Abstract
Grain filling is critical for determining yield and quality, raising the question of whether central coordinators exist to facilitate the uptake and storage of various substances from maternal to filial tissues. The duplicate NAC transcription factors ZmNAC128 and ZmNAC130 could potentially serve as central coordinators. By analyzing differentially expressed genes from zmnac128 zmnac130 mutants across different genetic backgrounds and growing years, we identified 243 highly and differentially expressed genes (hdEGs) as the core target genes. These 243 hdEGs were associated with storage metabolism and transporters. ZmNAC128 and ZmNAC130 play vital roles in storage metabolism, and this study revealed two additional starch metabolism-related genes, sugary enhancer1 and hexokinase1, as their direct targets. A key finding of this study was the inclusion of 17 transporter genes within the 243 hdEGs, with significant alterations in the levels of more than 10 elements/substances in mutant kernels. Among them, six out of the nine upregulated transporter genes were linked to the transport of heavy metals and metalloids (HMMs), which was consistent with the enrichment of cadmium, lead, and arsenic observed in mutant kernels. Interestingly, the levels of Mg and Zn, minerals important to biofortification efforts, were reduced in mutant kernels. In addition to their direct involvement in sugar transport, ZmNAC128 and ZmNAC130 also activate the expression of the endosperm-preferential nitrogen and phosphate transporters ZmNPF1.1 and ZmPHO1;2. This coordinated regulation limits the intake of HMMs, enhances biofortification, and facilitates the uptake and storage of essential nutrients.
1. Introduction
Grain filling encompasses intricate biochemical and physiological processes, including the uptake of macro- and micronutrients from maternal tissues to developing grains, along with the synthesis of storage compounds within endosperms or embryos. This phase is pivotal in determining grain yield and quality. The relatively large caryopsis of maize (Zea mays), combined with its agricultural and scientific importance, positions it as an optimal system for investigating seed development in the grass family [1].
Maize plants have developed a highly specialized maternal/filial interface structure in the basal kernel zone, consisting of the basal endosperm transfer layer (BETL), placenta-chalazal (PC), and pedicel (PED), to facilitate nutrient transport from the maternal plant to the developing endosperm [2]. To date, four sugar transporters (ZmSWEET4c [sugars will eventually be exported transporter 4c] [3], ZmSUGCAR1 [sucrose and glucose carrier 1] [4], ZmSUT1 [sucrose transporter 1] [5], ZmSUT7 [6]) and one cell wall invertase-encoding gene (ZmMN1 [miniature 1]) [7], in the BETL, have been shown to function in the uptake of carbohydrates/carbons (C). Recently, duplicate maize PHO1-type phosphate (P) transporters, ZmPHO1;2a and ZmPHO1;2b have been shown to function in P reallocation and grain filling [8,9]. Additionally, the Zn-NA (nicotianamine) transporter gene ZmYSL2 has been found to be involved in the zinc (Zn) upload to maize kernels [10]. Although progress has been made in understanding the uptake of C, P, and Zn in maize grains, the uptake of other essential nutrients such as nitrogen (N) and magnesium (Mg) has not been fully elucidated.
A second critical aspect of the grain-filling stage is the conversion of nutrients into seed-storage compounds such as starch, protein, and lipids. In grains, C is mainly stored in the form of starch within endosperm starch granules, which accounts for approximately 70% of the kernel weight [1]. While sugar metabolism is complex, the starch synthesis pathway in the maize endosperm is well understood. The key enzymes involved in starch synthesis in the maize endosperm include the following: sucrose synthases (such as shrunken1 [Sh1], SUS1, and SUS2); UDP-glucose pyrophosphorylase; ADP-glucose pyrophosphorylase (AGPase), which is composed of a small subunit (brittle2 [Bt2]) and a large subunit (shrunken2 [Sh2]); ADP-glucose (ADPG) transporter (brittle1 [Bt1]); soluble starch synthases (SS-I, -II, -III, -IV, and -V); granule-bound starch synthase (GBSS1, or waxy [Wx]); starch-branching enzymes (such as SBE-IIa and -IIb); and starch-debranching enzymes (maize pullulanase1 [Zpu1]) [11]. In terms of N, the maize seed storage proteins are primarily composed of endosperm prolamins (zeins) and embryo globulins. The α-, β-, γ-, and δ-zeins account for approximately 60% of the total seed proteins [12]. The abundance of zeins largely determines the amino acid composition of kernels, but their low lysine and tryptophan contents negatively impact protein quality [12].
During the filling stage, major starch metabolism genes and zeins are highly and specifically expressed in the endosperm [6,13], highlighting the essential role of transcriptional regulation in coordinating this complex process. Multiple transcription factors (TFs), including opaque2 (O2) [14,15,16,17], prolamin-box binding factor 1 (PBF1) [18,19], O2 heterodimerizing proteins (OHP1 and OHP2) [20,21], duplicate indeterminate domain (IDD) TFs (NKD1 and NKD2) [22], one MADS-type TF (ZmMADS47) [23], one bZIP-type TF (ZmbZIP22) [24], and duplicate NAC-type TFs (ZmNAC128 and ZmNAC130) [25,26], have been shown to directly regulate zein gene expression. Additionally, O2, PBF1, NKD1, NKD2, ZmNAC128, ZmNAC130, and opaque11 (O11) have been identified as regulators of starch metabolism genes [22,25,26,27,28]. Recently, ABSCISIC ACID INSENSITIVE 19 (ZmABI19) was shown to directly regulate multiple important TFs, such as O2, PBF1, ZmbZIP22, ZmNAC130, and O11, in the endosperm [29]. Furthermore, ZmABI19 interacts with the basic leucine zipper 29 to synergistically regulate O2 expression [30]. O2, serving as a core TF in maize endosperm filling [14,15,16,17], can genetically or molecularly interact with ZmNAC128/ZmNAC130, or NKD1/NKD2 to effectively coordinate endosperm filling [26,31]. Recently, two genes downstream of O2, namely, the GRAS domain-containing protein ZmGRAS11 [32] and the RNA polymerase common subunit ZmRPABC5b [33], have been characterized to play essential roles in endosperm filling development. However, the regulatory mechanisms underlying nutrient entry into grains are poorly understood, despite advancements in our understanding of nutrient storage metabolism.
ZmNAC128 and ZmNAC130 have emerged as key players in maize kernel development, as their mutations result in severe defects in grain filling, such as significant reductions in starch and protein content in mature kernels [25,26]. These two NACs not only act as core regulators of starch and zein synthesis but also participate in the regulation of sugar and zinc uptake. These intriguing findings prompted us to explore the existence of central regulators that coordinate both the uptake and storage of macro- and micronutrients from mother plants into filial grains. Therefore, it is imperative to investigate whether ZmNAC128 and ZmNAC130 fulfill this role as central regulators. This approach holds great potential for uncovering essential grain-filling mechanisms, which can ultimately contribute to crop yield and quality improvements.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The knockout lines of ZmNAC128 and ZmNAC130 in the KN5585 inbred background were previously generated [26]. This study utilized three sets of zmnac128 zmnac130 mutant materials, with the first and second sets derived from our previous study [26], and the third set specifically developed for this study. The zmnac128 zmnac130 mutant and the corresponding wild type in the B73 × KN5585 background were generated from the F1 cobs by crossing the inbred B73 with zmnac128 zmnac130 in the KN5585. The F1 plants from the cross of B73 with KN5585 zmnac128 zmnac130 were grown in the field. We selected self-pollinated cobs from these T1 plants to complete the subsequent experiments. For this study, endosperms of the wild type (WT) and zmnac128 zmnac130 mutant were genotyped and collected from the F1 cobs that had been pollinated at the same time. All maize materials were planted in a field located in Hefei (Anhui Province, China) from March to July in both 2022 and 2023. Given the complexity of the genetic background of B73 × KN5585, we used the zmnac128 zmnac130 mutant in the KN5585 background, as well as the KN5585 (wild type) itself, as the samples for all subsequent molecular experiments and various substances and element analysis experiments in this study.
2.2. RNA Sequencing (RNA-Seq)
The RNA-seq method employed in this study followed the same procedure outlined in our previous work [26]. Total RNA was extracted from more than three 16-days-after-pollination (16-DAP) endosperms from the WT and zmnac128 zmnac130 mutant using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Subsequently, libraries were prepared and sequenced on a DNBSEQ-T7 platform, generating 150 bp paired-end reads. The resulting clean reads were aligned to the B73 reference genome (RefGen_v5) using the HISAT2 program [34]. A differential expression analysis was performed using the R package DESeq [35], with differentially expressed genes defined as those exhibiting an absolute log2 fold change (FC) greater than 1 and an adjusted p-value below 0.05.
2.3. Quantification of Lipid Content in Mature Kernels
More than ten mature kernels were milled into flour and subsequently filtered using a 50-mesh stainless steel screen. The lipid content of the resulting milled flour was determined using a pre-constructed calibration model based on an MPA-type Fourier-transform near-infrared spectrophotometer (Bruker, Ettlingen, Germany). Sample spectra were collected on the MPA spectrophotometer, and the modeling algorithm employed was a partial least squares regression. Each type of mature kernel flour was divided into four portions for measurement purposes.
2.4. Analysis of Element Content in Mature Kernels
The element profiling was conducted using inductively coupled plasma–mass spectrometry (ICP-MS), following previously established methods [10]. For each sample, six dry mature kernels were ground into powder, and 2–5 mg of each sample was collected for the elemental profiling. To ensure consistency, all samples were normalized using a heuristic algorithm that utilized the best-measured elements, as described previously [36].
2.5. Determination of Phytohormone Content in 16-DAP Endosperm
Approximately 100 mg of frozen 16-DAP endosperm powder was extracted in 1 mL of ice-cold 50% aqueous ACN (v/v). The samples were sonicated for 3 min at 4 °C and subsequently extracted using a benchtop laboratory rotator for 30 min at 4 °C. After centrifugation (10 min, 10,000× g, 4 °C), the supernatant was transferred to clean plastic microtubes. All samples were purified using C18 reversed-phase, polymer-based, solid-phase extraction (RP-SPE) cartridges, that had been washed with 1 mL of MeOH and 1 mL of sterile deionized water, then equilibrated with 50% aqueous ACN (v/v). After loading a sample, the cartridge was then rinsed with 1 mL of 30% ACN (v/v) and this fraction was collected. After this single-step SPE, the samples were evaporated until dry under a gentle stream of nitrogen and stored at −20 °C until analysis. For UHPLC–ESI–MS/MS analysis, the samples were dissolved in 200 μg/mL of 30% ACN (v/v) and transferred to insert-equipped vials.
The sample extracts were analyzed using a UPLC-Orbitrap-MS system (UPLC, Vanquish, Thermo Fisher Scientific, Carlsbad, CA, USA). The HRMS data were recorded on a Q Exactive hybrid Q-Orbitrap mass spectrometer equipped with a heated ESI source (Thermo Fisher Scientific, Carlsbad, CA, USA) utilizing the SIM MS acquisition methods. Data were acquired on the Q-Exactive using Xcalibur 4.1 (Thermo Fisher Scientific, Carlsbad, CA, USA) and processed using TraceFinder™4.1 Clinical (Thermo Fisher Scientific, Carlsbad, CA, USA).
2.6. RNA Extraction and Quantitative RT-PCR
The total RNA was extracted from maize endosperms at 12- and 20-DAP using TRIzol reagent (Invitrogen). Subsequently, 2 µg of total RNA was utilized to synthesize the first-strand cDNA with the HiScript® II 1st Strand cDNA Synthesis Kit (R212, Vazyme, Nanjing, China). Using SYBR Green (Genstar, Beijing, China), the quantitative RT-PCR assay was performed on a CFX Connect real-time PCR system (Bio-Rad, Hercules, CA, USA) following the standard operating manual. We used the default 100% efficiency setting on the CFX Connect real-time PCR system.
To estimate the relative expression of the target gene between the WT and mutants, the comparative CT (ΔΔCT) method [37] was used with ZmActin serving as the reference. The primer sequences are listed in Table S3.
2.7. Dual-Luciferase Reporter (DLR) Assay
Leaves prepared for maize protoplasts were collected from more than 20 two-week seedlings of the inbred B73 cultivated in darkness. The process we followed for the DLR assay was in accordance with the method described previously [26]. The protoplast isolation involved the process of zymolysis using Cellulose R10 (Yakult, Tokyo, Japan) and Macerozyme R10 (Yakult). The recombinant vector pRI101 (Clontech, Takara Bio USA, Inc., San Jose, CA, USA) and pGreenII 0800-LUC were transfected into a protoplast using polyethylene glycol (PEG)-calcium. The protoplast was cultivated overnight for the further detection of the LUC/REN ratio using a dual-luciferase reporter assay system (Promega, Madison, WI, USA). The vector pRI101 (Clontech, Takara Bio USA, Inc., San Jose, CA, USA) was used for the expression of ZmNAC128 and ZmNAC130 under the control of the 35S promoter and the vector pGreenII 0800-LUC was used to generate the reporter constructs by cloning the promoters of different target genes upstream of LUC. The primers are listed in Table S3.
2.8. Phylogenetic Analysis
The protein sequence was aligned and analyzed by ClustalW with the MEGA11 software. The neighbor-joining tree was constructed with the MEGA11 software (http://megasoftware.net/, accessed on 24 June 2022) according to the protein sequence alignment.
2.9. Statistical Analysis
Data processing of means, standard deviations, and p-values was performed with Microsoft Excel (2019) using AVERAGE, STDEV.S, and Student’s t-test, respectively. Statistical tests involving a one-way ANOVA were performed with GraphPad Prism (version 8.4.3). The hierarchical clustering analysis using Pearson correlation and visualization was performed with the corrplot package (v 0.92). The raw data and detailed statistical analysis are in Dataset S2.
3. Results
3.1. Generation and Analysis of Transcriptomes from Endosperms of zmnac128 zmnac130 Mutants across Different Genetic Backgrounds and Growing Years
Our previous studies revealed that mutations in ZmNAC128 and ZmNAC130, which are specifically and highly expressed duplicate TFs in endosperm filling, result in severe defects in kernel filling [25,26]. We identified more than 10 genes as direct targets, which partly explains the role of ZmNAC128 and ZmNAC130 in endosperm filling, but these genes account for only a small fraction of the thousands of genes affected by the loss-of-function mutation of these two NAC TFs. Therefore, an essential question arises: how does their core regulatory network operate?
To explore the core genes in the regulatory network, we obtained three sets of transcriptomes from the endosperms of the zmnac128 zmnac130 mutants and the corresponding wild-type (WT) or nontransgenic (NT) siblings under different genetic backgrounds and growing years at 16 DAP. Two sets of data were generated in our previous study [26]: one from the KN5585 genetic background (referred to as “set-1”) and the other from the KN5585 × B73 background (referred to as “set-2”) grown on a farm in Hefei City, China, during the summer of 2022. Set-3 was generated on the same farm during the summer of 2023, also using the KN5585 × B73 background. Among all 14 transcriptomes from these three experiments, an average of 43,560,203 (92.93%) clean reads were uniquely mapped to the maize B73 reference genome (Zm-B73-REFERENCE-NAM-5.0) (Table S1). The fragments per kilobase of the exon model per million mapped fragments (FPKM) were used to quantify gene expression.
To evaluate the influence of genetic background and growing years on the transcriptome, we performed hierarchical clustering analysis using a Pearson correlation of gene expression among samples. The results revealed a distinct segregation between transcriptomes of the two genetic backgrounds, as well as a noticeable separation of transcriptomes from the same genetic background across two growing years (Figure 1A). Two possible explanations for these results are as follows: (1) The hybrid progeny of KN5585 and B73 introduced increased complexity in the transcriptomes of set-2 and set-3. (2) The genetic background had a stronger impact on transcriptome variation than growing years. However, further research is needed to test these hypotheses. Identifying highly and differentially expressed genes (hdEGs) under different genetic backgrounds and growing years will help to unravel the core regulatory mechanism of ZmNAC128 and ZmNAC130 with reduced interference.
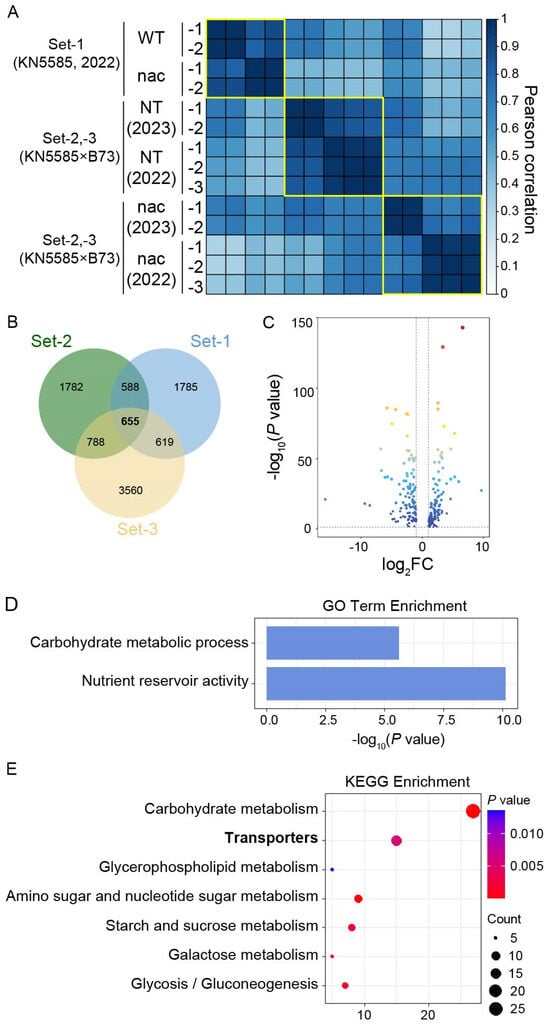
Figure 1.
An analysis of transcriptomes from 16-DAP endosperms of zmnac128 zmnac130 mutants with different genetic backgrounds and growing years. (A) A hierarchical clustering analysis of three sets of transcriptomes based on a Pearson correlation of gene expression. WT, wild type. NT, nontransgenic sibling segregated from crossing the inbred B73 with the zmnac128 zmnac130 mutant in the inbred KN5585. nac, zmnac128 zmnac130 mutant. -1, -2, and -3 represent three independent samples. 2022 and 2023 represent the two growing years. (B) A Venn diagram shows the dEGs in the three sets of transcriptomes. The number in each circle represents the number of dEGs. (C) A volcano plot displays the downregulated and upregulated genes among the hdEGs. (D,E) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the 243 hdEGs are shown in (C).
3.2. Identifying the hdEGs across Different Genetic Backgrounds and Growing Years as Core Members in the Regulatory Network of ZmNAC128 and ZmNAC130
Using a threshold of log2(fold change, FC) > 1 and p value < 0.05, we identified 3813, 3647, and 5622 differentially expressed genes (dEGs) in the transcriptomes of set-1, set-2, and set-3, respectively. A Venn diagram showed that 655 dEGs were shared among all three sets (Figure 1B). We further screened highly expressed genes with FPKM values > 10 among the 655 dEGs based on the data from the KN5585 background. This led to the identification of 243 hdEGs, including 130 upregulated genes with FPKM values > 10 in the zmnac128 zmnac130 mutant and 113 downregulated genes with FPKM values > 10 in the WT (Figure 1C; Dataset S1). These 243 hdEGs were considered core targets of ZmNAC128 and ZmNAC130 and were further investigated in this study.
Gene Ontology (GO) analysis of these 243 hdEGs revealed significant enrichment in two terms: ‘carbohydrate metabolic process’ and ‘nutrient reservoir activity’ (Figure 1D). These results support our previous findings on the regulatory roles of these genes in starch and zein synthesis. Interestingly, the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis also indicated that transporters were significantly enriched in these 243 hdEGs (Figure 1E). The results suggested that the core role of ZmNAC128 and ZmNAC130 was in nutrient uptake and storage. Our previous studies revealed the important roles of these TFs in the uptake of sugars and Zn, but further investigation is needed to determine the regulatory mechanisms involved in the transport of other nutrients and elements.
3.3. Identifying Starch Metabolism-Related Sugary Enhancer 1 (SE1) and Hexokinase 1 (HXK1) as Direct Targets of ZmNAC128 and ZmNAC130
Following our previous findings [25,26], we observed an enrichment of storage metabolism associated with carbon (C), nitrogen (N), and phosphate (P) in these 243 hdEGs (Figure 1D,E). This finding confirmed that storage metabolism is central to the regulatory network of ZmNAC128 and ZmNAC130.
Our previous studies demonstrated that ZmNAC128 and ZmNAC130 coordinate sugar uptake and starch metabolism during endosperm filling [25,26]. ZmNAC128 and ZmNAC130 directly regulate the expression of two crucial sugar transporter genes, ZmSWEET4c and ZmSUGCAR1, with ZmSWEET4c being one of the 243 hdEGs. Among the 243 hdEGs, we found two downregulated genes, SE1 (Zm00001eb115450) and HXK1 (Zm00001eb121400), which are related to endosperm starch metabolism (Dataset S1) [38,39]. A real-time PCR confirmed that the expression of the SE1 and HXK1 genes was also significantly downregulated in the 12- and 20-DAP endosperms of the zmnac128 zmnac130 mutant (Figure 2B). By analyzing the DNA affinity purification sequencing (DAP-Seq) data for ZmNAC128 and ZmNAC130 which were previously produced [26], we detected whether there were distinct binding peaks for two NACs in the SE1 and HXK1 promoters. The results indicated that there were distinct peaks bound by ZmNAC128 and ZmNAC130 within the two promoters with conserved NAC-binding motifs (Figure 2A). A dual-luciferase reporter (DLR) assay was further performed to detect the transactivation activities of the two promoters. The results indicated that, compared to that of the empty control, the luciferase (LUC) activity driven by the SE1 or HXK1 promoter was significantly increased by ZmNAC128 or ZmNAC130, but the two NACs together did not further enhance the LUC activity (Figure 2C). This finding is consistent with our previous findings that ZmNAC128 and ZmNAC130 have functional redundancy [25,26]. Thus, combined with our previous findings, up to eight starch metabolism-related genes are directly regulated by ZmNAC128 and ZmNAC130. These findings highlight the vital role of ZmNAC128 and ZmNAC130 in controlling sugar uptake and storage in filling endosperms.
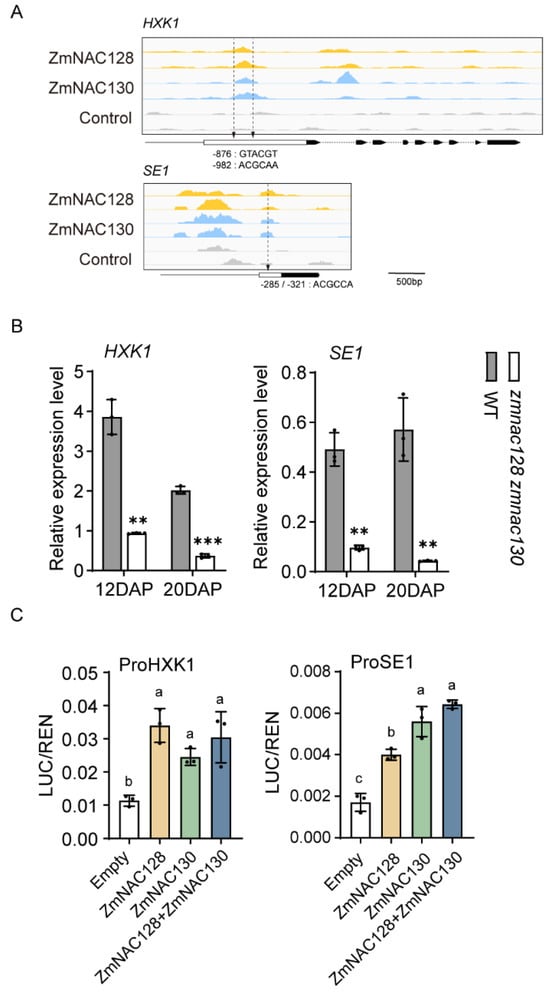
Figure 2.
The identification of two carbohydrate metabolism-related genes, HXK1 and SE1, as direct targets of ZmNAC128 and ZmNAC130. (A) DAP-seq analysis shows the binding peaks of ZmNAC128 and ZmNAC130 on these two gene promoters. The cis-elements (‘ACGCAA’ and ‘GTACGT’), which were previously identified [25,26], are marked by arrows in the promoters. (B) A real-time PCR indicates the relative expression levels of these genes in 12- and 20-DAP endosperms of the zmnac128 zmnac130 mutant compared to those of the WT. The ZmActin was used as a reference gene for normalization, employing the default 100% efficiency setting for detected genes on the CFX Connect real-time PCR system. (C) DLR assays detect LUC activities driven by the promoters in the presence of ZmNAC128, ZmNAC130, or both. Empty, the negative control. The data represent the mean ± standard deviation (SD) of three independent samples (B,C). Significant differences (** p < 0.01 and *** p < 0.001) were determined using a Student’s t-test (B). Different lowercase letters indicate significant differences according to a one-way ANOVA with Tukey’s multiple comparisons test (p < 0.05) (C).
3.4. Regulatory Roles of ZmNAC128 and ZmNAC130 on Toxic HMM Accumulation and Biofortification in Maize Kernels
The enrichment of ‘transporters’ in the 243 hdEGs led us to investigate their relationship with ZmNAC128 and ZmNAC130. This category included nine upregulated and eight downregulated transporter genes (Table 1). Among the nine upregulated transporters, six were closely associated with heavy-metal and metalloid (HMM) transport [40,41,42], including one C-type ABC transporter/multidrug resistance protein-related transporter ZmMRPA14, one natural resistance-associated macrophage protein ZmNRAMP5, and four multidrug and toxic compound extrusion transporters (ZmMATE4, ZmMATE11, ZmMATE17, and ZmMATE48). Therefore, we utilized inductively coupled plasma-mass spectrometry (ICP-MS) to measure the levels of various elements in mature kernels of the zmnac128 zmnac130 mutant. In addition to Zn [26], the levels of eight other elements were also significantly altered in zmnac128 zmnac130 kernels, including cadmium (Cd), lead (Pb), arsenic (As), boron (B), natrium (Na), phosphorus (P), magnesium (Mg), and molybdenum (Mo) (Figure 3A–D).

Table 1.
Information on the transporter genes in the 243 hdEGs.
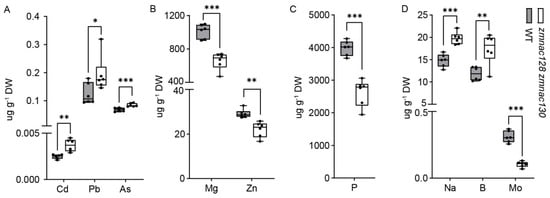
Figure 3.
ZmNAC128 and ZmNAC130 affect the accumulation of numerous elements in mature kernels. (A) The levels of three toxic HMMs were significantly increased in mature kernels of the zmnac128 zmnac130 mutant. (B) The levels of two biofortified elements were significantly decreased in mature kernels of the zmnac128 zmnac130 mutant. The Zn content data were from our previous publication [25,26]. (C) The P level was significantly decreased in mature kernels of the zmnac128 zmnac130 mutant. (D) The levels of Na, B, and Mo were significantly changed in mature kernels of the zmnac128 zmnac130 mutant. The data represent the mean ± SD of 5 independent samples (A–D). Significant differences (* p < 0.05, ** p < 0.01, and *** p < 0.001) were determined via Student’s t-test.
Notably, Cd, Pb, and As, which act as toxic HMMs closely related to grain safety [43], were significantly accumulated in zmnac128 zmnac130 kernels. OsNRAMP5, which is an orthologous gene of ZmNRAMP5 (Figure 4A), has been identified as the main Cd transporter in rice [44]. The expression of ZmNRAMP5 was significantly upregulated in the endosperm filling of the zmnac128 zmnac130 mutant compared to that in the WT (Figure 4C), indicating that this gene may be a major factor in the increased Cd content in the zmnac128 zmnac130 kernels. However, DAP-seq analysis indicated that there were not any apparent peaks bound by ZmNAC128 and ZmNAC130 in the ZmNRAMP5 promoter (Figure 4B). It is suggested that ZmNAC128 and ZmNAC130 seem to indirectly regulate the expression of ZmNRAMP5.
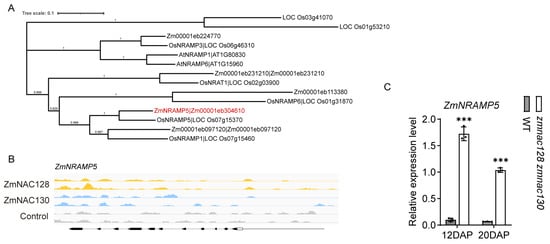
Figure 4.
The identification of ZmNRAMP5 as the direct target gene of ZmNAC128 and ZmNAC130. (A) A phylogenetic analysis depicting the relationship among NRAMP proteins from maize, rice, and Arabidopsis. The tree was generated by MEGA11 software using the blastp result of NRAMP protein sequences. Blastp is performed by phytozome v13 and the N-J method is used with 1000 bootstrap. (B) DAP-seq analysis shows the binding peaks of ZmNAC128 and ZmNAC130 on the promoter of ZmNRAMP5. (B) A real-time PCR indicating the relative expression levels of ZmNRAMP5 in 12- and 20-DAP endosperms of the zmnac128 zmnac130 mutant compared to those of the WT. ZmActin was used as a reference gene for normalization. (C) DLR assays detecting LUC activities driven by the ZmNRAMP5 promoter in the presence of ZmNAC128, ZmNAC130, or both. Empty, the negative control. The data represent the mean ± SD of 3 independent samples (B,C). Significant differences (*** p < 0.001) were determined using a Student’s t-test (B). Different lowercase letters indicate significant differences according to a one-way ANOVA with Tukey’s multiple comparisons test (p < 0.05) (C).
Moreover, the levels of two biofortified elements, Zn and Mg, significantly decreased in the mutant (Figure 3B). ZmNAC128 and ZmNAC130 were previously reported to control Zn accumulation in kernels by directly regulating ZmYSL2 expression [26]. Hence, ZmNAC128 and ZmNAC130 have dual functions: they restrict the accumulation of toxic HMMs while promoting the biofortification of Zn and Mg in the kernel filling.
3.5. ZmNAC128 and ZmNAC130 Directly Regulate the Expression of Grain-Filling-Controlling P Transporter ZmPHO1;2
Consistent with the significant reduction in P content (Figure 3C), two duplicate genes involved in maize grain phosphate transport, ZmPHO1;2a and ZmPHO1;2b, also exhibited significantly downregulated expression in the filing endosperms of the zmnac128 zmnac130 mutant (Figure 5B; Table 1). Both ZmPHO1;2a and ZmPHO1;2b are highly expressed in endosperm filling (Figure S3) and are involved in determining the P content in kernels [8]. Electron in situ hybridization revealed that ZmPHO1;2a is expressed in the BETL, while ZmPHO1;2b is exclusively expressed in endosperm tissues surrounding the embryo (Figure S3). It is suggested that the two ZmPHO1;2 genes function in the transfer of P at the interface of maternal/endosperm and endosperm/embryo.
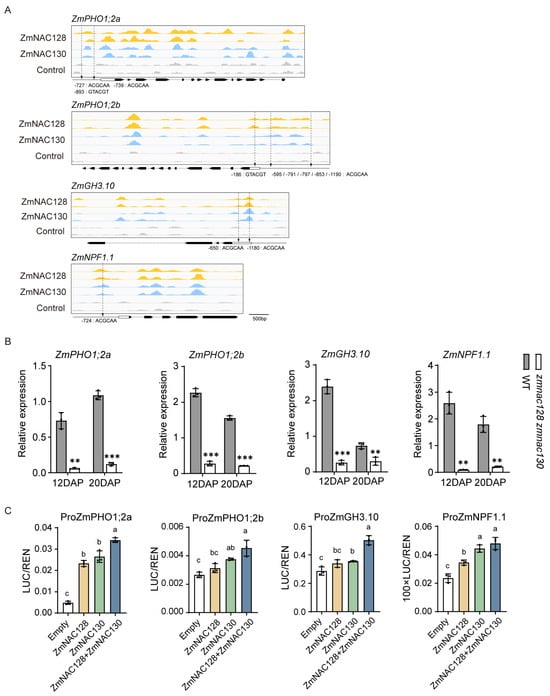
Figure 5.
The identification of N-/P-related transporter genes and one JA-Ile synthesis-related gene as the direct targets of ZmNAC128 and ZmNAC130. (A) DAP-seq analysis shows the binding peaks of ZmNAC128 and ZmNAC130 on these four gene promoters. The cis-elements (‘ACGCAA’ and ‘GTACGT’), which were previously identified [25,26], are marked by arrows in the promoters. (B) A real-time PCR indicates the relative expression levels of these four genes in 12- and 20-DAP endosperms of the zmnac128 zmnac130 mutant compared to those of the WT. ZmActin was used as a reference gene for normalization. (C) DLR assays detect LUC activities driven by the promoters in the presence of ZmNAC128, ZmNAC130, or both. Empty represents the negative control. The data represent the mean ± SD of 3 independent samples (B,C). Significant differences (** p < 0.01 and *** p < 0.001) were determined using a Student’s t-test (B). Different lowercase letters indicate significant differences according to a one-way ANOVA with Tukey’s multiple comparisons test (p < 0.05) (C).
To determine whether ZmNAC128 and ZmNAC130 directly regulate the expression of ZmPHO1;2, we performed a DAP-seq, which revealed that there were peaks bound by the two NACs in the promoters of ZmPHO1;2a and ZmPHO1;2b, with conserved NAC-binding motifs around the peaks (Figure 5A). Furthermore, DLR results indicated that LUC activity driven by the ZmPHO1;2a promoter was significantly greater in the presence of ZmNAC128 or ZmNAC130 than in the empty control. Moreover, the co-occurrence of two NACs further enhanced LUC activity compared to that of either one. In addition, LUC activity driven by the ZmPHO1;2b promoter was significantly increased in the presence of ZmNAC130 but not ZmNAC128 and the co-occurrence of the two NACs did not further enhance LUC activity compared to that of ZmNAC130 (Figure 5C). The results demonstrated that ZmNAC130 and ZmNAC128 play major roles in ZmPHO1;2a expression and minor roles in ZmPHO1;2b expression.
3.6. A Regulatory Mechanism of ZmNAC128 and ZmNAC130 in the Expression of the Endosperm-Specific Nitrate Transporter ZmNPF1.1
Among the downregulated transporter genes of these 243 hdEGs (Table 1), ZmNPF1.1 is particularly interesting because it is a N transporter gene that is preferentially expressed in the BETL (Figure S3). A real-time PCR confirmed that the expression of ZmNPF1.1 was significantly downregulated in the endosperm filling of the zmnac128 zmnac130 mutant (Figure 5B). To investigate whether ZmNAC128 and ZmNAC130 directly regulate the expression of ZmNPF1.1, DAP-seq analysis was performed, and the results revealed that there were apparent peaks bound by ZmNAC128 and ZmNAC130 in the ZmNPF1.1 promoter, with the presence of the conserved NAC-binding motif around the peaks (Figure 5A). DLR assays further indicated that, compared to that in the empty control, LUC activity driven by the ZmNPF1.1 promoter was significantly greater in the presence of ZmNAC128 or ZmNAC130, but the co-occurrence of two NACs did not significantly enhance the LUC activity (Figure 5C).
Furthermore, a previous study revealed that an exogenous jasmonic acid (JA) treatment strongly induces the expression of ZmNPF1.1 [45]. Interestingly, among these 243 hdEGs, ZmGH3.10 (Zm00001eb338800), which belongs to Group I of the GH3 (Gretchen Hagen3) family and is associated with JA-amino acid conjugation [46,47], was downregulated. As the conjugation of the amino acid isoleucine (Ile) to JA is critical for conferring major bioactivity and is central to JA signaling and responses [46], we utilized an ultraperformance liquid chromatography/tandem mass spectrometry (UPLC-MS/MS) system to measure the contents of JA and JA-Ile in the 20-DAP kernels of the zmnac128 zmnac130 mutant. The results showed a significant reduction in both the JA-Ile and JA contents in the zmnac128 zmnac130 mutant compared to those in the WT (Figure S4A), and the ratio of JA-Ile to JA was also significantly decreased in the mutant (Figure S4B). Since ZmGH3.10 is highly expressed in endosperm filling (Figure S3), it likely plays an important role in the JA-Ile synthesis in maize grains. DAP-seq analysis revealed the binding peaks of ZmNAC128 and ZmNAC130 in the promoter with conserved NAC-binding motifs around the peaks (Figure 5A). DLR further confirmed that, compared to that in the empty control, LUC activity driven by the ZmGH3.10 promoter was significantly greater in the presence of ZmNAC130 but not in the presence of ZmNAC128, while the co-occurrence of the two NACs further increased the LUC activity compared to that in the case of ZmNAC130 (Figure 5C).
Taken together, these results indicate that ZmNAC128 and ZmNAC130 not only directly regulate the expression of ZmNPF1.1, but also control the synthesis of active JA-Ile by regulating the expression of ZmGH3.10. This, in turn, modulated the expression of ZmNPF1.1 through JA signaling.
4. Discussion
It is now widely recognized that a TF can influence the expression of hundreds, or even thousands, of genes in the tissues where it is expressed and functions. However, only a limited number, typically a few or at most dozens, of downstream genes of a single TF have been identified. Understanding the core regulatory network of a TF is akin to solving a complex puzzle, necessitating continuous exploration of its downstream genes to complete the picture. For instance, the regulatory network of O2, a core TF involved in grain filling in maize, has been extensively studied for more than 30 years. Numerous studies have contributed to expanding the body of knowledge of O2 [14,15,16,17,26,27,48,49,50,51]. Therefore, exploring the core regulatory network of a TF is a complex and challenging endeavor.
The seed is one of the most important sink organs of plants; it not only absorbs and assimilates from mature plants but also transports various nutrients and toxic HMMs from underground root tissues. The transport of these nutrients or toxic HMMs across the maternal/filial interface of the seed is tightly regulated by a diverse array of transporters. Targeting these transporters could be a promising direction for crop improvement. A recent study on iron biofortification in maize kernels is enlightening, as it revealed that by manipulating the expression of ZmNAC78, researchers enhanced the expression of its downstream iron transporters in the BETL and thereby improved the iron content in kernels [52].
This study showed that ZmNAC128 and ZmNAC130, as versatile central coordinators, control the accumulation of numerous substances and the synthesis of three major storage compounds (starch, protein, and lipids) in maize grains (Figure 6). First, ZmNAC128 and ZmNAC130 inhibit the expression of many transporters (ZmNRAMP5, MATE genes, etc.) to limit the entry of toxic HMMs into kernels. Second, ZmNAC128 and ZmNAC130 enhance the biofortification of Zn and Mg. These genes directly regulate ZmYSL2 expression and thus promote Zn uptake. Third, ZmNAC128 and ZmNAC130 control the uptake of C-, N-, and P-related nutrients by directly regulating the expression of five crucial transporters, ZmSWEET4c, ZmSUGCAR1, ZmNPF1.1, and ZmPHO1;2. These two NAC TFs positively influence the phosphorus content of seeds by regulating the expression of ZmPHO1;2. Natural phosphorus reserves are limited, highlighting the importance of developing phosphorus-efficient crops. The rice phosphorus-starvation tolerance 1 (OsPSTOL1) gene plays a crucial role in grain yield under phosphorus-deficient soil conditions [53]. The homologous gene (Zm00001d049727, ZmPSTOL1) of OsPSTOL1 in maize is almost not expressed in developing kernels [6,54], suggesting that ZmNAC128 and ZmNAC130 are unlikely to directly regulate ZmPSTOL1 expression. So, overexpression of ZmPSTOL1, ZmNAC128, and ZmNAC130 in maize should be a potential strategy to significantly enhance the grain yield in phosphorus-deficient soil.
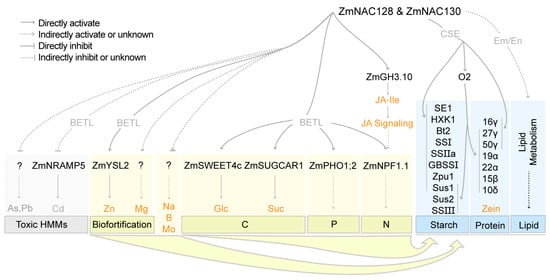
Figure 6.
A working model for ZmNAC128 and ZmNAC130 acting as central coordinators in limiting toxic HMMs, promoting biofortification, and regulating the uptake and storage metabolism of C, N, and P nutrients. All target genes of ZmNAC128 and ZmNAC130 in this model were identified by the present study and our previous studies [25,26].
Moreover, ZmNAC128 and ZmNAC130 also regulate ZmGH3.10 expression to control the active JA-Ile content and thus the JA signaling for ZmNPF1.1 expression and grain filling. Combined with the uptake of macro- and micronutrients, the two NACs collaborate with O2 to regulate the expression of all types of zein genes and 10 starch metabolism genes, as well as lipid synthesis in kernels.
Based on these findings, we plan to further increase the expression of ZmNAC128 and ZmNAC130 via the use of a strong BETL-specific promoter. This strategy should simultaneously inhibit the accumulation of toxic HMMs and promote biofortification in kernels. To further facilitate the synthesis of seed storage compounds, we also plan to genetically combine 27-kD γ-zein promoter-driving varieties with BETL-specific promoter-driving varieties. Ideally, the expression of ZmNAC128 and ZmNAC130 will be enhanced, especially in the BETL and CSE, without affecting other agronomic traits. Our ultimate goal is to breed maize varieties that exhibit high yield, superior quality, biofortification, and safety.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15060663/s1, Figure S1: The lipid content was significantly reduced in the mature kernels of the zmnac128 zmnac130 mutant; Figure S2: An electronic RNA in situ hybridization showing the spatial expression pattern of 18 transporter genes from Table 1 in 12-DAP kernels; Figure S3: The spatiotemporal expression pattern of ZmPHO1;2, ZmNPF1.1, and ZmGH3.10; Figure S4: The content and ratio of JA-Ile to JA in 20-DAP kernels of the zmnac128 zmnac130 mutant; Table S1: The information of mapped reads in three sets of RNA-seq; Table S2: The proteins used as blastp query sequences; Table S3: The primer sequences used in this article; Dataset S1: The 243 hdEGs are commonly present in the three sets of transcriptomes from 16-DAP endosperms of zmnac128 zmnac130 mutants; Dataset S2: The raw data and detailed statistical analysis used in this study.
Author Contributions
Conceptualization, D.P. and Z.Z.; methodology, D.P. and S.P.; software, X.D. and D.P.; validation, X.D., D.P., E.C., and J.H.; formal analysis, D.P. and Z.Z.; investigation, D.P. and Z.Z.; resources, D.P.; data curation, D.P.; writing—original draft preparation, D.P. and Z.Z.; writing—review and editing, D.P. and Z.Z.; visualization, D.P.; supervision, Z.Z.; project administration, Z.Z.; funding acquisition, Z.Z. and E.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers: 31671703 and 32072010 (to Z.Z.), and 32301729 (to E.C.).
Data Availability Statement
RNA-Seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accessed on 1 January 2024) under the accession number GSE252297.
Acknowledgments
We thank Daiyin Chao and Zhenfei Chao from the CAS Center for Excellence in Molecular Plant Sciences in Shanghai, China.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sabelli, P.A.; Larkins, B.A. The development of endosperm in grasses. Plant Physiol. 2009, 149, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.E.; Ma, F. Determinants of Kernel Sink Strength; CABI: Wallingford, UK, 2017. [Google Scholar]
- Sosso, D.; Luo, D.P.; Li, Q.B.; Sasse, J.; Yang, J.L.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, J.; Yu, M.; Zhang, M.; Zhong, Y.; Wang, T.; Liu, P.; Song, W.; Zhao, H.; Fastner, A.; et al. The sugar transporter ZmSUGCAR1 of the Nitrate Transporter 1/Peptide Transporter family is critical for maize grain filling. Plant Cell 2022, 34, 4232–4254. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.F.; Leach, K.A.; Boyer, N.R.; Swyers, M.J.; Benitez-Alfonso, Y.; Skopelitis, T.; Luo, A.; Sylvester, A.; Jackson, D.; Braun, D.M. Sucrose Transporter ZmSut1 Expression and Localization Uncover New Insights into Sucrose Phloem Loading. Plant Physiol. 2016, 172, 1876–1898. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xiao, W.; Tian, L.; Guo, L.; Ma, G.; Ji, C.; Huang, Y.; Wang, H.; Wu, X.; Yang, T.; et al. Spatial transcriptomics uncover sucrose post-phloem transport during maize kernel development. Nat. Commun. 2023, 14, 7191. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 1996, 8, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, L.; Gao, Q.F.; Wang, J.M.; Li, X.Y.; Wang, H.; Liu, Y.; Lin, H.; Liu, J.Y.; Wang, X.; et al. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 2021, 53, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Nieves, A.L.; Salazar-Vidal, M.N.; Torres-Rodríguez, J.V.; Pérez-Vázquez, L.M.; Massange-Sánchez, J.A.; Gillmor, C.S.; Sawers, R.J.H. The pho1;2a′-m1.1 allele of Phosphate1 conditions misregulation of the phosphorus starvation response in maize (Zea mays ssp. mays L.). Plant Direct 2022, 6, e416. [Google Scholar] [CrossRef] [PubMed]
- Chao, Z.-F.; Chen, Y.-Y.; Ji, C.; Wang, Y.-L.; Huang, X.; Zhang, C.-Y.; Yang, J.; Song, T.; Wu, J.-C.; Guo, L.-X.; et al. A genome-wide association study identifies a transporter for zinc uploading to maize kernels. EMBO Rep. 2023, 24, e55542. [Google Scholar] [CrossRef] [PubMed]
- Hannah, L.; Boehlein, S. Starch Biosynthesis in Maize Endosperm; CABI: Wallingford, UK, 2017. [Google Scholar]
- Larkins, B.; YongRui, W.Y.W.; RenTao, S.R.S.; Messing, J. Maize Seed Storage Proteins; CABI: Wallingford, UK, 2017. [Google Scholar]
- Chen, J.; Zeng, B.; Zhang, M.; Xie, S.; Wang, G.; Hauck, A.; Lai, J. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014, 166, 252–264. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Burr, F.A.; Burr, B. Transposon Tagging and Molecular Analysis of the Maize Regulatory Locus Opaque-2. Science 1987, 238, 960–963. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.J.; Burr, F.A.; Aukerman, M.J.; Burr, B. Maize Regulatory Gene Opaque-2 Encodes a Protein with a Leucine-Zipper Motif That Binds to Zein DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Li, C.B.; Qiao, Z.Y.; Qi, W.W.; Wang, Q.; Yuan, Y.; Yang, X.; Tang, Y.P.; Mei, B.; Lv, Y.D.; Zhao, H.; et al. Genome-Wide Characterization of cis-Acting DNA Targets Reveals the Transcriptional Regulatory Framework of Opaque2 in Maize. Plant Cell 2015, 27, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.P.; Li, G.S.; Ryu, C.H.; Ma, C.; Zhang, S.S.; Lloyd, A.; Hunter, B.G.; Larkins, B.A.; Drews, G.N.; Wang, X.F.; et al. Opaque-2 Regulates a Complex Gene Network Associated with Cell Differentiation and Storage Functions of Maize Endosperm. Plant Cell 2018, 30, 2425–2446. [Google Scholar] [CrossRef]
- VicenteCarbajosa, J.; Moose, S.P.; Parsons, R.L.; Schmidt, R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 1997, 94, 7685–7690. [Google Scholar] [CrossRef]
- Wu, Y.R.; Messing, J. Rapid Divergence of Prolamin Gene Promoters of Maize After Gene Amplification and Dispersal. Genetics 2012, 192, 507–519. [Google Scholar] [CrossRef]
- Pysh, L.D.; Aukerman, M.J.; Schmidt, R.J. OHP1—A Maize basic domain leucine zipper protein that interacts with Opaque2. Plant Cell 1993, 5, 227–236. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Wu, Y. Transcriptional Regulation of Zein Gene Expression in Maize through the Additive and Synergistic Action of Opaque2, Prolamine-Box Binding Factor, and O2 Heterodimerizing Proteins. Plant Cell 2015, 27, 1162–1172. [Google Scholar] [CrossRef]
- Gontarek, B.C.; Neelakandan, A.K.; Wu, H.; Becraft, P.W. NKD Transcription Factors Are Central Regulators of Maize Endosperm Development. Plant Cell 2016, 28, 2916–2936. [Google Scholar] [CrossRef]
- Qiao, Z.Y.; Qi, W.W.; Wang, Q.; Feng, Y.N.; Yang, Q.; Zhang, N.; Wang, S.S.; Tang, Y.P.; Song, R.T. ZmMADS47 Regulates Zein Gene Transcription through Interaction with Opaque2. PLoS Genet. 2016, 12, e1005991. [Google Scholar] [CrossRef]
- Li, C.; Yue, Y.; Chen, H.; Qi, W.; Song, R. The ZmbZIP22 Transcription Factor Regulates 27-kD γ-Zein Gene Transcription during Maize Endosperm Development. Plant Cell 2018, 30, 2402–2424. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Yu, H.; He, J.; Peng, D.; Zhu, P.; Pan, S.; Wu, X.; Wang, J.; Ji, C.; Chao, Z.; et al. The transcription factors ZmNAC128 and ZmNAC130 coordinate with Opaque2 to promote endosperm filling in maize. Plant Cell 2023, 35, 4066–4090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, X.; Yang, J.; Messing, J.; Wu, Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 10842–10847. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Qi, W.; Lv, Y.; Yan, S.; Xu, L.; Yang, W.; Yuan, Y.; Chen, Y.; Zhao, H.; Song, R. OPAQUE11 Is a Central Hub of the Regulatory Network for Maize Endosperm Development and Nutrient Metabolism. Plant Cell 2018, 30, 375–396. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Guo, L.; Ji, C.; Wang, H.; Wang, J.; Zheng, X.; Xiao, Q.; Wu, Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 2021, 33, 104–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, H.; Guo, L.; Wu, X.; Xiao, Q.; Wang, J.; Wang, Q.; Ma, G.; Wang, W.; Wu, Y. ABA-induced phosphorylation of basic leucine zipper 29, ABSCISIC ACID INSENSITIVE 19, and Opaque2 by SnRK2.2 enhances gene transactivation for endosperm filling in maize. Plant Cell 2022, 34, 1933–1956. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Galli, M.; Spears, C.J.; Zhan, J.; Liu, P.; Yadegari, R.; Dannenhoffer, J.M.; Gallavotti, A.; Becraft, P.W. NAKED ENDOSPERM1, NAKED ENDOSPERM2, and OPAQUE2 interact to regulate gene networks in maize endosperm development. Plant Cell 2023, 36, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, S.; Zhao, Q.; Lv, D.; Wang, B.; Xiao, K.; Zhu, J.; Li, S.; Yang, W.; Liu, X.; et al. ZmGRAS11, transactivated by Opaque2, positively regulates kernel size in maize. J. Integr. Plant Biol. 2021, 63, 2031–2037. [Google Scholar] [CrossRef]
- Chen, Q.; Guo, Y.; Zhang, J.; Zheng, N.; Wang, J.; Liu, Y.; Lu, J.; Zhen, S.; Du, X.; Li, L.; et al. RNA polymerase common subunit ZmRPABC5b is transcriptionally activated by Opaque2 and essential for endosperm development in maize. Nucleic Acids Res. 2023, 51, 7832–7850. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lahner, B.; Gong, J.; Mahmoudian, M.; Smith, E.L.; Abid, K.B.; Rogers, E.E.; Guerinot, M.L.; Harper, J.F.; Ward, J.M.; McIntyre, L.; et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 1215–1221. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Finegan, C.; Boehlein, S.K.; Leach, K.A.; Madrid, G.; Hannah, L.C.; Koch, K.E.; Tracy, W.F.; Resende, M.F.R. Genetic Perturbation of the Starch Biosynthesis in Maize Endosperm Reveals Sugar-Responsive Gene Networks. Front. Plant Sci. 2022, 12, 800326. [Google Scholar] [CrossRef]
- Zhang, X.; Mogel, K.; Lor, V.S.; Hirsch, C.N.; De Vries, B.; Kaeppler, H.F.; Tracy, W.F.; Kaeppler, S.M. Maize sugary enhancer1 (se1) is a gene affecting endosperm starch metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 20776–20785. [Google Scholar] [CrossRef]
- Song, W.Y.; Yamaki, T.; Yamaji, N.; Ko, D.; Jung, K.H.; Fujii-Kashino, M.; An, G.; Martinoia, E.; Lee, Y.; Ma, J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 2014, 111, 15699–15704. [Google Scholar] [CrossRef]
- Liu, X.; Chen, S.; Chen, M.; Zheng, G.; Peng, Y.; Shi, X.; Qin, P.; Xu, X.; Teng, S. Association Study Reveals Genetic Loci Responsible for Arsenic, Cadmium and Lead Accumulation in Rice Grain in Contaminated Farmlands. Front. Plant Sci. 2019, 10, 61. [Google Scholar] [CrossRef]
- Kar, D.; Pradhan, A.A.; Dutta, A.; Bhagavatula, L.; Datta, S. The Multidrug and Toxic Compound Extrusion (MATE) Family in Plants and Their Significance in Metal Transport. In Plant Metal and Metalloid Transporters; Kumar, K., Srivastava, S., Eds.; Springer Nature Singapore: Singapore, 2022; pp. 151–177. [Google Scholar]
- Clemens, S.; Ma, J.F. Toxic Heavy Metal and Metalloid Accumulation in Crop Plants and Foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef]
- Jia, L.; Hu, D.; Wang, J.; Liang, Y.; Li, F.; Wang, Y.; Han, Y. Genome-Wide Identification and Functional Analysis of Nitrate Transporter Genes (NPF, NRT2 and NRT3) in Maize. Int. J. Mol. Sci. 2023, 24, 12941. [Google Scholar] [CrossRef]
- Delfin, J.C.; Kanno, Y.; Seo, M.; Kitaoka, N.; Matsuura, H.; Tohge, T.; Shimizu, T. AtGH3.10 is another jasmonic acid-amido synthetase in Arabidopsis thaliana. Plant J. 2022, 110, 1082–1096. [Google Scholar] [CrossRef]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef]
- Kemper, E.L.; Neto, G.C.; Papes, F.; Moraes, K.C.; Leite, A.; Arruda, P. The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell 1999, 11, 1981–1994. [Google Scholar] [CrossRef]
- Deng, Y.T.; Wang, J.C.; Zhang, Z.Y.; Wu, Y.R. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020, 18, 1897–1907. [Google Scholar] [CrossRef]
- Lappe, R.R.; Baier, J.W.; Boehlein, S.K.; Huffman, R.; Lin, Q.; Wattebled, F.; Settles, A.M.; Hannah, L.C.; Borisjuk, L.; Rolletschek, H.; et al. Functions of maize genes encoding pyruvate phosphate dikinase in developing endosperm. Proc. Natl. Acad. Sci. USA 2018, 115, E24–E33. [Google Scholar] [CrossRef]
- Ji, C.; Xu, L.; Li, Y.; Fu, Y.; Li, S.; Wang, Q.; Zeng, X.; Zhang, Z.; Zhang, Z.; Wang, W.; et al. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize. Mol. Plant 2022, 15, 468–487. [Google Scholar] [CrossRef]
- Yan, P.; Du, Q.; Chen, H.; Guo, Z.; Wang, Z.; Tang, J.; Li, W.X. Biofortification of iron content by regulating a NAC transcription factor in maize. Science 2023, 382, 1159–1165. [Google Scholar] [CrossRef]
- Gamuyao, R.; Chin, J.H.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.M.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef]
- Kettenburg, A.T.; Lopez, M.A.; Yogendra, K.; Prior, M.J.; Rose, T.; Bimson, S.; Heuer, S.; Roy, S.J.; Bailey-Serres, J. PHOSPHORUS-STARVATION TOLERANCE 1 (OsPSTOL1) is prevalent in upland rice and enhances root growth and hastens low phosphate signaling in wheat. Plant Cell Environ. 2023, 46, 2187–2205. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).