Abstract
The low survival rate of transplanted plantlets, which has limited the utility of tissue-culture-based methods for the rapid propagation of tree peonies, is due to plantlet dormancy after rooting. We previously determined that the auxin response factor PsARF may be a key regulator of tree peony dormancy. To clarify the mechanism mediating tree peony plantlet dormancy, PsARF genes were systematically identified and analyzed. Additionally, PsARF16a was transiently expressed in the leaves of tree peony plantlets to examine its regulatory effects on a downstream gene network. Nineteen PsARF genes were identified and divided into four classes. All PsARF genes encoded proteins with conserved B3 and ARF domains. The number of motifs, exons, and introns varied between PsARF genes in different classes. The overexpression of PsARF16a altered the expression of NCED, ZEP, PYL, GA2ox1, GID1, and other key genes in abscisic acid (ABA) and gibberellin (GA) signal transduction pathways, thereby promoting ABA synthesis and decreasing GA synthesis. Significant changes to the expression of some key genes contributing to starch and sugar metabolism (e.g., AMY2A, BAM3, BGLU, STP, and SUS2) may be associated with the gradual conversion of sugar into starch. This study provides important insights into PsARF functions in tree peonies.
1. Introduction
The tree peony (Paeonia suffruticosa Andr.) is a famous traditional flower in China, known as the “national beauty” and “the king of flowers.” In recent years, the value of its oil and ornamental features (i.e., as a fresh-cut flower species) and its health benefits have gradually been revealed, making the tree peony an increasingly important species in the floral industry, with a growing market demand [1,2]. Traditionally, tree peonies were propagated from seeds or by plant division and grafting, but the associated methods have drawbacks, such as a long cycle, low reproduction efficiency, and separation of offspring traits [3]. Accordingly, there is a lack of efficient methods for developing improved tree peony varieties and seedlings to satisfy the increasing market demand, which has seriously restricted the industrial development of tree peonies. The short cycle and high reproduction efficiency of tissue culture are conducive to the rapid development of enhanced tree peony varieties. Although a few tree peony plants have been obtained through tissue culture, the current methods must be optimized before they can be applied on an industrial scale. More specifically, the apical buds of plantlets become dormant after rooting, which results in a low survival rate after transplanting [4,5,6].
We previously conducted substantial research on the mechanism underlying tree peony plantlet dormancy. In particular, we measured endogenous hormone contents and performed transcriptome and targeted metabolome analyses during the dormancy process of plantlets. We determined that tree peony plantlet dormancy is closely related to abscisic acid (ABA) and auxin contents and proportions [5]. Aux/IAA and auxin response factor (ARF) transcription factors are key regulators of the auxin signaling pathway [7,8]. On the basis of association analyses, ARF was revealed to be a potentially important regulator of tree peony plantlet dormancy [5]. We speculated that the accumulation of auxin in plantlets after rooting is initiated promotes ABA synthesis, eventually leading to dormancy. Previous studies have shown that auxin and ABA synergistically regulate seed dormancy, with the associated physiological effects caused by changes to ABA signaling pathways [9]. Auxin and ABA contents and signal transduction are reportedly positively correlated with Arabidopsis thaliana seed dormancy [10]. Auxin may contribute to ABA-mediated seed dormancy and germination in two ways (i.e., promoting ABA biosynthesis or activating ABA responses) [11]. ARF transcription factors are important for regulating auxin signaling [12]. The ABI3, ABI4, and ABI5 genes encode three crucial downstream components of the ABA signal transduction pathway, of which ABI3 is the main regulator of seed dormancy and germination [13]. The inductive effect of auxin on seed dormancy depends on ABI3. Earlier research indicated ARF transcription factors may recruit or activate other specific transcription factors to promote ABI3 expression, thereby promoting dormancy [9,11]. Among the 23 AtARF genes in A. thaliana [14], AtARF10, AtARF16, and AtARF17 encode important transcription factors affecting the auxin signaling pathway during auxin-induced dormancy [15,16,17]. Both AtARF10 and AtARF16 are expressed when auxin levels are high, which leads to the activation of ABI3 transcription and the maintenance of seed dormancy in A. thaliana [9]. In wheat, TaARF15-A1 may down-regulate the expression of some genes that induce leaf senescence, while up-regulating the expression of genes that delay senescence, ultimately promoting dormancy [18]. Auxin and ARF10/16 are required for the maintenance of ABI3 expression in seeds. Moreover, ABI3, AtARF10/16, and bZIP67 are transcription factors that bind to RY and GBL elements in the DOG1 promoter region to regulate A. thaliana seed dormancy and germination [19]. Therefore, auxin signaling pathways are necessary for plant dormancy, with AtARF10 and AtARF16 functioning as key downstream components.
The completely sequenced tree peony genome [20,21] is useful for identifying and functionally characterizing tree peony genes. However, there are currently no reports of studies on tree peony PsARF genes. In the current study, tree peony PsARF gene family members were systematically identified on the basis of genome data and full-length transcriptome sequencing data. In addition, we cloned PsARF16a, which is a homolog of the A. thaliana gene (AtARF16) associated with dormancy, and overexpressed it in tree peonies using a transient expression system to explore its regulatory effects on a downstream gene network. The results preliminarily revealed the regulatory effect of PsARF16a on ABA synthesis, which promotes plantlet dormancy. The study findings provide a theoretical basis for future research conducted to modulate tree peony plantlet dormancy.
2. Materials and Methods
2.1. Materials
P. suffruticosa cultivar ‘Daojin’ plantlet roots, stems, leaves, and apical buds were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until used for the full-length transcriptome sequencing analysis. Total RNA was extracted from ‘Daojin’ plantlets for the gene cloning experiment. At approximately 3 months after transplantation, the leaves of the ‘Daojin’ plantlets were injected for the transient transformation and quantitative real-time PCR (qRT-PCR) analysis.
2.2. Full-Length Transcriptome Sequencing
Full-length transcriptome sequencing was conducted by Biomarker Technologies Co., Ltd. (Beijing, China). Total RNA was extracted from the root, stem, leaf, and apical bud samples collected from tree peony plantlets. RNA quality was examined using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). High-quality RNA was used to construct the cDNA library. PacBio long reads were filtered, with redundant sequences removed using CD-HIT-EST. Sequences shorter than 50 bp and those with a sequence accuracy <0.9 were removed. The filtered full-length transcripts were functionally annotated using the NCBI non-redundant protein sequences (NR; https://www.ncbi.nlm.nih.gov/, accessed on 12 November 2023), Swiss-Prot (https://www.uniprot.org/, accessed on 12 November 2023), Gene Ontology (GO; https://www.geneontology.org/, accessed on 12 November 2023), Clusters of Orthologous Genes (COG; https://www.ncbi.nlm.nih.gov/research/cog-project/, accessed on 12 November 2023), EuKaryotic Orthologous Groups (KOG; https://ftp.ncbi.nlm.nih.gov/pub/COG/KOG/, accessed on 12 November 2023), Protein family (Pfam; https://pfam.janelia.org, accessed on 12 November 2023), and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/, accessed on 12 November 2023) databases.
2.3. Identification of Tree Peony PsARF Genes
The published genomes [20,21] of P. suffruticosa ‘Luoshenxiaochun’ (CNP0000281) and Paeonia ostii (CNP0003098) as well as the full-length transcriptome data of P. suffruticosa ‘Daojin’ generated by the Institute of Horticulture of Henan Academy of Agricultural Sciences were used to build a local database. Using “auxin response factor” as the keyword, the annotation information for the sequencing results was screened to select ARF family members and identify PsARF genes containing the Auxin_resp domain. Sequence information for the PsARF genes (e.g., nucleic acid sequence, protein sequence, coding sequence, and genome location) was obtained using genome data as the reference.
2.4. Phylogenetic Analysis of PsARF Genes
Tree peony PsARF protein sequences were compared with A. thaliana AtARF and rice OsARF protein sequences using ClustalX (http://www.clustal.org/, accessed on 20 December 2023). AtARF and OsARF protein sequences were downloaded from online databases (https://www.arabidopsis.org/, accessed on 21 December 2023 and https://www.ricedata.cn/, accessed on 21 December 2023, respectively). A phylogenetic tree was constructed according to the maximum likelihood method using MEGA11 (https://www.megasoftware.net/, accessed on 25 December 2023), with the validation parameter (bootstrap) set to 500. The identified tree peony ARF genes were named according to their similarities to A. thaliana ARF genes in the phylogenetic tree.
2.5. Bioinformatics Analysis of PsARF Genes
On the basis of the PsARF sequences, the isoelectric point, molecular weight, and other physicochemical properties of the encoded proteins were predicted using ProtParam (http://web.expasy.org/protparam/, accessed on 2 December 2023). The PsARF domain structures were predicted using NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 5 December 2023), whereas conserved motifs were analyzed using MEME 5.5.4 (http://meme-suite.org/tools/meme, accessed on 13 December 2023). The PsARF gene structures were analyzed using GSDS 2.0 (http://gsds.cbi.pku.edu.cn, accessed on 29 December 2023).
2.6. Gene Cloning and Vector Construction
On the basis of the transcriptome data for dormancy after the rooting process of tree peony plantlets and the phylogenetic relationships with A. thaliana genes, PsARF16a, which may be related to tree peony plantlet dormancy, was selected for cloning. Total RNA was extracted from tree peony plantlets using the MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). RNA quality was assessed using the NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). High-quality RNA was reverse-transcribed to cDNA using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa). The PsARF16a cDNA sequence was amplified by PCR using PrimeSTAR® GXL DNA Polymerase, a high-fidelity Taq enzyme (Baori Medical Biological Technology (Beijing) Co., Ltd.), and the following primers (5′→3′): Fw: GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCCTCTGGTGAACTCAAAG; Rv: GGGGACCACTTTGTACAAGAAAGCTGGGTATTACGCAAATATGCTCAAATG. The amplified product was purified and inserted into the pDONR207 vector via a BP reaction (Gateway™ BP Clonase™ II; Thermo Fisher Scientific, Waltham, MA, USA) and then used to construct the overexpression vector pK2GW7 via an LR reaction (Gateway™ LR Clonase™ II; Thermo Fisher Scientific). The resulting recombinant plasmid was inserted into Agrobacterium tumefaciens AGL1 cells for the subsequent analysis of transient gene expression [22].
2.7. Subcellular Localization
To verify that PsARF16a is a transcription factor, we examined its subcellular localization. Specifically, PsARF16a was cloned into the p221-GFP vector using the ClonExpress® II One Step Cloning Kit C112-V9.1 (Vazyme, Nanjing, China) to obtain the p221-PsARF16a-GFP recombinant plasmid, which was used for the transfection of A. thaliana mesophyll protoplasts according to the PEG method. Protoplasts were extracted and transfected as described by Yoo et al. [23]. After a 16 to 20 h incubation, the subcellular distribution of the recombinant protein was detected using a TCS SP8 fluorescence microscope (Leica, Wiltsburg, Germany) with a 40× magnification objective lens. For the subcellular localization analysis, p221-AtWRKY40-mCherry (Shanghai Plantshop Biotechnology Co., Ltd., Shanghai, China), which was used as a positive control, was co-expressed with PsARF16a-GFP. The GFP and mCherry signals were detected using excitation wavelengths of 488 and 560 nm, respectively, and emission wavelengths of 500–550 and 600–650 nm, respectively. In total, 4 × 105 protoplasts were used for each transfection. Three independent experiments were performed, with similar results. Microscopy images were prepared using Leica Application Suite X software, version 1.4.6, (Leica, Wiltsburg, Germany).
2.8. RNA Sequencing and Transcriptome Analysis
The recombinant plasmid carrying PsARF16a was injected into the leaves of transplanted tree peony plantlets, whereas MS (Duchefa Biochemie, Haarlem, The Netherlands) liquid medium was injected into the control leaves. For each leaf, the region surrounding the injection site was collected at 24, 36, 48, and 60 h after the injection for the subsequent transcriptome sequencing analysis. Total RNA was extracted using an EASYspin Plus Plant RNA Mini Kit (Beijing Adelai Biological Technology Co., Ltd., Beijing, China). RNA purity and concentration were determined using the NanoDrop ultra-microspectrophotometer 2000 (NanoDrop Technologies, Wilmington, DE, USA). A cDNA library was constructed and sequenced by Beijing Baimike Biotechnology Co., Ltd. The cDNA library quality was evaluated using the 2100 Bioanalyzer (Agilent Technologies), whereas the effective concentration of the cDNA library was determined by qRT-PCR. An Illumina HiSeq 2000 system (Illumina, CA, USA) was used for the paired-end sequencing of the high-quality cDNA library.
The differentially expressed genes (DEGs) among treatments were detected using DEseq [24]. The resulting p-values were adjusted using the Benjamini and Hochberg approach for controlling the false-discovery rate (FDR). An FDR < 0.01 and fold-change ≥ 2 were set as the criteria for identifying DEGs. A KEGG pathway enrichment analysis was performed using KOBAS 2.0 [25]. A gene expression heat map was constructed using the Biomarker cloud platform (https://international.biocloud.net/zh/software/tools/list, accessed on 16 February 2024).
2.9. Validation by Quantitative Real-Time PCR
To verify the reliability of the transcriptome sequencing data, qRT-PCR analysis was conducted to determine the relative expression levels of 10 DEGs associated with hormone signal transduction and starch and sucrose metabolism. Total RNA was extracted using the MiniBEST Plant RNA Extraction Kit (TaKaRa). The quality of the extracted RNA was evaluated using the NanoDrop 2000 spectrophotometer. High-quality RNA was reverse-transcribed using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa), after which the qRT-PCR analysis was completed using the SYBR Green PCR Master Mix Kit (Takara Bio, Shiga, Japan) and the Bio-Rad CFX instrument (Bio-Rad, Hercules, CA, USA). Relative expression levels were calculated according to the 2−ΔΔCt method, with a tree peony Pstubulin gene (Gene.20610::PB-F_transcript_12290/psu.T.00016986.1) serving as an internal control. The expression of all selected genes was analyzed using three replicates. The qRT-PCR data were analyzed using SPSS (SPSS, Inc., Chicago, IL, USA). Information regarding the qRT-PCR primers is listed in Table 1.

Table 1.
Primers used for quantitative real-time PCR analyses of differentially expressed genes.
3. Results
3.1. Identification and Physicochemical Characteristics of PsARF Genes
In total, 19 PsARF genes were identified on the basis of the genome data and full-length transcriptome data of Paeonia ostii, P. suffruticosa cultivar ‘Luo shen xiao chun’, and P. suffruticosa cultivar ‘Daojin’. The PsARF genes in tree peonies were compared with the AtARF genes in A. thaliana to identify homologs. The tree peony PsARF genes were named according to their similarities to AtARF genes. Bioinformatics analyses were conducted to obtain basic information regarding the physicochemical properties of PsARF genes (Table 2). The PsARF proteins were revealed to contain the B3 and ARF domains, making them true ARF proteins. The Aux/IAA domain was also detected in the PsARF proteins, except for PsARF3, PsARF10, PsARF11b, PsARF16a, PsARF16b, PsARF17, and PsARFx. Sixteen of the PsARF genes were distributed on five chromosomes, with chromosome 2 carrying the most (i.e., six PsARF genes). The remaining PsARF genes were distributed on other chromosomes. The PsARF sequences (367–1005 amino acids) had a predicted molecular weight of 40.17–112.46 kDa, with a theoretical isoelectric point of 4.47–10.11. Most of the PsARF proteins had an isoelectric point less than 7 (i.e., acidic proteins).

Table 2.
Basic information regarding the ARF gene family in tree peonies.
3.2. Evolutionary Relationships among PsARF Genes
To reveal the evolutionary relationships of tree peony PsARF genes, the PsARF protein sequences and the sequences of 46 ARF proteins in A. thaliana and rice were used to construct a phylogenetic tree (Figure 1). Eighteen PsARF genes were divided into four classes (Classes I, II, III, and IV), with each class consisting of AtARF, OsARF, and PsARF genes. However, the PsARF genes were significantly more closely related to A. thaliana ARF genes than to rice ARF genes. Similar to the rice gene OsARFx/BGIOSGA025276, PsARFx was clustered in a branch separate from the other A. thaliana and rice ARF genes, suggesting the functions of the proteins encoded by these two genes differ from those of the other ARF proteins. Class I was the largest, with 12 AtARF, 6 OsARF, and 6 PsARF genes, but these genes likely evolved relatively independently. Notably, AtARF12–15 and AtARF20–22 as well as PsARF11a and PsARF11b were clustered together, separate from the other ARF genes, with no highly homologous sequences detected. Class II included two pairs of A. thaliana and tree peony ARF genes (i.e., orthologs). In contrast, Classes III and IV included both orthologous and paralogous pairs of A. thaliana and tree peony ARF genes, with relatively diverse evolutionary patterns.
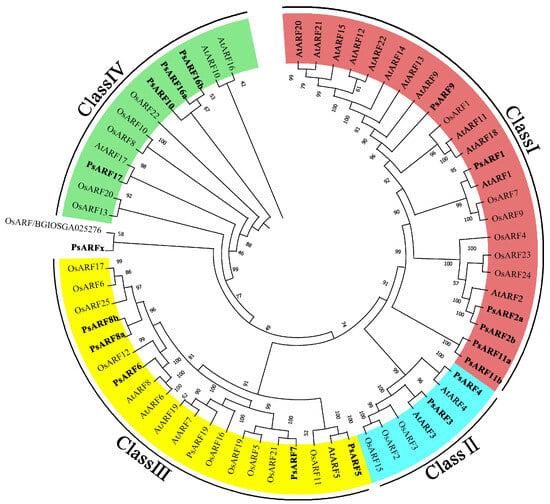
Figure 1.
Phylogenetic tree of the ARF gene families in tree peonies, A. thaliana, and rice. Classes I, II, III, and IV are indicated in red, blue, yellow, and green, respectively. Tree peony ARF genes in different classes are in bold.
3.3. Conserved Domains and Motifs in PsARF Proteins
According to the conserved domain analysis (Figure 2A), most of the PsARF proteins contained the B3, Auxin_resp, and Aux/IAA3 domains. All of the Class III proteins included the B3, Auxin_resp, and Aux/IAA3 domains. These domains were also detected in the proteins in Class I, except for PsARF11b, which lacked the C-terminal Aux/IAA domain. All of the Class IV proteins lacked the C-terminal Aux/IAA domain but contained the B3 and ARF domains. Similarly, PsARFx also contained only the B3 and Auxin_resp domains. Further analyses of the conserved motifs in PsARF proteins (Figure 2B) revealed differences in the types and number of motifs among PsARF sequences, which may reflect the functional diversity of these proteins. The B3 domain consists of motifs 1, 2, 8, and 12; the Auxin_resp domain consists of motifs 4, 7, 9, 13, and 15; and the Aux/IAA domain consists of motifs 6, 10, and 14. The Auxin_resp domain of PsARFx contained motif 7 but lacked motifs 4, 9, 13, and 15, which may explain why PsARFx was included in a separate branch of the phylogenetic tree. The amino acid sequences of each motif (Figure 2C) confirmed that the domains B3, Auxin_resp, and Aux/IAA were highly conserved, as were several amino acid sites in the domains.
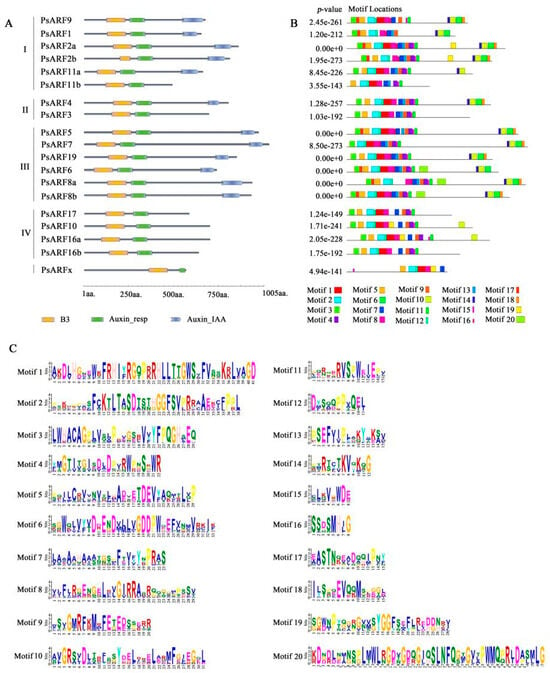
Figure 2.
Conserved domains and motifs in the ARF proteins in tree peonies. (A) Conserved domains in PsARF proteins. (B) Conserved motifs in PsARF proteins. (C) Motif logos and amino acid compositions.
3.4. PsARF Gene Structures
The structures of 16 PsARF genes in Paeonia ostii [21] were analyzed on the basis of the available PsARF gene annotation information (Figure 3). All 16 PsARF genes contained exons and introns, with similar exon–intron structures in the PsARF genes belonging to the same class. The number of exons varied between PsARF genes in different classes. Specifically, Class I genes had 13–16 exons, Class II genes had 10 exons, Class III genes had 11–19 exons, and Class IV genes had 3–7 exons. The PsARFx gene contained only two exons and no predicted untranslated region. These results suggest that structural differences among PsARF genes may be associated with their functional diversity.
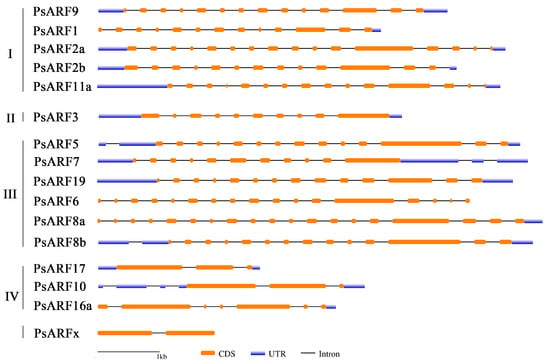
Figure 3.
Structural characteristics of ARF genes in tree peonies. Orange and blue squares and black lines represent the coding sequence (CDS), untranslated region (UTR), and intron of PsARF genes, respectively. The length of 1 kb is indicated at the bottom.
3.5. Gene Cloning and Subcellular Localization
Basic information regarding PsARF16a is provided in Table 2. Sequencing results indicated that PsARF16a (2025 bp) encodes a protein comprising 674 amino acids, which was in accordance with the transcriptome data. During the subcellular localization analysis involving A. thaliana mesophyll protoplasts, the fluorescent signals of the PsARF16a-GFP fusion protein and mCherry (positive control) were detected in the nucleus (Figure 4), indicating that PsARF16a is a typical transcription factor that functions mainly in the nucleus.
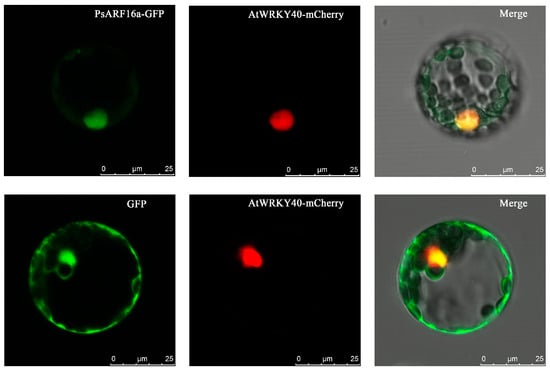
Figure 4.
Transient expression and subcellular localization of PsARF16a-GFP. p221-AtWRKY40-mCherry is a nuclear localization marker that was included in the co-transfection of A. thaliana mesophyll protoplasts for the transient expression analysis. Fluorescence was detected using a Leica fluorescence microscope. The merged GFP (green) and mCherry (red) signals suggest that PsARF16a was localized in the nucleus. Bar = 25 µm.
3.6. KEGG Pathway Enrichment Analysis of DEGs
The comparison between the control and samples transiently expressing PsARF16a revealed 1644, 605, 881, and 692 DEGs at 24, 36, 48, and 60 h, respectively, with more up-regulated genes than down-regulated genes at all time points. This suggested that many genes in the leaves were involved in a stress response, with significant changes to their expression levels following the transient expression of PsARF16a. The number of DEGs was highest at 24 h. Notably, the number of DEGs decreased significantly after 36 h, which indicated that most of metabolic activities in the leaves recovered and stabilized. The significantly enriched KEGG metabolic pathways among the DEGs included the following: plant hormone signal transduction, MAPK signaling pathway-plant, starch and sucrose metabolism, phenylpropanoid biosynthesis, glutathione metabolism, and carbon metabolism (Figure 5). These results may reflect the importance of the regulatory effects of PsARF16a on these metabolic pathways.
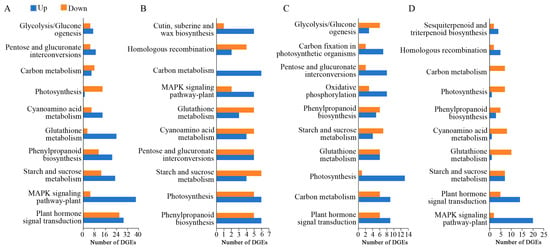
Figure 5.
Enriched KEGG pathways among the genes differentially expressed between the treatment and control groups at different post-injection time points. (A) Treatment vs. control at 24 h post-injection. (B) Treatment vs. control at 36 h post-injection. (C) Treatment vs. control at 48 h post-injection. (D) Treatment vs. control at 60 h post-injection.
3.7. DEG Screening and qRT-PCR Verification
Plant dormancy is closely related to endogenous hormones. Moreover, changes to sugar and starch metabolism are an important indicator of plant dormancy. The DEGs identified at different time points suggested the transient expression of PsARF16a modulated endogenous hormone signal transduction as well as sugar and starch metabolism. Hence, some key genes involved in endogenous hormone signal transduction or sugar and starch metabolism were screened on the basis of sequence similarity and homology (Figure 6), including IAA signaling pathway-related genes (e.g., AUX22D, IAA29, YUCCA10, SAUR23, and LAX5), gibberellin (GA) signaling pathway-associated genes (e.g., GA2ox1, GA2ox8, GAI1, and GID1), ABA signaling pathway-related genes (e.g., NCED3, ZEP, ABI1, PP2C38, PP2C55, and PYL4), and ethylene signaling pathway-related genes (e.g., CTR1, ERF1, and ETR2). Furthermore, AMY2A, BAM3, BGLU12, BGLU24, STP, and SUS2 were the identified genes involved in pathways related to starch and sucrose metabolism.
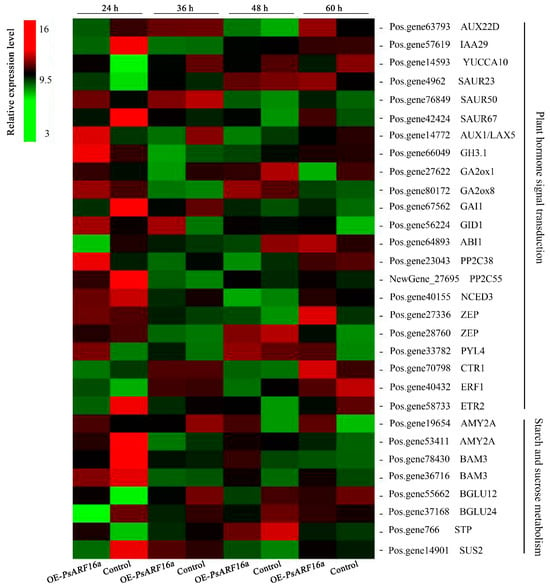
Figure 6.
Heat map of differentially expressed genes associated with plant hormone signal transduction as well as starch and sucrose metabolism. According to the transcriptome analysis, the relative expression levels of various genes at different time points were clustered. OE-PsARF16a represents samples transiently overexpressing PsARF16a. Red and green indicate high and low relative expression levels, respectively.
To validate the reliability of the RNA-seq results, the following 10 DEGs predominantly associated with plant hormone signal transduction or starch and sucrose metabolism according to the unigene annotations were selected for the qRT-PCR analysis conducted to verify their expression patterns at different time points: Pos.gene4962, NewGene_27695, Pos.gene28760, Pos.gene55662, Pos.gene41152, Pos.gene70798, Pos.gene47097, Pos.gene25516, Pos.gene23350, and Pos.gene64704 (Figure 7). The comparison with the control detected significant changes in the expression levels of most of these genes at 24 h after samples were injected with PsARF16a, but the expression levels recovered and stabilized after 24 h (i.e., no significant differences with the corresponding control levels). The Pos.gene4962, NewGene_27695, NewGene_28760, and Pos.gene47097 expression levels were significantly down-regulated at 24 h after the injection with PsARF16a, which contrasted with the significantly up-regulated expression of the other genes. The qRT-PCR data were in accordance with the RNA-seq results, indicating that the transcriptome sequencing data were accurate and reliable.
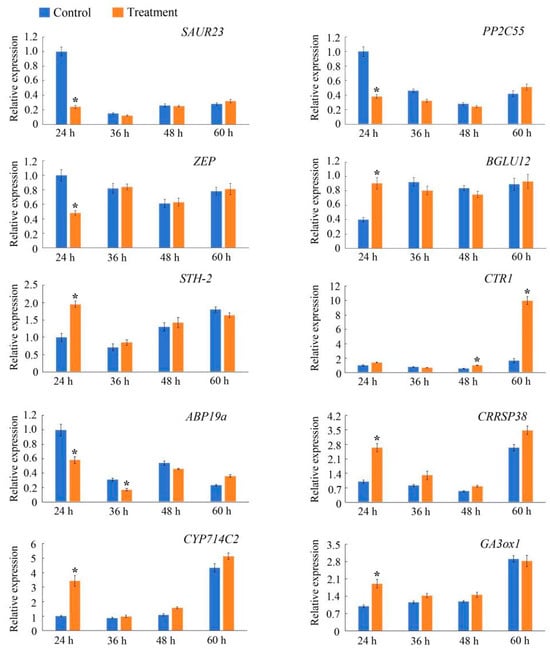
Figure 7.
Quantitative real-time PCR analysis of 10 differentially expressed genes in the treatment and control groups at different time points after samples were injected with PsARF16a. Error bars represent the standard error of three biological replicates. The asterisks indicate a significant difference (Student’s t-test: p < 0.05) between samples at different post-injection time points.
4. Discussion
In plants, ARF genes encode regulators of multiple critical biological processes related to growth and development [26]. In recent years, fully sequenced genomes of many plant species have been used to identify and analyze ARF gene families in different species, including A. thaliana, rice, poplar, tomato, corn, soybean, apple, barley, millet, and Dendrobium officinale [27]. In the current study, 19 PsARF genes were systematically identified by analyzing the genome and transcriptome data of three tree peony cultivars. Although there were similarities in the PsARF genes in the examined cultivars, there were also some differences. For example, PsARF4 was detected only in the genome of P. suffruticosa, whereas PsARF17 was exclusive to P. ostii, and PsARF16b was unique to ‘Daojin’. Phylogenetic analyses are important for elucidating gene family functions. Because of the relatively limited molecular-biology-based research on tree peonies, phylogenetic trees are critical for classifying tree peony gene functions. The 19 PsARF genes identified in this study were divided into four classes, which was consistent with the results of related research on pears [28] and grapes [29]. According to the constructed phylogenetic tree, PsARFx and the rice gene OsARFx/BGIOSGA025276 were distantly related to the other ARF genes in tree peonies, A. thaliana, and rice. Additionally, the Auxin_resp domain of PsARFx was incomplete (i.e., missing motifs 4, 9, 13, and 15), suggestive of incomplete gene transcription. Moreover, only two exons were detected in PsARFx, which may help to explain why this gene belonged to a separate branch of the phylogenetic tree. Compared with other PsARF proteins, PsARFx may have a different function because of its incomplete structure. Most of the examined PsARF proteins contained three conserved domains. The PsARF proteins in the same class had similar conserved domains and motifs. Similarly, the PsARF genes belonging to the same class were similar in terms of their exon–intron structures. These results reflect the reliability of the phylogenetic tree constructed on the basis of a multiple sequence alignment. Interestingly, the Class IV PsARF proteins lacked the Aux/IAA (CTD) domain. A previous study showed ARF activators and Aux/IAA suppressors can inhibit gene expression via the dimerization of the CTD domain [30]. The functions of ARF proteins lacking the CTD domain may not involve auxin signaling pathways [26], ultimately resulting in functional diversification. The PsARF genes in Class IV had fewer exons than the PsARF genes in the other classes, which was consistent with the findings of earlier related studies on apples [31] and pears [28]. Although the tree peony genome is larger than the A. thaliana genome, the PsARF gene family is smaller than the AtARF gene family, mainly because the tree peony has considerably fewer Class I genes than A. thaliana. This phenomenon was also observed in other plant species, including barley [32], apple [31], sweet orange [33], Eucalyptus grandis [34], and litchi [35]. Hence, similar to these species, the tree peony may have lost some PsARF genes during its evolution.
Earlier studies showed ARF genes are important for several processes related to plant growth and development, including flower and cotyledon formation, plant embryogenesis, leaf organ senescence, fruit maturation, and dormancy [36,37]. In the present study, our analysis of 65 ARF genes from A. thaliana, rice, and tree peonies showed that PsARF10 and PsARF16 are highly homologous to AtARF10 and AtARF16. Both AtARF10 and AtARF16, which are essential for maintaining ABI3 expression in seeds, can activate ABI3 transcription and promote seed dormancy [8]. Therefore, PsARF10 and PsARF16 may have similar functions and regulate tree peony plantlet dormancy. Dormancy induction and release are regulated by various signaling molecules, including sugars and plant hormones. Specifically, hormones act as the first signal in the dormancy-related signal transduction pathway, inhibiting or activating the activities of intracellular protein factors and enzymes by binding to receptors on the target cell membrane, thereby regulating gene expression and metabolism [38]. Endogenous ABA, GA, auxin (e.g., IAA), and zeanoside are the four major hormones that regulate plant dormancy and germination [39]. Of these hormones, ABA induces plant dormancy. Decreases in the GA content inhibit plant growth but do not induce dormancy [40]. Auxin is the most important regulator of bud dormancy, with IAA modulating P450 monooxygenase gene expression, while also controlling the ABA content, to induce bud dormancy [41]. In the current study, the significantly enriched pathways among the DEGs in PsARF16a-overexpressing tree peony plantlet leaves were related to plant hormone signal transduction as well as starch and sugar metabolism. The expression levels of most DEGs changed significantly at 24 h after PsARF16a was injected into the leaves, after which they recovered and stabilized, with no significant difference from the corresponding control level. Zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED) are two enzymes that regulate ABA biosynthesis [42]. In the current study, the overexpression of PsARF16a may have promoted the transcription of ABI, and ABI1 expression was significantly up-regulated at 60 h, which activated the ABA signal transduction pathway, resulting in the up-regulated expression of NCED3 at 36 and 60 h. In addition, ZEP expression was significantly down-regulated at 24 h, whereas ABA synthesis increased. Furthermore, the expression of the ABA receptor gene PYL was significantly up-regulated. Earlier research showed that PYL/PYR can initiate ABA signal transduction by inhibiting the activity of the protein phosphatase PP2C [43]. The PP2C protein HONSU is a key factor regulating the ABA/GA balance, which influences seed dormancy [44]. In our study, the PYL expression level was significantly up-regulated, which may have inhibited PP2C activities and significantly down-regulated PP2C expression at 24 h, thereby initiating ABA signal transduction and altering the ABA/GA balance. Consequently, the genes involved in GA metabolism were activated. The expression levels of GA2ox1 (GA synthase) and GID1 (GA receptor factor) were significantly up-regulated at 24 h. In tree peony plantlets, the GA3 content decreases significantly after rooting, whereas GID1 expression is significantly up-regulated [5]. Changes in carbohydrate content are important indicators of plant dormancy and germination, with the conversion of sugar and starch directly affecting the dormancy process. Dormancy release is the result of a shift in glucose metabolism from the glycolytic/tricarboxylic acid cycle to the pentose phosphate pathway [45]. During dormancy release and germination, starch hydrolysis is accelerated, with the associated increase in soluble sugar contents (sucrose, fructose, and glucose) enhancing the activities of a series of starch-degrading enzymes and increasing the expression of genes encoding the relevant enzymes [46,47]. However, the opposite changes occur during dormancy induction. Specifically, various metabolic activities are weakened or completely inhibited, energy consumption decreases substantially, and energy storage increases after plants enter dormancy. In our study, the expression levels of some key genes associated with starch and sugar metabolism were significantly affected by the overexpression of PsARF16a. For example, AMY2A, BAM3, and BGLU expression levels were significantly up-regulated at 24 h, which was in contrast to the significant decrease in the expression of STP and SUS2. A previous study indicated BGLU is a key gene in the sugar synthesis pathway, and its expression is closely associated with sugar contents [48]. These results imply that PsARF16a overexpression promoted leaf dormancy, with weakened metabolic activities, accelerated starch synthesis, and the gradual conversion of sugar into starch. On the basis of the study findings, we speculate that after plantlets were transferred to the rooting medium, the accumulation of a large amount of auxin enhanced the response to PsARF16a expression, which activated ABI transcription and induced the expression of some key genes that increased ABA synthesis and decreased GA synthesis, ultimately leading to imbalanced ABA and GA contents and plantlet dormancy.
5. Conclusions
In this study, 19 PsARF genes in three tree peony cultivars were identified and analyzed in terms of their physicochemical properties, gene structures, and protein structures. Our analysis of transient expression in tree peony leaves revealed the importance of PsARF16a for regulating ABA signal transduction and ABA synthesis. Moreover, PsARF16a may be a critical regulator of tree peony plantlet dormancy. The study data may form the basis of future investigations conducted to functionally characterize PsARF genes and further clarify the molecular mechanism underlying tree peony plantlet dormancy.
Author Contributions
Conceptualization and methodology, Z.F.; investigation, X.Y., Y.Z., H.W., Y.L., J.G. and L.W.; resources, X.W. and L.L.; formal analysis, H.Z.; writing—original draft, Z.F. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Funds of Distinguished Young Scholars of Henan Academy of Agricultural Sciences (grant no. 2022JQ03) and The Innovation Team project of Henan Academy of Agricultural Sciences (grant no. 2024TD36).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are included within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wen, S.S.; He, R.R.; Zheng, J.K.; Tian, R.N. Research advances in tissue culture of tree peony. Chin. For. Sci. 2018, 54, 143–155. [Google Scholar]
- Zhang, Y.; Gao, L.; Wang, Y.; Niu, D.; Yuan, Y.; Liu, C.; Zhan, X.; Gai, S. Dual functions of PsmiR172b-PsTOE3 module in dormancy release and flowering in tree peony (Paeonia suffruticosa). Hortic. Res. 2023, 10, uhad033. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Cheng, F.; Zhong, Y. Micropropagation of tree peony (Paeonia × lemoinei ‘High Noon’) and the assessment of genetic stability by SSR analysis. Propag. Ornam. Plants 2016, 16, 19–27. [Google Scholar]
- Zhu, X.T.; Li, X.Q.; Ding, W.J.; Jin, S.H.; Wang, Y. Callus induction and plant regeneration from leaves of peony. Hortic. Environ. Biotechnol. 2018, 59, 575–582. [Google Scholar] [CrossRef]
- Fu, Z.Z.; Xu, M.L.; Wang, H.J.; Wang, E.Q.; Li, Y.M.; Wang, L.M.; Gao, J.; Zhang, J.; Yuan, X. Analysis of the transcriptome and related physiological indicators of tree peony (Paeonia suffruticosa Andr.) plantlets before and after rooting in vitro. Plant Cell Tissue Organ Cult. 2021, 147, 529–543. [Google Scholar] [CrossRef]
- Liu, L. Studies on Tissue Culture and Establishment of the Regeneration System of Tree Peonies. Master’s Thesis, Southwest University, Chongqing, China, 2009. [Google Scholar]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Jürgens, G. Funneling auxin action: Specificity in signal transduction. Curr. Opin. Plant Biol. 2004, 7, 687–693. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.; He, Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Song, S.Q.; Liu, J.; Tang, C.F.; Zhang, W.H.; Xu, H.H.; Zhang, Q.; Gao, J.D. Metabolism and signaling of auxins and their roles in regulating seed dormancy and germination. Chin. Sci. Bull. 2020, 65, 3924–3943. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. J. Plant Growth Regul. 2001, 20, 281–291. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. Book 2008, 6, e0119. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the auxin response factor gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Li, H.F.; Liu, H.; Hao, C.Y.; Li, T.; Liu, Y.C.; Wang, X.L.; Yang, Y.X.; Zheng, J.; Zhang, X.Y. The auxin response factor TaARF15-A1 negatively regulates senescence in common wheat (Triticum aestivum L.). Plant Physiol. 2023, 191, 1254–1271. [Google Scholar] [CrossRef]
- Bryant, F.M.; Hughes, D.; Hassani-Pak, K.; Eastmond, P.J. Basic leucine zipper transcription factor 67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in Arabidopsis. Plant Cell 2019, 31, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Cheng, S.; Wang, Z.; Li, S.; Jin, X.; Lan, L.; Yang, B.; Yu, K.; Ni, X.; Li, N.; et al. Draft genome of the famous ornamental plant Paeonia suffruticosa. Ecol. Evol. 2020, 10, 4518–4530. [Google Scholar] [CrossRef]
- Yuan, J.; Jiang, S.; Jian, J.; Liu, M.; Yue, Z.; Xu, J.; Li, J.; Xu, C.; Lin, L.; Jing, Y.; et al. Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii. Nat. Commun. 2022, 13, 7328. [Google Scholar] [CrossRef]
- Russell, D.W.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.Q.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.D.; White, J.; Cheung, F.; Parvizi, B. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identifcation using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Liu, Z.H.; Yu, Y.C.; Xiang, F.N. Auxin response factors and plant growth and development. Hereditas 2011, 33, 1335–1346. [Google Scholar] [CrossRef]
- Bai, Y.W.; Ma, Y.J.; Chang, Y.T.; Zhang, W.B.; Deng, Y.Y.; Zhang, N.; Zhang, X.; Fan, K.K.; Hu, X.M.; Wang, S.H.; et al. Identification and transcriptome data analysis of ARF family genes in five Orchidaceae species. Plant Mol. Biol. 2023, 112, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.Q.; Jiang, S.L.; Wang, F.; Zhao, Y.N. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in pear. Sci. Agric. Sin. 2018, 51, 327–340. [Google Scholar]
- Wan, S.; Li, W.; Zhu, Y.; Liu, Z.; Huang, W.; Zhan, J. Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 2014, 33, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.; Jurgens, G.; De Smet, I. The evolving complexity of the auxin pathway. Plant Cell 2008, 20, 1738–1746. [Google Scholar] [CrossRef]
- Luo, X.C.; Sun, M.H.; Xu, R.R.; Shu, H.R.; Wang, J.W.; Zhang, S.Z. Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef]
- Huseyin, T. Genome-wide analysis of the auxin response factors(ARF) gene family in barley (Hordeum vulgare L.). J. Plant Biochem. Biotechnol. 2018, 28, 14–24. [Google Scholar]
- Li, S.B.; OuYang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front Plant Sci. 2015, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Soler, M.; Mila, I.; San Clemente, H.; Savelli, B.; Dunand, C.; Paiva, J.A.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J. Genome-wide characterization and expression profiling of the auxin response factor(ARF) gene family in Eucalyptus grandis. PLoS ONE 2014, 9, e108906. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Wei, Y.Z.; Wang, Y.; Zheng, X.W.; Li, W.C. Transcriptome-wide identification and expression profiling of the auxin response factor(ARF)gene family in Litchi chinensis Sonn. Chin. J. Trop. Crops 2017, 38, 1485–1491. [Google Scholar]
- Wu, J.; Wang, F.; Cheng, L.; Kong, F.; Peng, Z.; Liu, S.; Yu, X.; Lu, G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059–2073. [Google Scholar] [CrossRef]
- Yang, J.; Tian, L.; Sun, M.X.; Huang, X.Y.; Zhu, J.; Guan, Y.F.; Jia, Q.S.; Yang, Z.N. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 2013, 162, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, J.Y.; Zhou, Y.W. Research progress on molecular regulation mechanisms in bulbous plants at dormancy. Acta Prataculturae Sin. 2013, 22, 295–304. [Google Scholar]
- Yan, X.N.; Tian, M.; Liu, F.; Wang, C.X.; Zhang, Y. Hormonal and morphological changes during seed development of Cypripedium japonicum. Protoplasma 2017, 254, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Su, W.C.; Jia, Y.L.; Sun, L.L.; Xu, H.L.; Wu, R.H. Correlation analysis of abscisic acid change and dormancy in volunteer wheat. Chin. Plant Physiol. J. 2022, 58, 402–414. [Google Scholar]
- Vanstraelen, M.; Benková, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef]
- Li, J.; Li, C.Y. Seventy-year major research progress in plant hormones by Chinese scholars. Sci. Sin. Vitae 2019, 49, 1227–1281. [Google Scholar]
- Hu, S.; Wang, F.Z.; Liu, Z.N.; Liu, Y.P.; Yu, X.L. ABA signaling mediated by PYR/PYL/RCAR in plants. Hereditas 2012, 34, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Okubo, H.; Saitou, K. Increase in the expresson of an alpha-amylase gene and sugar accumulation induced during cold period reflects shoot elongation in hyacinth bulbs. J. Am. Soc. Hortic. Sci. 2006, 131, 185–191. [Google Scholar] [CrossRef]
- Shin, K.S.; Chakrabarty, D.; Paek, K.Y. Sprouting rate, change of carbohydrate contents and related enzymes during cold treatment of lily bulblets regenerated in vitro. Sci. Hortic. 2002, 96, 195–204. [Google Scholar] [CrossRef]
- Cochrane, M.P.; Duffers, C.M. The effect of low temperature storage on the activities of α-and β-amylase and α-glucosidase in potato tubers. Potato Res. 1991, 34, 333–341. [Google Scholar] [CrossRef]
- Stassen, M.J.J.; Stringlis, I.A. Decoupling Sugar and Spice in Soybean Rhizosphere Depends on BGLU Activity. Plant Cell Physiol. 2023, 64, 451–453. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).