Genome-Wide Identification and Characterization of the RWP-RK Proteins in Zanthoxylum armatum
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Properties of ZaRWP-RK Genes
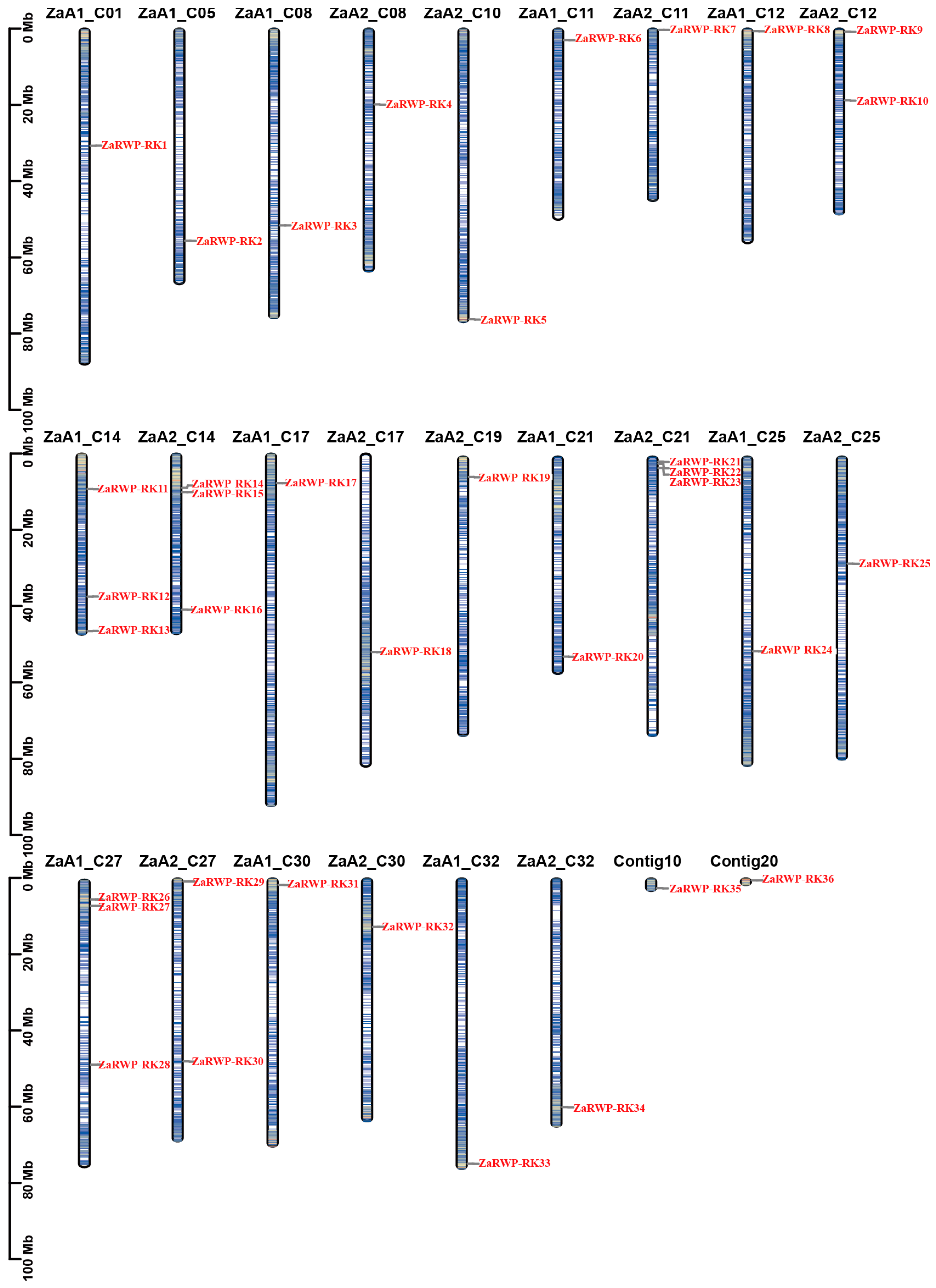
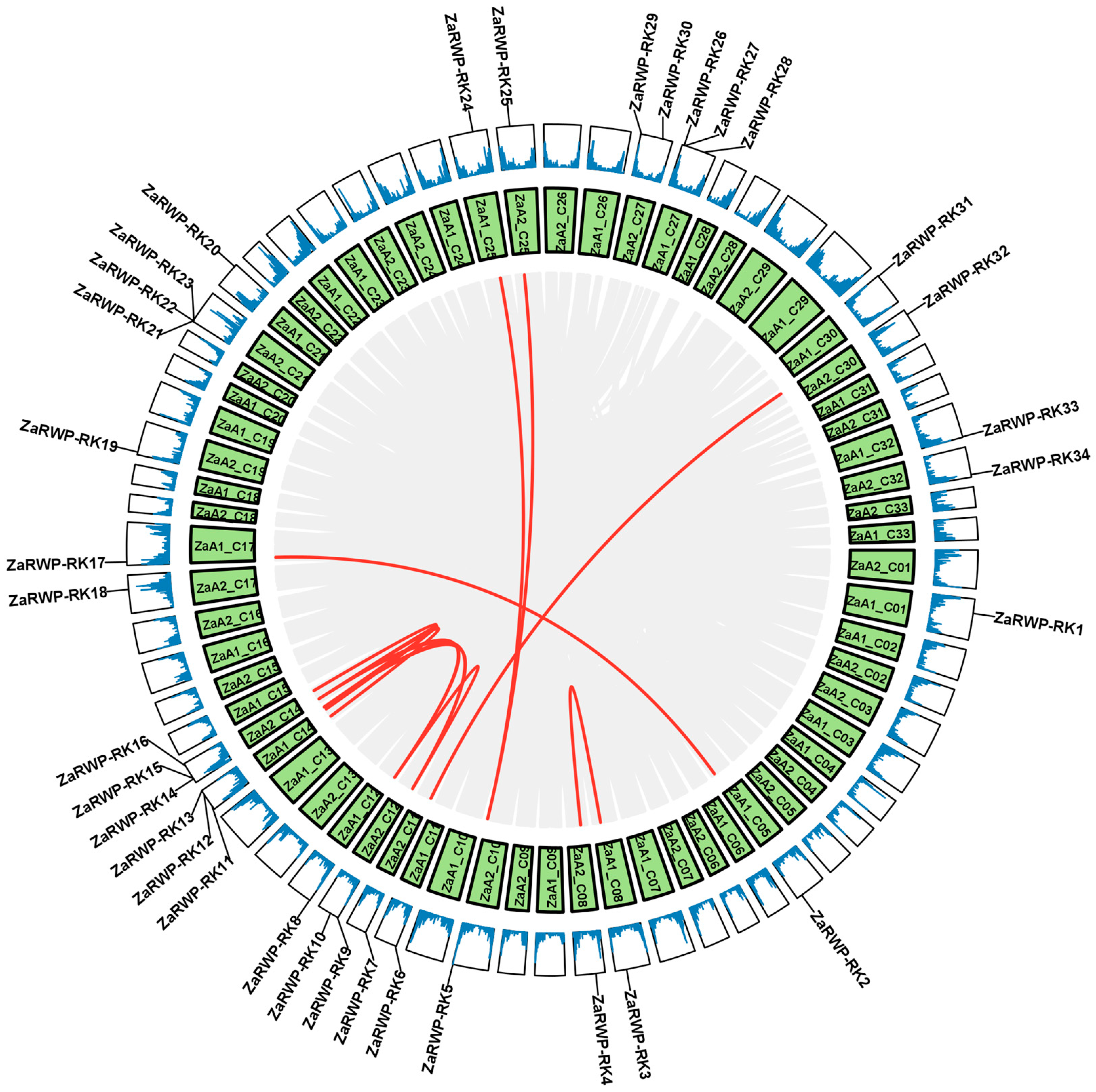
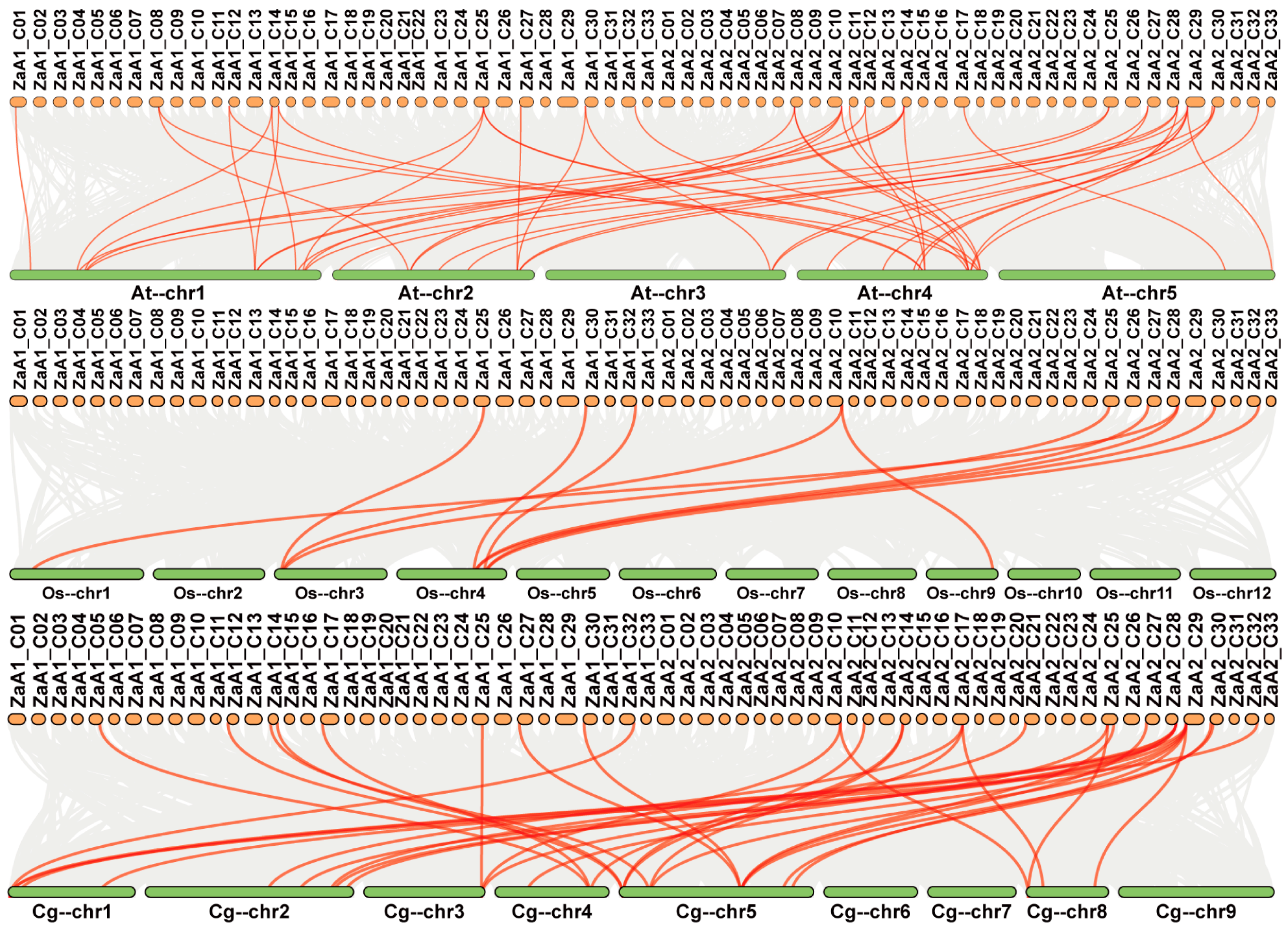
2.2. Chromosomal Location and Gene Duplication Events
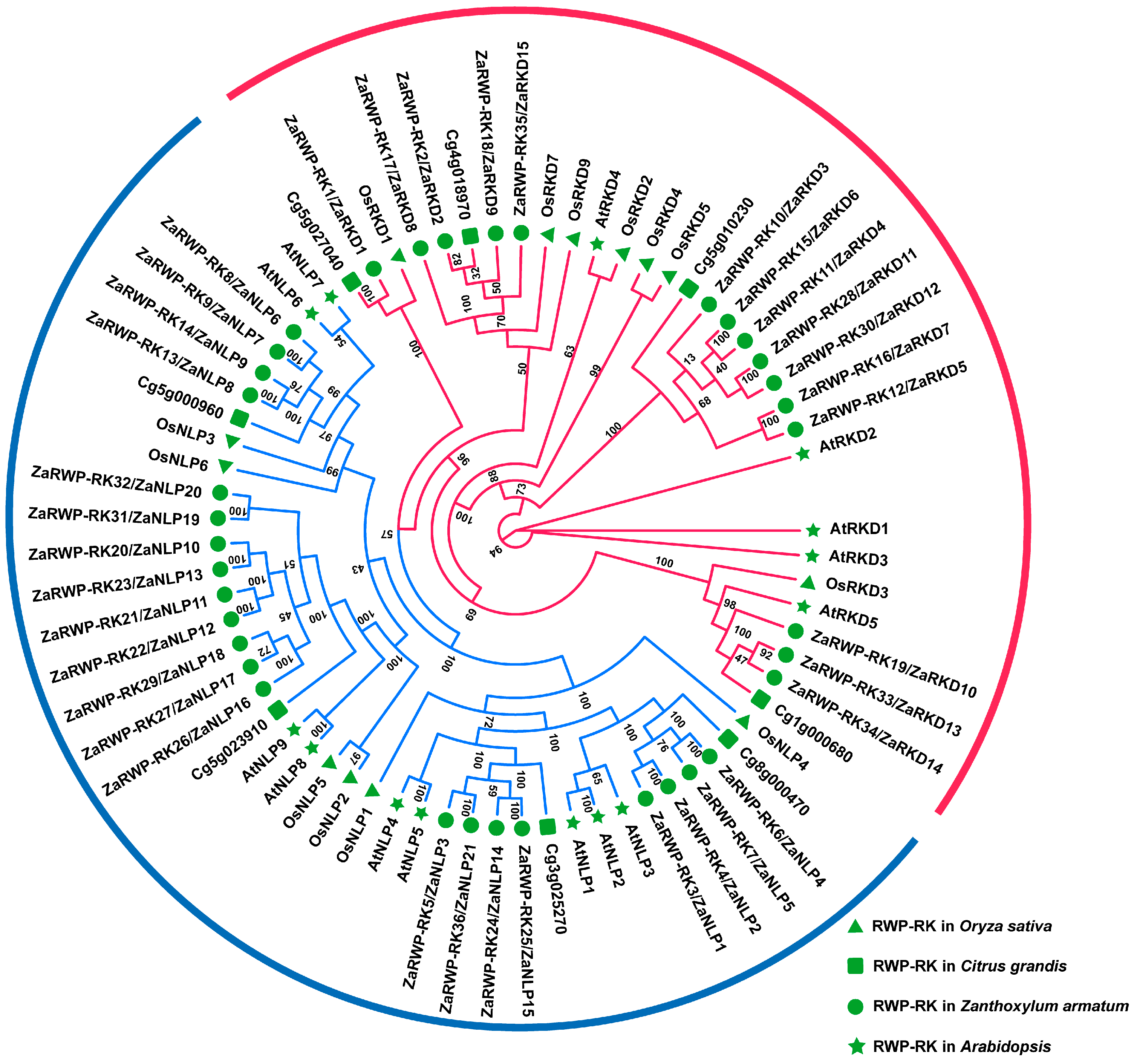
2.3. Phylogenetic Analysis of ZaRWP-RK Genes
2.4. Conserved Structural Domains, Conserved Motifs, and Cis-Element Analyses of ZaRWP-RK Genes
2.5. Plant Materials, RNA Extraction, and qRT-PCR Analysis
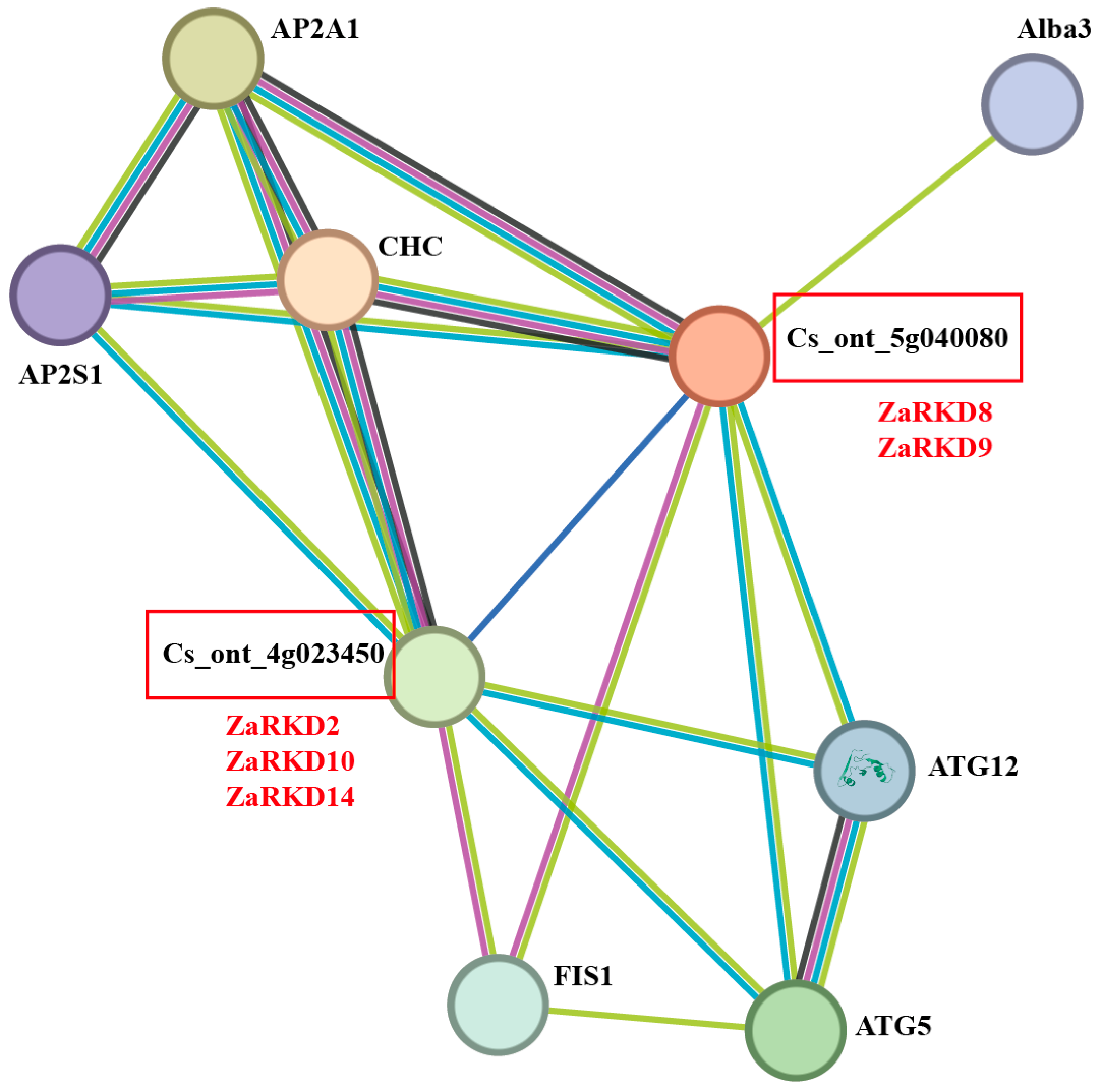
2.6. Protein–Protein Interaction Network Prediction of ZaRWP-RK Proteins
3. Results
3.1. Genome-Wide Identification and Physicochemical Properties of ZaRWP-RK Genes
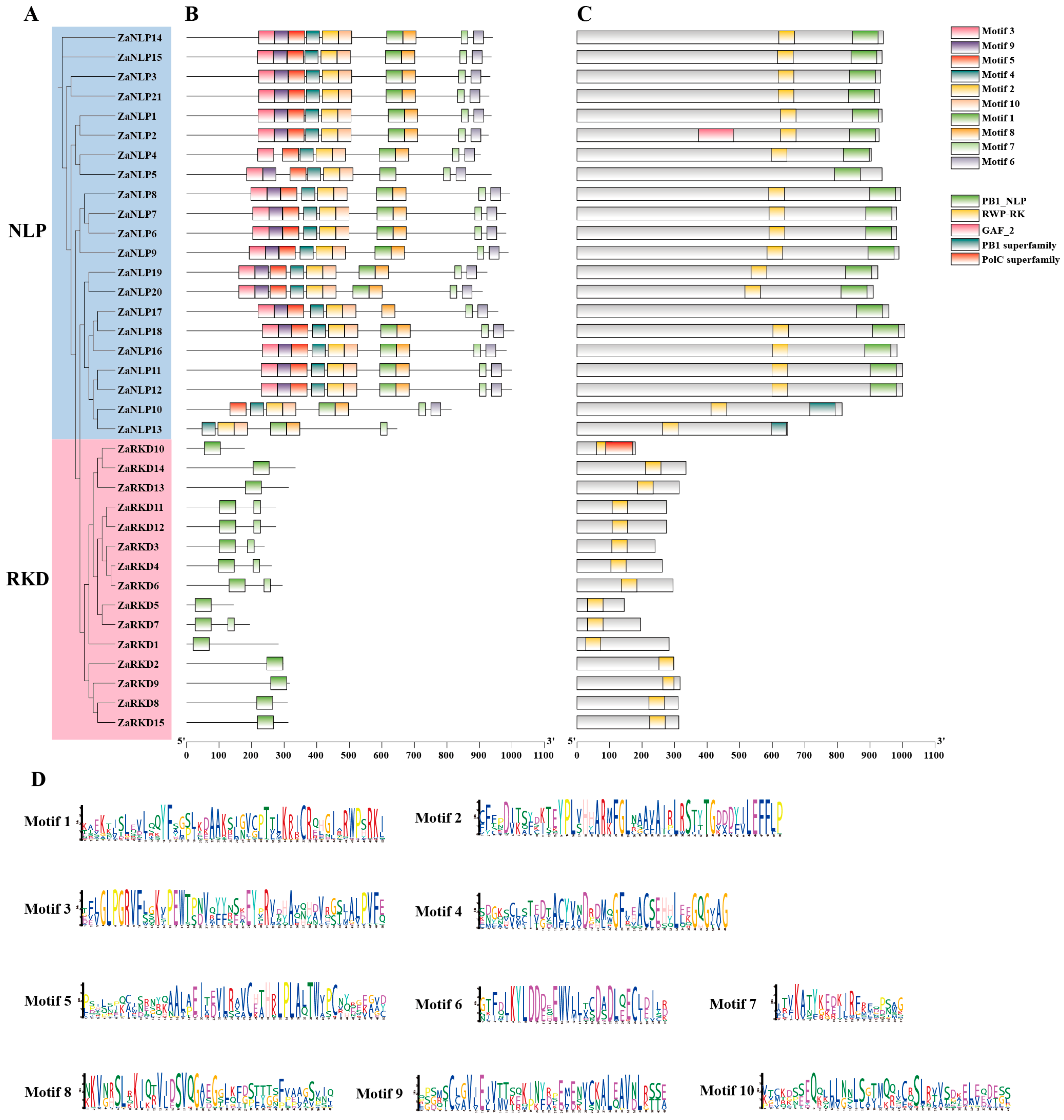
3.2. Phylogenetic Relationship, Conserved Structural Domains, and Motifs Analysis of ZaRWP-RK Proteins
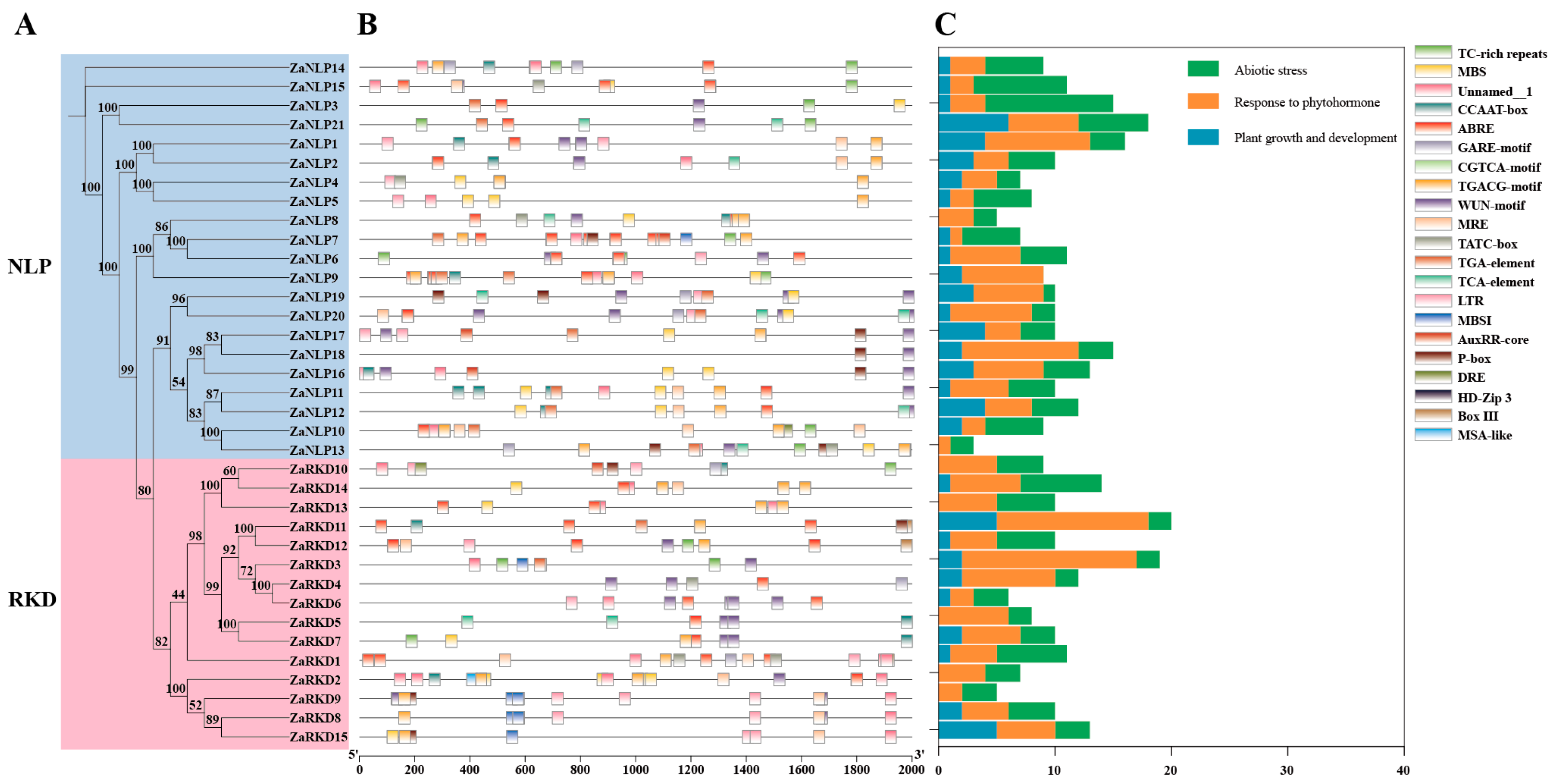
3.3. Cis-Element Analysis of ZaRWP-RK Promoters
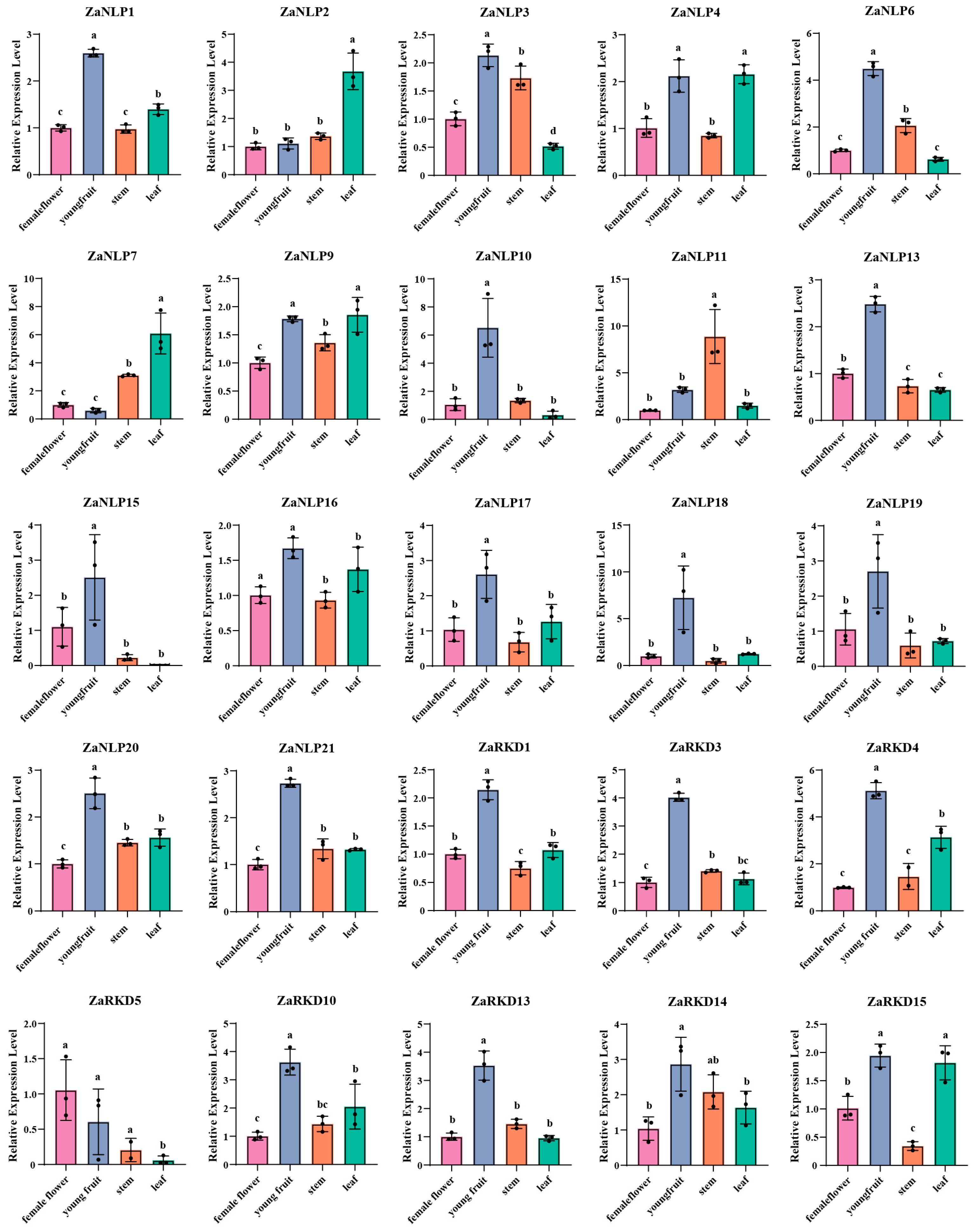
3.4. Tissue-Specific Expression of ZaRWP-RK Genes
3.5. Correlation Analysis of Apomixis-Related ZaRWP-RK Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, T.; Zhu, X.; Ye, Z.; Wang, Y.F.; Yao, C.; Xu, N.; Zhou, M.; Ma, J.; Qin, Y.; Shen, Y.; et al. Lupus enhancer risk variant causes dysregulation of IRF8 through cooperative lncRNA and DNA methylation machinery. Nat. Commun. 2022, 13, 1855. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Zhuo, M.; Yanagisawa, S. RWP-RK domain-containing transcription factors in the Viridiplantae: Biology and phylogenetic relationships. J. Exp. Bot. 2022, 73, 4323–4337. [Google Scholar] [CrossRef]
- Yin, M.; Zhang, Z.; Xuan, M.; Feng, H.; Ye, W.; Zheng, X.; Wang, Y. Conserved subgroups of the plant-specific RWP-RK transcription factor family are present in oomycete pathogens. Front. Microbiol. 2020, 11, 1724. [Google Scholar] [CrossRef]
- Mu, X.; Luo, J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell. Mol. Life Sci. 2019, 76, 3753–3764. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.; Ahmad, N.; Khalifa, M.A.S.; Du, Y.; Mandozai, A.; Khattak, A.N.; Piwu, W. Identification and molecular characterization of RWP-RK transcription factors in soybean. Genes 2023, 14, 369. [Google Scholar] [CrossRef] [PubMed]
- Waki, T.; Hiki, T.; Watanabe, R.; Hashimoto, T.; Nakajima, K. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr. Biol. 2011, 21, 1277–1281. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, F.; Rizzo, P.; Rutten, T.; Altschmied, L.; Bäumlein, H. RWP-RK domain-containing transcription factors control cell differentiation during female gametophyte development in Arabidopsis. New Phytol. 2017, 213, 1909–1924. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, H.; Huang, W.; Yi, L.; Qin, E.; Yang, T.; Wang, J.; Qin, R. Genome-wide identification, characterization, and regulation of RWP-RK gene family in the nitrogen-fixing clade. Plants 2020, 9, 1178. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Liu, M.; Lin, Z.; Wang, Z.F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef]
- Jiang, S.; Jardinaud, M.F.; Gao, J.; Pecrix, Y.; Wen, J.; Mysore, K.; Xu, P.; Sanchez-Canizares, C.; Ruan, Y.; Li, Q.; et al. NIN-like protein transcription factors regulate leghemoglobin genes in legume nodules. Science 2021, 374, 625–628. [Google Scholar] [CrossRef]
- Yan, H.; Sun, M.; Zhang, Z.; Jin, Y.; Zhang, A.; Lin, C.; Wu, B.; He, M.; Xu, B.; Wang, J.; et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat. Genet. 2023, 55, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Underwood, C.J. Apomixis. Curr. Biol. 2023, 33, R293–R295. [Google Scholar] [CrossRef] [PubMed]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Vijverberg, K.; Rigola, D.; Okamoto, S.; Oplaat, C.; Camp, R.H.O.D.; Radoeva, T.; Schauer, S.E.; Fierens, J.; Jansen, K. A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat. Genet. 2022, 54, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liang, Y.; Xie, Y.; Rao, Y.; Xiong, J.; Liu, C.; Wang, C.; Wang, X.; Qian, Q.; Wang, K. Efficient haploid induction via egg cell expression of dandelion PARTHENOGENESIS in foxtail millet (Setaria italica). Plant Biotechnol. J. 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Endo, T.; Fujii, H.; Nakano, M.; Sugiyama, A.; Daido, G.; Ohta, S.; Yoshioka, T.; Omura, M. MITE insertion-dependent expression of CitRKD1 with a RWP-RK domain regulates somatic embryogenesis in citrus nucellar tissues. BMC Plant Biol. 2018, 18, 166. [Google Scholar] [CrossRef]
- Purwestri, Y.A.; Lee, Y.S.; Meehan, C.; Mose, W.; Susanto, F.A.; Wijayanti, P.; Fauzia, A.N.; Nuringtyas, T.R.; Hussain, N.; Putra, H.L.; et al. RWP-RK Domain 3 (OsRKD3) induces somatic embryogenesis in black rice. BMC Plant Biol. 2023, 23, 202. [Google Scholar] [CrossRef]
- Liu, J.; Wan, J.; Zhang, Y.; Hou, X.; Shen, G.; Li, S.; Luo, Q.; Li, Q.; Zhou, M.; Liu, X.; et al. The establishment of comprehensive quality evaluation model for flavor characteristics of green Sichuan pepper (Zanthoxylum armatum DC.) in Southwest China. Food Chem. X 2023, 18, 100721. [Google Scholar] [CrossRef]
- Tang, N.; Cao, Z.; Wu, P.; Zhang, X.; Lou, J.; Liu, Y.; Wang, Q.; Hu, Y.; Si, S.; Sun, X.; et al. Genome-wide identification, interaction of the MADS-box proteins in Zanthoxylum armatum and functional characterization of ZaMADS80 in floral development. Front. Plant Sci. 2022, 13, 1038828. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, J.; Li, J.; Li, Y.; Zeng, X. Genome-wide identification and analysis of the growth-regulating factor family in Zanthoxylum armatum DC and functional analysis of ZaGRF6 in leaf size and longevity regulation. Int. J. Mol. Sci. 2022, 23, 9043. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Pullaiah, T. Apomixis in Angiosperms: Mechanisms, Occurrences, and Biotechnology, 1st ed.; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Wang, M.; Tong, S.; Ma, T.; Xi, Z.; Liu, J. Chromosome-level genome assembly of Sichuan pepper provides insights into apomixis, drought tolerance, and alkaloid biosynthesis. Mol. Ecol. Resour. 2021, 21, 2533–2545. [Google Scholar] [CrossRef]
- Song, L.; Huang, Y.; Zuo, H.; Tang, N.; Li, Z.; Jiao, W.; Xu, F.; Xu, Q.; Chen, Z. Chromosome-level assembly of triploid genome of Sichuan pepper (Zanthoxylum armatum). Hortic. Plant J. 2024, 10, 437–449. [Google Scholar] [CrossRef]
- Hu, L.; Xu, Z.; Fan, R.; Wang, G.; Wang, F.; Qin, X.; Yan, L.; Ji, X.; Meng, M.; Sim, S.; et al. The complex genome and adaptive evolution of polyploid Chinese pepper (Zanthoxylum armatum and Zanthoxylum bungeanum). Plant Biotechnol. J. 2023, 21, 78–96. [Google Scholar] [CrossRef]
- Fei, X.; Shi, Q.; Qi, Y.; Wang, S.; Lei, Y.; Hu, H.; Liu, Y.; Yang, T.; Wei, A. ZbAGL11, a class D MADS-box transcription factor of Zanthoxylum bungeanum, is involved in sporophytic apomixis. Hortic. Res. 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jin, Y.; Lu, Q.H.; Ren, N.; Wang, Y.Q.; Li, Q.S. Genome-wide identification and expression analysis of NIN-like protein (NLP) genes: Exploring their potential roles in nitrate response in tea plant (Camellia sinensis). Plant Physiol. Biochem. 2024, 207, 108340. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chang, W.; Fan, Y.; Sun, W.; Qu, C.; Zhang, K.; Liu, L.; Xu, X.; Tang, Z.; Li, J.; et al. Genome-wide identification and characterization of NODULE-INCEPTION-Like protein (NLP) family genes in Brassica napus. Int. J. Mol. Sci. 2018, 19, 2270. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. RaxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res 2023, 51, 384–388. [Google Scholar] [CrossRef]
- Chow, C.N.; Yang, C.W.; Wu, N.Y.; Wang, H.T.; Tseng, K.C.; Chiu, Y.H.; Lee, T.Y.; Chang, W.C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Wieloch, W.; Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005, 60, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Liu, Z.; Cheng, J.; Li, Z.; Tian, L.; Liu, M.; Yang, T.; Liu, Y.; Liu, Y.; Dai, H.; et al. Zanthoxylum-specific whole genome duplication and recent activity of transposable elements in the highly repetitive paleotetraploid Z. bungeanum genome. Hortic. Res. 2021, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Chardin, C.; Girin, T.; Roudier, F.; Meyer, C.; Krapp, A. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014, 65, 5577–5587. [Google Scholar] [CrossRef] [PubMed]
- Koszegi, D.; Johnston, A.J.; Rutten, T.; Czihal, A.; Altschmied, L.; Kumlehn, J.; Wüst, S.E.; Kirioukhova, O.; Gheyselinck, J.; Grossniklaus, U.; et al. Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 2011, 67, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Luo, J.; Yang, Y.; Jia, J.; Sun, M.; Wang, X.; Khan, I.; Huang, D.; Huang, L. The evolution and expansion of RWP-RK gene family improve the heat adaptability of elephant grass (Pennisetum purpureum Schum.). BMC Genom. 2023, 24, 510. [Google Scholar] [CrossRef]
- Kiehn, M.; Lorence, D.H. Chromosome counts on angiosperms cultivated at the National Tropical Botanical Garden, Kaua’i, Hawai’i. Pac. Sci. 1996, 50, 317–323. [Google Scholar]
- Huang, C.C. Rutaceae. In Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1997. [Google Scholar]
- Luo, X.; Liu, J.; Wang, J.; Gong, W.; Chen, L.; Wan, W. FISH analysis of Zanthoxylum armatum based on oligonucleotides for 5S rDNA and (GAA)6. Genome 2018, 61, 699–702. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ma, L.; Chen, X.; Fei, X.; He, B.; Luo, Y.; Liu, Y.; Wei, A. Genome-wide identification of the NAC gene family in Zanthoxylum bungeanum and their transcriptional responses to drought stress. Int. J. Mol. Sci. 2022, 23, 4769. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Song, Y.; Zhang, Z.S.; Wang, J.X.; Zhang, X.; Zang, J.Y.; Bai, M.Y.; Yu, L.H.; Xiang, C.B. GAF domain is essential for nitrate-dependent AtNLP7 function. BMC Plant Biol. 2022, 22, 366. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.J.; Davies, D.R.; Bullard, J.M.; Christensen, J.; Green, L.S.; Guiles, J.W.; Pata, J.D.; Ribble, W.K.; Janjic, N.; Jarvis, T.C. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc. Natl. Acad. Sci. USA 2008, 105, 20695–20700. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Arbiza, L. Cis-regulatory elements and human evolution. Curr. Opin. Genet. Dev. 2014, 29, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Duan, L.; Pruneda-Paz, J.L.; Oh, D.H.; Pound, M.; Kay, S.; Dinneny, J.R. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol. 2018, 177, 1650–1665. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2021, 35, 199–214. [Google Scholar] [CrossRef]
- Danso, K.; Elgba, W. Optimisation of somatic embryogenesis in cassava. In Biotechnologies for Plant Mutation Breeding; Jankowicz-Cieslak, J., Tai, T.H., Kumlehn, J., Bradley, J.T., Eds.; Springer Nature: Berlin/Heidelberg, Germany, 2017; pp. 73–90. [Google Scholar]
- Li, M.; Wrobel-Marek, J.; Heidmann, I.; Horstman, A.; Chen, B.; Reis, R.; Angenent, G.C.; Boutilier, K. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 2022, 188, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Oh, D.H.; Dassanayake, M.; Nguyen, K.T.; Ogas, J.; Choi, G.; Sun, T.P. Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions. Plant Physiol. 2017, 173, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Rövekamp, M.; Bowman, J.L.; Grossniklaus, U. Marchantia MpRKD regulates the gametophyte-sporophytet ransition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 2016, 26, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhao, P.; Sun, M.X. Autophagy in sexual plant reproduction: New insights. J. Exp. Bot. 2021, 72, 7658–7667. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jin, H.; Chen, J.; Li, C.; Wang, J.; Luo, J.; Wang, Z. Identification and analysis of the AP2 subfamily transcription factors in the pecan (Carya illinoinensis). Int. J. Mol. Sci. 2021, 22, 13568. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Hao, H.; Xue, Y.; Zhang, L.; Song, K.; Ding, Z.; Botella, M.A.; Wang, H.; Lin, J. Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 2013, 140, 3826–3837. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Z.; Tian, D.; Xu, M.; Pan, J.; Wu, H.; Wang, C.; Otegui, M.S. AP1/2β-mediated exocytosis of tapetum-specific transporters is required for pollen development in Arabidopsis thaliana. Plant Cell 2022, 34, 3961–3982. [Google Scholar] [CrossRef] [PubMed]
- Tsukimoto, R.; Isono, K.; Kajino, T.; Iuchi, S.; Shinozawa, A.; Yotsui, I.; Sakata, Y.; Taji, T. Mitochondrial fission complex is required for long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 2022, 63, 296–304. [Google Scholar] [CrossRef]
- Náprstková, A.; Malínská, K.; Záveská Drábková, L.; Billey, E.; Náprstková, D.; Sýkorová, E.; Bousquet-Antonelli, C.; Honys, D. Characterization of ALBA family expression and localization in Arabidopsis thaliana generative organs. Int. J. Mol. Sci. 2021, 22, 1652. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Duan, Y.; Zheng, H.; Tang, H.; Zheng, L.; Yu, X. Genome-Wide Identification and Characterization of the RWP-RK Proteins in Zanthoxylum armatum. Genes 2024, 15, 665. https://doi.org/10.3390/genes15060665
Zheng X, Duan Y, Zheng H, Tang H, Zheng L, Yu X. Genome-Wide Identification and Characterization of the RWP-RK Proteins in Zanthoxylum armatum. Genes. 2024; 15(6):665. https://doi.org/10.3390/genes15060665
Chicago/Turabian StyleZheng, Xianzhe, Yanling Duan, Huifang Zheng, Hao Tang, Liumeng Zheng, and Xiaobo Yu. 2024. "Genome-Wide Identification and Characterization of the RWP-RK Proteins in Zanthoxylum armatum" Genes 15, no. 6: 665. https://doi.org/10.3390/genes15060665
APA StyleZheng, X., Duan, Y., Zheng, H., Tang, H., Zheng, L., & Yu, X. (2024). Genome-Wide Identification and Characterization of the RWP-RK Proteins in Zanthoxylum armatum. Genes, 15(6), 665. https://doi.org/10.3390/genes15060665





