia 1990, 55, 315–319. [Google Scholar] [CrossRef]
- Kitagawa, M. A Contribution to the Flora of Manchuria. Bot. Mag. Tokyo 1937, 51, 153–154. [Google Scholar]
- Scholz, H. Die Unkraut-Hirse (Panicum miliaceum subsp. ruderale)—Neue Tatsachen und Befunde. Osterr. Bot. Z. 1983, 143, 233–244. [Google Scholar] [CrossRef]
- Haroun, S.A. Cytological Abnormality Control Seed Set in Panicum repens L. in Egypt. Cytologia 1995, 60, 347–351. [Google Scholar] [CrossRef][Green Version]
- Pessim, C.; Pagliarini, M.S.; Silva, N.; Jank, L. Chromosome Stickiness Impairs Meiosis and Influences Reproductive Success in Panicum Maximum (Poaceae) Hybrid Plants. Genet. Mol. Res. 2015, 14, 4195–4202. [Google Scholar] [CrossRef]
- Lovell, J.T.; Jenkins, J.; Lowry, D.B.; Mamidi, S.; Sreedasyam, A.; Weng, X.; Barry, K.; Bonnette, J.; Campitelli, B.; Daum, C.; et al. The Genomic Landscape of Molecular Responses to Natural Drought Stress in Panicum hallii. Nat. Commun. 2018, 9, 5213. [Google Scholar] [CrossRef]
- Lovell, J.T.; MacQueen, A.H.; Mamidi, S.; Bonnette, J.; Jenkins, J.; Napier, J.D.; Sreedasyam, A.; Healey, A.; Session, A.; Shu, S.; et al. Genomic Mechanisms of Climate Adaptation in Polyploid Bioenergy Switchgrass. Nature 2021, 590, 438–444. [Google Scholar] [CrossRef]
- Triplett, J.K.; Wang, Y.; Zhong, J.; Kellogg, E.A. Five Nuclear Loci Resolve the Polyploid History of Switchgrass (Panicum virgatum L.) and Relatives. PLoS ONE 2012, 7, e38702. [Google Scholar] [CrossRef]
- Hunt, H.V.; Badakshi, F.; Romanova, O.; Howe, C.J.; Jones, M.K.; Heslop-Harrison, J.S.P. Reticulate Evolution in Panicum (Poaceae): The Origin of Tetraploid Broomcorn Millet, P. Miliaceum. J. Exp. Bot. 2014, 65, 3165–3175. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, S.; Yang, Z.; Lai, J.; Gao, X.; Shi, J. A High-Quality, Phased Genome Assembly of Broomcorn Millet Reveals the Features of Its Subgenome Evolution and 3D Chromatin Organization. Plant Commun. 2023, 4, 100557. [Google Scholar] [CrossRef]
- Hamoud, M.A.; Haroun, S.A.; MacLeod, R.D.; Richards, A.J. Cytological Relationships of Selected Species of Panicum L. Biol. Plant. 1994, 36, 37–45. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Shi, J.; Wang, L.; Liang, C.; Yang, J.; Chen, J.; Chen, M. Biased Mutations and Gene Losses Underlying Diploidization of the Tetraploid Broomcorn Millet Genome. Plant J. 2023, 113, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Fuller, D.Q.; Huan, X.; Perry, L.; Li, Q.; Li, Z.; Zhang, J.; Ma, Z.; Zhuang, Y.; Jiang, L.; et al. Barnyard Grasses Were Processed with Rice around 10000 Years Ago. Sci. Rep. 2015, 5, 16251. [Google Scholar] [CrossRef]
- Yoshizaki, M. Emergence of Cultivated Plant Species in Japan. Archaeol. Q. 1995, 50, 18–24. [Google Scholar]
- Yoshizaki, M. Domesticated Plants of the Jomon Period. Quat. Quat. Res. 1997, 36, 343–346. [Google Scholar]
- Nasu, H. Domestrication of Plants during the Jomon Period. Quat. Res. 2018, 57, 109–126. [Google Scholar] [CrossRef]
- Seetharam, A.; Riley, K.W.; Harinarayana, G. (Eds.) Small Millets in Global Agriculture; Intercept: Wimborne, UK, 1989; ISBN 9788120404342. [Google Scholar]
- Damalas, C.A.; Dhima, K.V.; Eleftherohorinos, I.G. Morphological and Physiological Variation among Species of the Genus Echinochloa in Northern Greece. Weed Sci. 2008, 56, 416–423. [Google Scholar] [CrossRef]
- Wu, D.; Shen, E.; Jiang, B.; Feng, Y.; Tang, W.; Lao, S.; Jia, L.; Lin, H.-Y.; Xie, L.; Weng, X.; et al. Genomic Insights into the Evolution of Echinochloa Species as Weed and Orphan Crop. Nat. Commun. 2022, 13, 689. [Google Scholar] [CrossRef]
- Chu, Q.; Gong, X.; Hu, Y.; Zhu, Q.-H.; Fan, L.; Ye, C.-Y. The DNA Methylomes of Echinochloa Species Provide Insights into Polyploidization-Driven Adaptation and Orphan Crop Domestication. Plant J. 2025, 121, e70033. [Google Scholar] [CrossRef]
- Arrigo, N.; Barker, M.S. Rarely Successful Polyploids and Their Legacy in Plant Genomes. Curr. Opin. Plant Biol. 2012, 15, 140–146. [Google Scholar] [CrossRef]
- Salman-Minkov, A.; Sabath, N.; Mayrose, I. Whole-Genome Duplication as a Key Factor in Crop Domestication. Nat. Plants 2016, 2, 16115. [Google Scholar] [CrossRef]
- He, F.; Pasam, R.; Shi, F.; Kant, S.; Keeble-Gagnere, G.; Kay, P.; Forrest, K.; Fritz, A.; Hucl, P.; Wiebe, K.; et al. Exome Sequencing Highlights the Role of Wild-Relative Introgression in Shaping the Adaptive Landscape of the Wheat Genome. Nat. Genet. 2019, 51, 896–904. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, X.; Li, Y.; Xu, J.; Bi, A.; Kang, L.; Xu, D.; Chen, H.; Wang, Y.; Wang, Y.-G.; et al. Triticum Population Sequencing Provides Insights into Wheat Adaptation. Nat. Genet. 2020, 52, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, W.; Xie, X.; Wang, Y.; Yang, Z.; Peng, H.; Xin, M.; Yao, Y.; Hu, Z.; Liu, J.; et al. Dispersed Emergence and Protracted Domestication of Polyploid Wheat Uncovered by Mosaic Ancestral Haploblock Inference. Nat. Commun. 2022, 13, 3891. [Google Scholar] [CrossRef]
- Dempewolf, H.; Hodgins, K.A.; Rummell, S.E.; Ellstrand, N.C.; Rieseberg, L.H. Reproductive Isolation during Domestication. Plant Cell 2012, 24, 2710–2717. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.; Goldberg, E.E.; Igić, B. Comparative Evidence for the Correlated Evolution of Polyploidy and Self-Compatibility in Solanaceae. Evolution 2011, 65, 139–155. [Google Scholar] [CrossRef]
- Shimizu, K.K.; Tsuchimatsu, T. Evolution of Selfing: Recurrent Patterns in Molecular Adaptation. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 593–622. [Google Scholar] [CrossRef]
- Landry, C.R.; Lemos, B.; Rifkin, S.A.; Dickinson, W.J.; Hartl, D.L. Genetic Properties Influencing the Evolvability of Gene Expression. Science 2007, 317, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Des Marais, D.L.; Rausher, M.D. Escape from Adaptive Conflict after Duplication in an Anthocyanin Pathway Gene. Nature 2008, 454, 762–765. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene Duplication and Evolutionary Novelty in Plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative Genomic and Transcriptomic Analysis Suggests the Evolutionary Dynamic of GH3 Genes in Gramineae Crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef]
- Kong, W.; Gong, Z.; Zhong, H.; Zhang, Y.; Zhao, G.; Gautam, M.; Deng, X.; Liu, C.; Zhang, C.; Li, Y. Expansion and Evolutionary Patterns of Glycosyltransferase Family 8 in Gramineae Crop Genomes and Their Expression under Salt and Cold Stresses in Oryza Sativa ssp. Japonica. Biomol. 2019, 9, 188. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-Box Genes and Crop Domestication: The Jack of All Traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, L.-L.; Xu, Z.-S.; Fu, L.; Pang, H.-X.; Ma, Y.-Z.; Min, D.-H. Genome-Wide Analysis of MADS-Box Genes in Foxtail Millet (Setaria italica L.) and Functional Assessment of the Role of SiMADS51 in the Drought Stress Response. Front. Plant Sci. 2021, 12, 659474. [Google Scholar] [CrossRef]
- Lai, D.; Yan, J.; He, A.; Xue, G.; Yang, H.; Feng, L.; Wei, X.; Li, L.; Xiang, D.; Ruan, J.; et al. Genome-Wide Identification, Phylogenetic and Expression Pattern Analysis of MADS-Box Family Genes in Foxtail Millet (Setaria italica). Sci. Rep. 2022, 12, 4979. [Google Scholar] [CrossRef]
- Gao, H.; Suo, X.; Zhao, L.; Ma, X.; Cheng, R.; Wang, G.; Zhang, H. Molecular Evolution, Diversification, and Expression Assessment of MADS Gene Family in Setaria italica, Setaria viridis, and Panicum virgatum. Plant Cell Rep. 2023, 42, 1003–1024. [Google Scholar] [CrossRef]
- Ronald, P.C. Resistance Gene Evolution. Curr. Opin. Plant Biol. 1998, 1, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.; Kreitman, M.; Stahl, E.A.; Tian, D. Evolutionary Dynamics of Plant R-Genes. Science 2001, 292, 2281–2285. [Google Scholar] [CrossRef]
- Andersen, E.J.; Nepal, M.P. Genetic Diversity of Disease Resistance Genes in Foxtail Millet (Setaria italica L.). Plant Gene 2017, 10, 8–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, M.; Shen, J.; Song, X.; Dong, S.; Wen, Y.; Yuan, X.; Guo, P. Comparative Genomics Analysis in Grass Species Reveals Two Distinct Evolutionary Strategies Adopted by R Genes. Sci. Rep. 2019, 9, 10735. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kawase, M.; Kato, K. Structural Variation in the Waxy Gene and Differentiation in Foxtail Millet [Setaria italica (L.) P. Beauv.]: Implications for Multiple Origins of the Waxy Phenotype. Mol. Genet. Genom. 2002, 268, 214–222. [Google Scholar] [CrossRef]
- Araki, M.; Numaoka, A.; Kawase, M.; Fukunaga, K. Origin of Waxy Common Millet, Panicum miliaceum L. in Japan. Genet. Resour. Crop Evol. 2012, 59, 1303–1308. [Google Scholar] [CrossRef]
- Hoshino, T.; Nakamura, T.; Seimiya, Y.; Kamada, T.; Ishikawa, G.; Ogasawara, A.; Sagawa, S.; Saito, M.; Shimizu, H.; Nishi, M.; et al. Production of a Fully Waxy Line and Analysis of Waxy Genes in the Allohexaploid Crop, Japanese Barnyard Millet. Plant Breed. 2010, 129, 349–355. [Google Scholar]
| Species | Number of Chromosomes | Median Ploidy Level | Genome |
|---|---|---|---|
| S. italica | 18, 36 | 2n = 2x = 18 | AA |
| S. viridis | 18 | 2n = 2x = 18 | AA |
| S. leucopila | 18, 54, 68, 72, 108 | 2n = 2x = 18 | AA? |
| S. queenslandica | 18, 36 | 2n = 2x = 18 | AA |
| S. faberi | 36 | 2n = 4x = 36 | AAAA? AABB? |
| S. verticillata | 18, 27, 36, 54 | 2n = 4x = 36 | AABB |
| S. adhaerens | 18 | 2n = 2x = 18 | BB |
| S. grisebachii | 18 | 2n = 2x = 18 | CC |
| S. lachnea | 36 | 2n = 4x = 36 | CCC’C’ |
| S. glauca | 36 | 2n = 4x = 36 | DDDD |
| S. parviflora | 36 | 2n = 4x = 36 | DDDD |
| S. plicata | 18, 36, 54, 72 | 2n = 4x = 36 | EEEE? |
| S. palmifolia | 34, 36, 54 | 2n = 6x = 54 | EEEEEE? |
| S. arenaria | 54 | 2n = 6x = 54 | FFFFFF? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satomura, K. Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta. Genes 2025, 16, 426. https://doi.org/10.3390/genes16040426
Satomura K. Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta. Genes. 2025; 16(4):426. https://doi.org/10.3390/genes16040426
Chicago/Turabian StyleSatomura, Kazuhiro. 2025. "Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta" Genes 16, no. 4: 426. https://doi.org/10.3390/genes16040426
APA StyleSatomura, K. (2025). Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta. Genes, 16(4), 426. https://doi.org/10.3390/genes16040426








