Abstract
Genomics has revolutionised the study of the biology of parasitic diseases. The first Eukaryotic parasite to have its genome sequenced was the malaria parasite Plasmodium falciparum. Since then, Plasmodium genomics has continued to lead the way in the study of the genome biology of parasites, both in breadth—the number of Plasmodium species’ genomes sequenced—and in depth—massive-scale genome re-sequencing of several key species. Here, we review some of the insights into the biology, evolution and population genetics of Plasmodium gained from genome sequencing, and look at potential new avenues in the future genome-scale study of its biology.
1. Introduction
Since the genome of the malaria parasite Plasmodium falciparum was published in 2002 [1], alongside that of its mosquito vector [2] and its human host (Consortium and International Human Genome Sequencing Consortium, 2001) [3], malaria genomics has led the way in the study of eukaryotic pathogens. Since then, a growing number of Plasmodium species’ genomes have been sequenced and large-scale population resequencing studies have been carried out in P. falciparum and several other species. These efforts have allowed the evolutionary and population genomics of malaria parasites to be studied in unprecedented detail and provide a model for the application of genomics to the study of other parasite species. The application of other genome-wide sequencing analyses (e.g., transcriptomics and methylomics) broadens our knowledge of Plasmodium biology and presents new frontiers to study. Here we consider some of these and how they can be studied at scale. This review presents some of the key advances in the study of the evolutionary genomics of malaria parasites made possible by the application of genomic sequencing technologies.
2. Expanding Horizons: Sequencing Across the Genus Plasmodium
The genus Plasmodium contains a huge number of species that parasitise a wide range of vertebrate hosts (Figure 1). For a broad view of the phylogenetic relationships among the malaria parasites, see [4]. At the time of writing, 5-6 Plasmodium species are known to infect humans. Four of these have been known for many years: Plasmodium falciparum and Plasmodium vivax are the most widespread and clinically important species, with Plasmodium malariae and Plasmodium ovale both less common and less well studied. More recently, these have been joined by Plasmodium knowlesi, a zoonotic malaria that primarily infects macaques, and P. ovale has been recognised as not one but two (probable) species: P. ovale curtisi and P. ovale wallikeri [5].
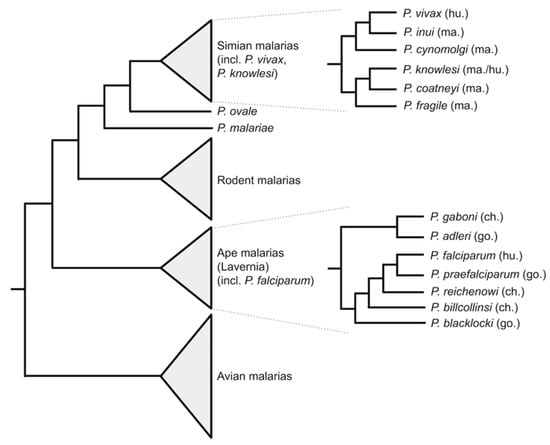
Figure 1.
Phylogeny of malaria parasites, showing the relationships among human-infecting species. The tree is a schematic diagram indicating topology (branch lengths are not to scale). The phylogenetic positions of human-infecting Plasmodium species are indicated. More detailed subtrees are shown for major lineages of simian malaria parasites and for the Laverania subgenus of ape parasites. Host species are indicated in brackets (hu = human; ma = macaque; ch = chimpanzee; go = gorilla). Tree topologies are after [4], and after [10] for the Laverania.
Plasmodium genomes are, in some ways, rather attractive genomes to sequence: haploid in the vertebrate host (and in vitro culture), not too large (around 23 Mb, encoding around 5500 genes for P. falciparum) and with 14 well-defined chromosomes. In other ways they present technical challenges: they are extremely AT-rich, the resulting lowered complexity of parts of the genome increasing the challenge in sequencing and assembly. However, these challenges have been overcome and as a result, a large and growing number of Plasmodium species’ genomes have been fully sequenced and assembled, to varying levels of completeness. For some reference genomes, initial sequencing and assembly efforts using first- and second-generation sequencing technologies have subsequently been improved using third-generation, long read sequencing technology. Additional strains have also been independently sequenced and assembled for several species [6,7,8,9].
Alongside Plasmodium falciparum [1], the other human malaria parasites P. vivax [6,11], P. malariae, P. ovale curtisi and P. ovale wallikeri [12,13,14] have been sequenced. Other members of the subgenus Laverania, parasites of African great apes, have been sequenced including the chimpanzee parasites P. reichenowi and P. adleri and gorilla parasites P. gaboni, P. blacklocki and P. billcollinsi [10,15]. A P. vivax-like parasite of great apes has also been sequenced [13]. The simian parasite of macaques and zoonotic human pathogen P. knowlesi has been sequenced [16,17], as well as other simian parasites including P. coatneyi [18] and P. cynomolgi [8,9]. Outside of the primate malarias, the genomes of rodent malaria parasites P. berghei, P. chabaudi chabaudi and P. yoelii yoelii, important model species for the study of malaria biology, have been sequenced [7,19,20,21,22]. Two avian malaria species, P. gallinaceum and P. relictum, have been sequenced [23]. Additional strains have also been independently sequenced and assembled for P. falciparum [24], P. vivax [6,11] and P. cynomolgi [8,9]. Together, this represents an enviably richly sequenced genus.
Having so many species’ genomes provides a rich resource for studying evolution in the genus. For example, an early study of comparative structural genomics between P. falciparum and rodent malaria parasites showed extensive chromosome structural rearrangement and that lineage-specific genes could often be found at the breakpoints of these translocations [25]. More recently, third-generation long read sequencing has provided further insights into the genomic architecture of Plasmodium. The central region of each chromosome, around the centromere, comprises a relatively stable ‘core genome’ characterised by broad co-linearity and one-to-one homology of the genes among genomes. An estimated 90% [26] or more [24,26] of the genome has been suggested to be ‘core genome’. The outer ‘subtelomeric regions’ are much more variable and are enriched for large and hypervariable gene families such as the variant surface antigens var, rifin and stevor [25,27,28,29,30], likely due to increased activity of processes like slipped-strand mispairing and ectopic recombination generating new genes and segmental duplications. The repetitive nature of the subtelomeric regions made them particularly difficult to study until long read sequencing technologies became more advanced [16,24]. The insight afforded by these advances is important because of the key roles played by some of these gene families in the infection biology of the parasite, such as immune evasion and sequestration of infected red blood cells. Large multi-gene families show different distributions among Plasmodium species; for example, the kir and SICAvar expanded gene families of P. knowlesi are not confined to the subtelomeres but occur throughout the genome [16,17]. Links between the structural genomics and the biology of different Plasmodium species will be an interesting area of study made possible by the advances in sequencing technologies.
A densely sequenced genus has other advantages. Comparative genomics among more closely related species allows the dating of gene family expansions [10,31] and the identification of processes such as introgression and horizontal gene transfer [10,32,33], both of which can play major roles in adaptation and virulence. Among members of the Laverania, one introgression event is of particular interest. It transferred orthologues of members of a key erythrocyte invasion complex (rh5 and cyrpa) from an ancestor of the gorilla parasite P. adleri to the lineage leading to P. falciparum and its sister species, the gorilla parasite P. praefalciparum [10,32,33]. These genes, along with the proteins P113 and RIPR form a protein complex that is essential for red blood cell invasion [34,35,36,37]. RH5 binds the erythrocyte surface protein Basigin and displays differential binding efficiency to human, gorilla and chimpanzee Basigin, implicating it as a key determinant of host specificity [38,39,40] and a vaccine candidate [41].
Identification of cryptic or incipient species is the point where inter-specific comparative genomics meets population genomics, discussed in the next section. Two notable examples here are the human parasite Plasmodium ovale and the simian parasite Plasmodium knowlesi. Plasmodium ovale is now recognised to be two distinct, non-recombining lineages, called Plasmodium ovale curtisi and Plasmodium ovale wallikeri [5]. Recognition of this previously cryptic diversity is evolutionarily significant, but also relevant to the clinical manifestations of infection. For example, Plasmodium ovale wallikeri displays a shorter latency period (time to onset of symptoms, linked to the dormant liver hypnozoite life-cycle stages) and deeper thrombocytopaenia (low blood platelet levels) than Plasmodium ovale curtisi [42,43,44,45]. In addition, where malaria transmission is seasonal, as it is in much of West Africa, latency periods are significantly lower during transmission season [42,43]. Plasmodium knowlesi, which naturally infects macaques but can be transmitted to humans, is found across southeast Asia following the distribution of its macaque hosts. Population genomics indicates both putative cryptic lineages and large-scale introgression, discussed in the next section.
3. The Variable Genome: Genomic Diversity within Plasmodium Species
Population level re-sequencing of Plasmodium genomes has allowed population genomics to be studied in great detail, both to identify geographical population structure and genome regions under various selection pressures including anti-malarial drugs and the host’s adaptive immune response (Figure 2). For an excellent review of malaria population genomics, see [46]. The large-scale data-sharing Malaria Genomic Epidemiology Network (MalariaGEN) has made available thousands of parasite genomes—7000 P. falciparum genomes from 28 countries at the time of writing—to the research community [47,48]. The data provide an unprecedented snapshot of P. falciparum population genetics and the associated advances in sample preparation, sequencing technology, analysis methods and data sharing lay the foundations to more fully integrate genomics into disease epidemiology, though technical challenges remain [49,50].
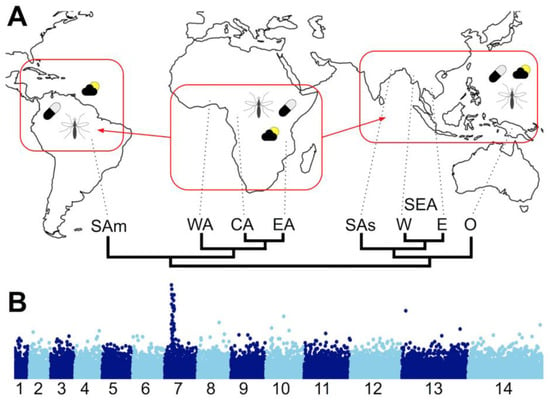
Figure 2.
Plasmodium falciparum population genomics. (A) Map representing spread of the globally distributed species P. falciparum from sub-Saharan Africa to South America and Asia and the differing selection pressures that may drive local adaptation to different settings within and between continents: climatic and environmental conditions (sun and cloud); different vector species (the mosquito); differing treatment regimes (the pill). Below the map is a schematic population structure tree based on genome-wide variation data, after [48]. (Regional populations are indicated at the tips of the tree: SAm = South America; WA = West Africa; CA = Central Africa; EA = East Africa; SAs = South Asia; SEA,W = South East Asia, West; SEA,W = South East Asia, East; O = Oceania). (B) Representation of a genome-wide scan for signatures of selection. Indices (e.g., homozygosity or linkage disequilibrium within a population, FST among populations, etc.) are plotted on the y-axis across all chromosomes on the x-axis (here, for P. falciparum’s 14 chromosomes) to identify outlier regions, such as the chloroquine resistance transporter gene (crt) region on chromosome 7 shown here. Data points could be individual variant sites, genes or other chromosomal regions, depending upon the index being plotted. By highlighting outliers compared to a genomic average, confounding effects of population histories can be avoided.
P. falciparum arose in Africa. Until recently, it was thought to have evolved from an ancestor that infected the common ancestor of humans and chimpanzees, and to have speciated with its hosts (into P. falciparum in humans and P. reichenowi in chimpanzees). More recent sampling of the Laverania subgenus suggests a likely cross-species transmission from gorillas, as its closest relative is the gorilla parasite P. praefalciparum [51,52]. Perhaps due to this recent host switch the historical P. falciparum populations underwent a population bottleneck that is evident today in the low level of genetic diversity present in the species, much lower than in the other Laverania [10,52,53], or in P. vivax [54].
Before genomic data were available for P. falciparum, microsatellite genotyping showed a population structure reflecting current transmission intensities in different parts of the world [55]. For example, extensive outcrossing occurs in many parts of sub-Saharan Africa where transmission intensity is high, as the obligately sexual parasite more frequently encounters gametes from other parasites in the mosquito host. This leads to extremely low linkage disequilibrium (LD) even between genes in close proximity on a chromosome and to low divergence (as measured by FST) among populations. In other regions where transmission rates are lower, mixed-genotype infections are rarer and selfing is more the norm leading to LD over a greater range on chromosomes and to higher FST among populations [55]. The increased resolution of whole-genome population resequencing allowed the identification of fine population structure and the recognition of different populations [56]. For example, across sub-Saharan Africa, genome-wide genetic diversity defines East, West, Central and other ancestral parasite populations [48,56,57], including a highly divergent population in the Horn of Africa [57].
Alongside whole genome resequencing, still relatively expensive despite falling costs, the development of other ‘post-genomic’ tools such as SNP ‘barcoding’ panels [58,59,60] has allowed parasite population structures to be studied. This approach has been used in a number of studies of Plasmodium vivax [61,62,63,64,65,66]. P. vivax, like P. falciparum, may have originated in Africa and be of gorilla origin [52], though some lines of evidence support a simian origin in Asia [67]. Today Plasmodium vivax is the most geographically widespread of the human malarias, and accounts for an increasing proportion of malaria cases in many endemic areas [68]. The species is more genetically diverse than P. falciparum [54,69] which may partly reflect low diversity and a more recent bottleneck in P. falciparum, but may also partly reflect its distinctive population structure. In co-endemic regions, the population structures of P. vivax have often been seen to differ from P. falciparum, being more genetically diverse and less geographically structured [65,69,70,71]. The population genetics of P. vivax varies across its range. In Latin America high levels of differentiation are seen among sites, suggested to indicate multiple independent introductions of the parasite [72,73,74,75]. In southeast Asia, genetic diversity tends to be high, with lower differentiation among different regions and countries [72,73,74,75,76]. Little population structure and high genetic diversity is seen in the south Pacific [69,77,78]. P. vivax is rare, though not absent, across much of Africa [79] due to the prevalence of the Duffy antigen negative blood group in many African populations; the Duffy antigen being the primary receptor required for erythrocyte invasion [79,80]. African endemic foci are found in Madagascar and the Horn of Africa in the east [79,80,81] and Mauritania in the west [61,62,63], with patterns of genetic diversity and divergence from geographically distinct populations indicating long-lived, endemic populations [61,62,63]. The contrast between P. vivax and P. falciparum in co-endemic regions (e.g., [65,71]) is significant because the fracturing of parasite populations into smaller, distinct, isolated subpopulations, fewer polyclonal infections, less outcrossing, increased linkage disequilibrium and reduced genetic diversity are all indicators of falling endemicity due to successful disease control. Therefore, the absence of these indicators in P. vivax in some settings suggests a greater resilience of the species to malaria control interventions. In some cases P. vivax appears to retain, somewhat paradoxically, high genetic diversity even with high LD [82] or declining transmission [83], whether as a result of more recent populations fracturing, epidemic transmission patterns, the effects of selection or the effects of relapse from dormant hypnozoites is not fully determined.
The population structure of the simian parasite and human zoonosis Plasmodium knowlesi has been best studied in Bornean and Peninsular Malaysia. The parasite shows highly differentiated subgroups, broadly associated with their natural vertebrate host species: long tailed (Macaca fascicularis) and pig tailed (Macaca nemestrina) macaques [84]. Both are found in human infections in Bornean Malaysia [85,86,87,88]. A third divergent population is documented in both humans and macaques on Peninsular Malaysia [89,90,91]. Further substructure is seen within this Peninsular Malaysia population [90]. Moreover, whole genome sequencing has shown evidence of introgression and a mosaic structure of the P. knowlesi genome, indicating a breakdown of genetic isolation between divergent groups [85,86,87,88]. This complex population structure should be studied further as it may provide important insights into transmission and pathogenesis in this zoonotic parasite.
In addition to population structure, another area of major interest in Plasmodium genome biology is the identification of genes evolving under selective pressures favouring novel alleles or maintaining diversity within populations. The effects and, importantly, the targets of specific forces like anti-malarial drug exposure or protective adaptive immunity can be identified. Prior to large-scale genomic re-sequencing, considerable efforts were expended in disentangling the effects of selection and population processes (such as population subdivision or population expansion) on population genetic indices, such as Tajima’s D, based on population allele frequencies (for a review see [92]). Genome-scale data allows population effects (that will tend to affect the whole genome) and selection effects (the effects of which are more localised) to be separated more easily, as gene or genome region indices can be compared to the distribution of the index across the genome in order to identify outliers. Such whole genome scans for selective sweeps—genome regions with reduced levels of genetic diversity or heterozygosity, or elevated LD—pinpoint chromosomal regions associated with drug-resistance, clearly visible in continent-wide patterns of P. falciparum genomic diversity [57]. These characteristic signatures mark loci associated with resistance to different antimalarial drugs. The dhfr and dhps genes, encoding targets of sulfadoxine and pyrimethamine (used in combination as sulfadoxine-pyrimethamine; SP) show clear signatures of recent strong selection in both P. falciparum [93,94] and P. vivax [95,96,97], even where SP is not a first line treatment of vivax malaria, possibly showing an ‘off-target’ effect of falciparum malaria treatment. The chloroquine resistance gene crt shows similar signatures of selection on resistance alleles in P. falciparum [93], while gene amplification has been reported at the crt and multidrug resistance gene mdr1 loci [96,98,99]. In addition to this, recent selection on the region upstream of crt has also been reported in Ethiopian P. vivax [100].
In the case of resistance to the artemisinin, antimalarials the picture is complex. The first descriptions of slower P. falciparum parasite clearance in response to artemisinin were from Cambodia, and subsequently the phenotype spread across southeast Asia [101,102,103]. Numerous studies have associated resistance with mutations in the kelch13 gene on P. falciparum chromosome 13 [101,104,105,106,107], though a large number of different kelch13 mutations exist [108] and cases of slow parasite clearance have been described in Africa in the absence of known kelch13 resistance makers [109]. The precise role of kelch13 in resistance is unclear and is the subject of intense research. Indeed, the mechanism of resistance to artemisinins may differ from that to other drugs in being broader and involving changes to many cellular processes affecting the cell cycle and cellular housekeeping and stress-response processes including the unfolded protein response [110,111,112,113]. Such a broad response can complicate the search for reliable genetic markers of resistance, markers that are vitally important for monitoring the spread of the phenotype from southeast Asia to sub-Saharan Africa. Alongside kelch13 variants, large scale genomic re-sequencing of southeast Asian parasite populations associated the resistance phenotype with variants in several other genes (fd, mdr2, crt, arps10) [105]. Another large-scale re-sequencing study showed 155 SNPs within kelch13 of which 128 were found in African and 62 in southeast Asian parasites [47]. Analysis of their genetic backgrounds, a key benefit of the whole genome sequencing approach, indicated that the African variants arose independently within Africa, rather than being transferred from southeast Asia, yet they do not appear to be associated with artemisinin resistance in Africa [47]. It has been suggested that certain genetic backgrounds, common in southeast Asia but not Africa, may predispose for kelch13 resistance mutations and explain the association seen with variants in several other genes [105].
Genome-scale scans of indices of divergence among populations can pinpoint genes under differential selection among these populations. For example, P. falciparum populations from countries with differing transmission patterns show unusually high divergence (as measured by the fixation index FST) coinciding with a gene (gdv1) associated with sexual differentiation [114,115]. In addition to this, apparent differential selection in crt was seen between two west African countries: Gambia and Guinea [114,115]. Selection driven by other pressures can also be detected in the genomes of parasites. For example, the globalisation of P. falciparum required adaptation to different vector species. Consistent with this, genes expressed in the mosquito stages of the parasite’s life cycle are among the most geographically divergent in the genome [48] and signatures of adaptation are visible in a number of these genes [116].
4. The Dynamic Genome: Methylomics in Plasmodium
Genomics has allowed us to understand Plasmodium population level processes in unprecedented detail. However, other ‘omes’ also offer useful windows into aspects of the parasite’s biology that cannot always be studied using genetic variation alone. Understanding how malaria parasites regulate their gene expression is crucial to comprehend Plasmodium biology, from the radical changes between life-cycle stages to the expression of alternative invasion pathway proteins and other host-parasite interaction proteins. One way to study this is by transcriptomic quantification of expressed RNA [117,118,119,120,121,122]; another is by epigenomics: mapping of epigenetic marks in the DNA or chromatin structure of the genome [123,124,125,126,127,128]. DNA methylation in Plasmodium represents a potential new frontier in Plasmodium genomics, yet it remains far from being fully understood and its existence has been somewhat controversial. Here, we consider DNA methylation in P. falciparum.
During erythrocyte infection the P. falciparum genome is mostly euchromatic [129,130]. Nevertheless, epigenetic mechanisms to control gene expression involving non-coding RNAs, the use of alternative histone variants and histone post-translational modifications are well documented [123,124,125]. Whether the Plasmodium epigenetic machinery also included DNA methylation, and in particular 5-methylcytosine (5mC), was a question that awaited an answer for a long time. The apparent lack of 5mC added to other characteristics of Plasmodium: lack of a functional RNA interference system [131] and linker histone H1 [131,132] and a mainly euchromatic genome [129,130]. Moreover, Plasmodium would not be the only eukaryote dispensing with 5mC: the fruit fly Drosophila melanogaster presents an extremely low level (less than 0.01%) of 5mC [133] and Caenorhabditis elegans, Trichoplax adhaerens, Schmidtea mediterranea, the urochordate Oikopleura dioica, the rotifer Adineta vaga have all dispensed with 5mC [134,135].
However, in eukaryotes 5mC plays a major role in many different processes including genomic imprinting, DNA repair, response to stress and the environment, regulation of gene expression and immune response [133,136,137,138,139]. The AT-rich Plasmodium genome [1] represents an obstacle for mass spectrometry-based methods to detect 5mC [140,141] and early analyses did not detect it [140]. Even though approaches that utilize methylation sensitive restriction enzymes (cutting at CG rich sites) and immunoprecipitation are not ideal to properly interrogate the AT-rich Plasmodium genome, a methylation-sensitive restriction approach identified a signature of 5mC in the Dihydrofolate Reductase Thymidylate Synthase (DHFR-TS) gene [142].
Recent advances in the sensitivity of mass spectrometry and the sequencing of bisulfite treated DNA have allowed better investigation of 5mC in Plasmodium [126,127,128]. Bisulfite treatment converts unmethylated cytosines into uracils and permits differentiation between methylated and unmethylated sites [143]. It is possible, therefore, sequencing the bisulfite converted DNA, to discern between methylated-cytosines (read as C) and unmethylated-cytosine (read as T). Comparing the sequence of the bisulfite treated genome with a reference genome it is possible to identify methylated and unmethylated cytosines and the proportion of cytosines in the genome that are methylated. In addition, at every cytosine site the proportions of reads containing methylated and unmethylated cytosine can be seen, indicating the proportion of cells in a sample that are methylated at a given position. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), Ponts and collaborators were able to clearly identify signatures of 5mC in the P. falciparum genome and suggested between 0.36% and 1.31% of all cytosines were methylated, depending on the stage analysed (ring, trophozoite or schizont) [128]. This was corroborated by deep sequencing of bisulfite treated P. falciparum DNA recovered from an asynchronous population that returned 0.58% methylated cytosines [128]. McInroy and colleagues found a remarkably similar pattern of DNA methylation in the rodent parasite Plasmodium berghei [127]. They used LC-MS/MS and a PCR-free bisulfite sequencing method (‘recovery after bisulfite treatment’; ReBuilT) to investigate the P. berghei genome for signatures of 5mC. Mass spectrometry and ReBuilT indicated, respectively, 0.33% and 1.87% of all cytosines were methylated.
Recent findings suggest that the majority of modified cytosines in P. falciparum may not in fact be 5mC, but 5-hydroxymethylcytosine (5hmC) [126]. This modified cytosine is derived from the Ten-Eleven Translocation methylcytosine dioxygenase (TET) mediated oxidation of the methyl group (-CH3 -> CH2OH) of 5mC [144] (Figure 3). TET is part of the pathway to restore unmethylated cytosine: TET catalyzes the conversion of 5mC to 5-carboxylcytosine (5caC), via the intermediates 5hmC and 5-formylcytosine (5fC) [145,146,147,148]; then, uracil-DNA glycosylase and base-excision repair (BER) excises and repairs 5caC back to unmethylated cytosine [145,146,147,149] (Figure 3A). Hammam and colleagues used a variety of methods to query methylation levels in P. falciparum, including ELISA, bisulfite sequencing (BS-seq) and oxidative bisulfite sequencing (oxBS-seq), hydroxymethylated DNA immunoprecipitation (hmeDIP) and LC-MS mass spectrometry [126]. Using anti-5mC or anti-5hmC antibodies in different Plasmodium life cycle stages (Ring, Trophozoite, Schizont) and ELISA, they estimated 0.19–0.38% cytosine to be 5hmC and only 0.01–0.02% to be 5mC [126]. These proportions were corroborated using a combination of BS-seq and oxBS-seq (0.2–0.26% 5hmC-like and 0.01–0.02% 5mC) [126]. The bisulfite reaction that is part of the BS-seq protocol is unable to convert 5mC or 5hmC into uracil, therefore BS-seq cannot distinguish between these 2 cytosine modifications [150,151]. However, in oxBS-seq 5hmC is first oxidised to 5fC that bisulfite treatment can convert to uracil allowing, by comparing BS-seq and oxBS-seq results, the level of 5hmC to be quantified [150,151] (Figure 3B). Hydroxymethylated DNA immunoprecipitation (hmeDIP) followed by next generation sequencing [152,153] showed the vast majority (98–98.5%) of 5hmC occurred in genic regions while only around 1–1.5% was intergenic, and that most was found in a CHH context (H representing any nucleotide but G) [126]. Further, hydroxymethylated genes had high levels of mRNA expression in all life-cycle stages analysed in the study [126]. Oddly, LC-MS did not identify 5hmC but gave a similar signal that could belong to a more hydrophobic positional isomer (5hmC-like modification) [126]. Overall, Hammam et al.’s [126] results corroborated previous findings on the level and distribution of DNA methylation in Plasmodium [127,128], but suggest also that the predominant cytosine modification is 5hmC (or 5hmC-like).
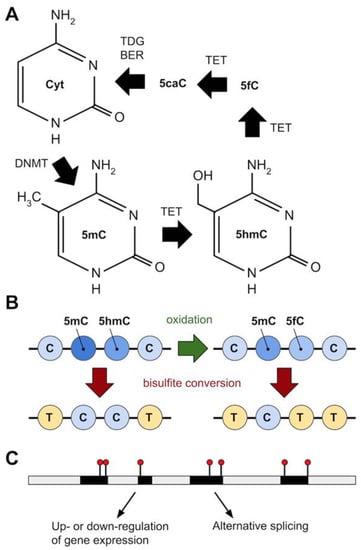
Figure 3.
Epigenomics in Plasmodium. (A) Cytosine can be converted to 5-Methylcytosine (5mC) by DNA methyltransferase (DNMT), and 5mC to 5-Hydroxymethylcytosine (5hmC) by Ten-eleven translocation methylcytosine dioxygenase (TET), which also converts 5hmC to 5-formylcytosine (5fC) and 5fC to 5-carboxylcytosine (5caC). Uracil-DNA glycosylase (TDG) and base-excision repair (BER) converts 5caC back to (unmethylated) cytosine. (B) The standard genomic method to detect 5mC (bisulfite treatment to convert unmethylated C to uracil, seen as a T in the final sequencing library, followed by whole genome sequencing) cannot distinguish 5mC from 5hmC. The addition of an oxidation step, converting 5hmC to 5fC (which bisulfite treatment does convert to uracil) can allow 5hmC to be detected and quantified. This approach suggests the majority of methylated C in Plasmodium is 5hmC. (C) Epigenetic marks (black lines with red circles) are shown on a stretch of DNA (grey = intergenic/intronic; black = exonic). In Plasmodium, exonic sequence near to intron-exon boundaries is enriched for these modified cytosines. Possible functional consequences include gene silencing or upregulation, and alternative splicing. Genome-wide patterns of methylation could act as alternative markers for gene expression.
The mechanistic role of 5mC in Plasmodium is not clear (Figure 3C). In mammals, most 5mC are found in CG duplexes; however, that is not the case in P. falciparum where the vast majority of 5mC are found in CHH contexts [128]. This is also seen in P. berghei (CAH is particularly common), where only a small proportion was associated with CG, CC or CHG [127]. The distribution of methylation in P. falciparum, with more methylation in the gene body than in intergenic regions and demarcating exon-intron boundaries, is also seen in other lineages [135] and may suggest a role in regulating alternative splicing [154] or RNA polymerase II DNA binding affinity [155]. In P. berghei, methylation seems to be equally distributed between genic and intergenic regions though within genes methylation seems to be mostly exonic and to mark intron–exon boundaries [127].
There seems to be a correlation between the distribution of 5mC and specific histone modifications in P. falciparum. Heterochromatin is characterized by H3K9me3 while transcriptionally active euchromatin is marked by H3K9ac and H4K20me3, at least in var genes [123,124,125]. H3K9ac and H3K4me3 associate to the coding region of actively expressed genes while H3K36me2 is a marker of repression. Cytosine methylation levels increase downstream of histone markers (H3K9ac and H3K4me3) linked to higher expression, while methylation levels do not change around silencing histone markers (H3K9me3 and H4K20me3) [128]. It is important to note here, that in many metazoans, H3K4me3 anticorrelates with 5mC due to the reduced affinity of DNMT3 ADD domain for this histone modification [156,157].
The fact that methylation profiles are different between DNA strands and that methylation levels change among life cycle stages suggest most if not all 5mC in Plasmodium is de novo methylation [128]. This implies that Plasmodium must express enzymes to add and remove 5mC. The eukaryotic methylation toolkit includes six DNA Methyltransferases (DNMTs) with different domains and different phylogenetic relationships, three Ten-Eleven Translocation methylcytosine dioxygenase (TET) and eleven Methyl-CpG Binding Domain (MBD) readers [135,145,147]. Some eukaryotic lineages have duplicated or lost some of the main DNMTs and other components of the methylation toolkit [135]. Initial investigations identified only one DNMT2 in the Plasmodium genome [128]. In mammals and other eukaryotes, different DNMTs have dedicated roles. DNMT1 seems to be the dedicated ‘maintenance methyltransferase’, replicating the methylation signature to newly formed DNA strands after cell division [135,145,147]. DNMT3 is the dedicated ‘de novo methyltransferase’, adding new methyl markers, often asymmetrically, to DNA strands. DNMT2 is evolutionarily highly conserved, but its role seems to be associated to tRNA methylation and species that only express this methylases often lack 5mC [145,158]. Indeed, Plasmodium aspartic tRNA is methylated by Plasmodium DNMT2 (TRDMT1) [159]. Importantly, Hammam and colleagues, using ELISA, demonstrated TET hydroxylase activity in Plasmodium nuclear extracts however they could not identify a TET homologous in the P. falciparum genome [126].
Plasmodium methylation context (mostly CHH) may suggest possible components of its methylation system. Similarities and differences between eukaryotic lineages suggest a great diversity and plasticity in the evolution of mechanisms of 5mC and its function [135]. For example, animals have well conserved DNMT1 and DNMT3 genes but display substantial differences in the distribution and level of 5mC, ranging from hypermethylated to sparsely methylated genomes in mammals and hymenoptera respectively [135]. DNMT5 mediates nucleosomal linkers methylation in some marine algae, but this pattern is not replicated in DNMT5-expressing fungi [160,161] and in some plants DNMT3 only methylates CHH context [162]. This suggests that the particular pattern of methylation observed in an organism may be the results of the activity of modulators (e.g., MDB proteins) that control DNMT activity. It is also possible that methylated cytosines in CHH context serve as binding sites for MDB proteins [147]. In mammal brains MeCP2 (an MDB protein family member) binds 5mC in non-CpG context and regulates gene expression [163,164,165,166].
The recent discovery that the more abundant cytosine modification in P. falciparum is indeed 5hmC may have implications for how this epigenetic marker modulates gene expression. The 5hmC signature is read by specific members of the MBD protein family such as methyl-CpG binding protein2 (Mecp2), methyl-CpG binding domain proteins 4 (Mbd4), Kaiso and SRA (reviewed in [167]). Mbd4 was also implicated in the process of demethylation converting 5hmC in 5hmUracil by deamination [168]. The 5hmC was also found to be particularly enriched in embryonic stem cells and brain tissues in euchromatic regions and in gene bodies, where its level correlated with gene expression [153,168,169,170,171,172].
It is becoming apparent that Plasmodium epigenetic machinery does include DNA methylation but much remains unknown. Bioinformatic approaches have at times been insufficient to identify Plasmodium orthologs [1] and these approaches failed to clearly identify the components of Plasmodium methylation/demethylation pathway. It is possible that MDB proteins play fundamental roles in modulating DNA methylation/demethylation in Plasmodium and that may even be the crucial component of this pathway. The role of 5hmC, as well as 5fC, needs further consideration. 5fC was found at promoters of active genes and in association with H3K4me3 in ES cells, again underlining the connection between cytosine modifications, histone modifications and gene expression [173,174,175]. Methodological advances are helping to overcome some of the limitations imposed by Plasmodium’s AT-rich genome on fully investigating the role of cytosine modification. For example, the PCR-free ‘ReBuilT’ approach used by McInroy et al. [127] recovers many sequence fragments that are usually lost during bisulfite sequencing library preparation and avoids PCR amplification biases (favouring GC-rich DNA).
5. Conclusions
Recent years have seen huge advances in Plasmodium genomics. Through intensive and extensive study of the genomes of these parasites, we have learned a great deal about their origins, evolution and biology. Future prospects include the further embedding of genomics in malaria epidemiology as well as advances in, and possible large-scale analysis of, the other ‘omics of malaria to gain further insights into this deadly, fascinating parasite.
Author Contributions
M.P. and G.D.W. wrote and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.C.; Wides, R.; et al. The Genome Sequence of the Malaria Mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Galen, S.C.; Borner, J.; Martinsen, E.S.; Schaer, J.; Austin, C.C.; West, C.J.; Perkins, S.L. The polyphyly of Plasmodium: Comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict. R. Soc. Open Sci. 2018, 5, 171780. [Google Scholar] [CrossRef]
- Sutherland, C.J.; Tanomsing, N.; Nolder, D.; Oguike, M.; Jennison, C.; Pukrittayakamee, S.; Dolecek, C.; Hien, T.T.; Rosário, V.E.D.; Arez, A.P.; et al. Two Nonrecombining Sympatric Forms of the Human Malaria Parasite Plasmodium ovale Occur Globally. J. Infect. Dis. 2010, 201, 1544–1550. [Google Scholar] [CrossRef]
- Auburn, S.; Böhme, U.; Steinbiss, S.; Trimarsanto, H.; Hostetler, J.; Sanders, M.; Gao, Q.; Nosten, F.; Newbold, C.I.; Berriman, M.; et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 2016, 1, 4. [Google Scholar] [CrossRef]
- Otto, T.D.; Böhme, U.; Jackson, A.P.; Hunt, M.; Franke-Fayard, B.; Hoeijmakers, W.A.M.; Religa, A.A.; Robertson, L.; Sanders, M.; Ogun, S.A.; et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Pasini, E.M.; Böhme, U.; Rutledge, G.G.; Der Wel, A.V.-V.; Sanders, M.; Berriman, M.; Kocken, C.H.; Otto, T.D. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res. 2017, 2, 42. [Google Scholar] [CrossRef]
- Tachibana, S.-I.; Sullivan, S.A.; Kawai, S.; Nakamura, S.; Kim, H.R.; Goto, N.; Arisue, N.; Palacpac, N.M.Q.; Honma, H.; Yagi, M.; et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat. Genet. 2012, 44, 1051–1055. [Google Scholar] [CrossRef]
- Otto, T.D.; Gilabert, A.; Crellen, T.; Böhme, U.; Arnathau, C.; Sanders, M.; Oyola, S.O.; Okouga, A.P.; Boundenga, L.; Willaume, E.; et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat. Microbiol. 2018, 3, 687–697. [Google Scholar] [CrossRef]
- Carlton, J.M.; Adams, J.H.; Silva, J.C.; Bidwell, S.L.; Lorenzi, H.; Caler, E.; Crabtree, J.; Angiuoli, S.V.; Merino, E.F.; Amedeo, P.; et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 2008, 455, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Ansari, H.R.; Templeton, T.J.; Subudhi, A.K.; Ramaprasad, A.; Tang, J.; Lu, F.; Naeem, R.; Hashish, Y.; Oguike, M.C.; Benavente, E.D.; et al. Genome-scale comparison of expanded gene families in Plasmodium ovale wallikeri and Plasmodium ovale curtisi with Plasmodium malariae and with other Plasmodium species. Int. J. Parasitol. 2016, 46, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Gilabert, A.; Otto, T.D.; Rutledge, G.G.; Franzon, B.; Ollomo, B.; Arnathau, C.; Durand, P.; Moukodoum, N.D.; Okouga, A.; Ngoubangoye, B.; et al. Plasmodium vivax—Like genome sequences shed new insights into Plasmodium vivax biology and evolution. PLoS Biol. 2018, 16, e2006035. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, G.G.; Böhme, U.; Sanders, M.; Reid, A.J.; Cotton, J.A.; Maiga-Ascofare, O.; Djimdé, A.A.; Apinjoh, T.O.; Amenga-Etego, L.; Manske, M.; et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature 2017, 542, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.D.; Rayner, J.C.; Böhme, U.; Pain, A.; Spottiswoode, N.; Sanders, M.; Quail, M.; Ollomo, B.; Renaud, F.; Thomas, A.W.; et al. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat. Commun. 2014, 5, 4754. [Google Scholar] [CrossRef]
- Benavente, E.D.; De Sessions, P.F.; Moon, R.W.; Grainger, M.; Holder, A.A.; Blackman, M.J.; Roper, C.; Drakeley, C.J.; Pain, A.; Sutherland, C.J.; et al. A reference genome and methylome for the Plasmodium knowlesi A1-H.1 line. Int. J. Parasitol. 2018, 48, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Pain, A.; Böhme, U.; Berry, A.E.; Mungall, K.; Finn, R.; Jackson, A.P.; Mourier, T.; Mistry, J.; Pasini, E.M.; Aslett, M.A.; et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 2008, 455, 799–803. [Google Scholar] [CrossRef]
- Chien, J.-T.; Pakala, S.B.; Geraldo, J.A.; Lapp, S.A.; Humphrey, J.C.; Barnwell, J.W.; Kissinger, J.; Galinski, M.R. High-Quality Genome Assembly and Annotation for Plasmodium coatneyi, Generated Using Single-Molecule Real-Time PacBio Technology. Genome Announc. 2016, 4, e00883-16. [Google Scholar] [CrossRef]
- Brugat, T.; Reid, A.J.; Lin, J.-W.; Cunningham, D.; Tumwine, I.; Kushinga, G.; McLaughlin, S.; Spence, P.; Böhme, U.; Sanders, M.; et al. Antibody-independent mechanisms regulate the establishment of chronic Plasmodium infection. Nat. Microbiol. 2017, 2, 16276. [Google Scholar] [CrossRef]
- Carlton, J.M.; Angiuoli, S.V.; Suh, B.B.; Kooij, T.W.; Pertea, M.; Silva, J.C.; Ermolaeva, M.D.; Allen, J.; Selengut, J.D.; Koo, H.L.; et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 2002, 419, 512–519. [Google Scholar] [CrossRef]
- Fougère, A.; Jackson, A.P.; Paraskevi Bechtsi, D.; Braks, J.A.; Annoura, T.; Fonager, J.; Spaccapelo, R.; Ramesar, J.; Chevalley-Maurel, S.; Klop, O.; et al. Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole. PLoS Pathog. 2016, 12, e1005917. [Google Scholar] [CrossRef]
- Hall, N.; Karras, M.; Raine, J.D.; Carlton, J.M.; Kooij, T.W.; Berriman, M.; Florens, L.; Janssen, C.S.; Pain, A.; Christophides, G.K.; et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 2005, 307, 82–86. [Google Scholar] [CrossRef]
- Böhme, U.; Otto, T.D.; Cotton, J.A.; Steinbiss, S.; Sanders, M.; Oyola, S.O.; Nicot, A.; Gandon, S.; Patra, K.P.; Herd, C.; et al. Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Res. 2018, 28, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.D.; Böhme, U.; Sanders, M.J.; Reid, A.J.; Bruske, E.I.; Duffy, C.W.; Bull, P.C.; Pearson, R.D.; Abdi, A.I.; Dimonte, S.; et al. Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res. 2018, 3, 52. [Google Scholar] [CrossRef]
- Kooij, T.W.; Carlton, J.M.; Bidwell, S.L.; Hall, N.; Ramesar, J.; Janse, C.J.; Waters, A.P. A Plasmodium whole-genome synteny map: Indels and synteny breakpoints as foci for spe-cies-specific genes. PLoS Pathog. 2005, 1, e44. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.; Iqbal, Z.; Vauterin, P.; Pearson, R.; Campino, S.; Theron, M.; Gould, K.; Mead, D.; Drury, E.; O’Brien, J.; et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016, 26, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Cloonan, N.; Fischer, K.; Thompson, J.; Waine, G.; Lanzer, M.; Saul, A. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 1998, 97, 161–176. [Google Scholar] [CrossRef]
- Fernandez, V.; Hommel, M.; Chen, Q.; Hagblom, P.; Wahlgren, M. Small, Clonally Variant Antigens Expressed on the Surface of the Plasmodium falciparum–Infected Erythrocyte Are Encoded by the rif Gene Family and Are the Target of Human Immune Responses. J. Exp. Med. 1999, 190, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Rubio, J.P.; Thompson, J.K.; Cowman, A.F. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 1996, 15, 4069–4077. [Google Scholar] [CrossRef]
- Su, X.; Heatwole, V.M.; Wertheimer, S.P.; Guinet, F.; Herrfeldt, J.A.; Peterson, D.S.; Ravetch, J.A.; Wellems, T.E. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of plasmodium falciparum-infected erythrocytes. Cell 1995, 82, 89–100. [Google Scholar] [CrossRef]
- Larremore, D.; Sundararaman, S.A.; Liu, W.; Proto, W.R.; Clauset, A.; Loy, D.E.; Speede, S.; Plenderleith, L.J.; Sharp, P.M.; Hahn, B.H.; et al. Ape parasite origins of human malaria virulence genes. Nat. Commun. 2015, 6, 8368. [Google Scholar] [CrossRef]
- Plenderleith, L.J.; Liu, W.; Learn, G.H.; Loy, D.E.; Speede, S.; Sanz, C.M.; Morgan, D.B.; Bertolani, P.; Hart, J.A.; Hart, T.B.; et al. Ancient Introgression between Two Ape Malaria Parasite Species. Genome Biol. Evol. 2019, 11, 3269–3274. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, S.A.; Plenderleith, L.J.; Liu, W.; Loy, D.E.; Learn, G.H.; Li, Y.; Shaw, K.S.; Ayouba, A.; Peeters, M.; Speede, S.; et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat. Commun. 2016, 7, 11078. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lopaticki, S.; Riglar, D.T.; Dekiwadia, C.; Uboldi, A.D.; Tham, W.-H.; O’Neill, M.T.; Richard, D.; Baum, J.; Ralph, S.A.; et al. An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by Plasmodium falciparum. PLOS Pathog. 2011, 7, e1002199. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, A.M.; Matile, H.; Papastogiannidis, P.; Kamber, J.; Favuzza, P.; Voss, T.S.; Wittlin, S.; Pluschke, G. Passive Immunoprotection of Plasmodium falciparum-Infected Mice Designates the CyRPA as Candidate Malaria Vaccine Antigen. J. Immunol. 2012, 188, 6225–6237. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.S.; Amlabu, E.; Pandey, A.K.; Mitra, P.; Chauhan, V.S.; Gaur, D. Multiprotein complex between the GPI-anchored CyRPA with PfRH5 and PfRipr is crucial for Plasmodium falciparum erythrocyte invasion. Proc. Natl. Acad. Sci. USA 2015, 112, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Volz, J.C.; Yap, A.; Sisquella, X.; Thompson, J.K.; Lim, N.T.; Whitehead, L.W.; Chen, L.; Lampe, M.; Tham, W.; Wilson, D.; et al. Essential Role of the PfRh5/PfRipr/CyRPA Complex during Plasmodium falciparum Invasion of Erythrocytes. Cell Host Microbe 2016, 20, 60–71. [Google Scholar] [CrossRef]
- Galaway, F.; Yu, R.; Constantinou, A.; Prugnolle, F.; Wright, G.J. Resurrection of the ancestral RH5 invasion ligand provides a molecular explanation for the origin of P. falciparum malaria in humans. PLoS Biol. 2019, 17, e3000490. [Google Scholar] [CrossRef]
- Wanaguru, M.; Liu, W.; Hahn, B.H.; Rayner, J.C.; Wright, G.J. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2013, 110, 20735–20740. [Google Scholar] [CrossRef]
- Wright, K.E.; Hjerrild, K.A.; Bartlett, J.; Douglas, A.D.; Jin, J.; Brown, R.E.; Illingworth, J.J.; Ashfield, R.; Clemmensen, S.B.; de Jongh, W.A.; et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature 2014, 515, 427–430. [Google Scholar] [CrossRef]
- Payne, R.O.; Silk, S.E.; Elias, S.; Miura, K.; Diouf, A.; Galaway, F.; De Graaf, H.; Brendish, N.J.; Poulton, I.D.; Griffiths, O.J.; et al. Human vaccination against RH5 induces neutralizing antimalarial antibodies that inhibit RH5 invasion complex interactions. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Joste, V.; Bailly, J.; Hubert, V.; Pauc, C.; Gendrot, M.; Guillochon, E.; Madamet, M.; Thellier, M.; Kendjo, E.; Argy, N.; et al. Plasmodium ovale wallikeri and P. ovale curtisi Infections and Diagnostic Approaches to Imported Malaria, France, 2013–2018. Emerg. Infect. Dis. 2021, 27, 372–384. [Google Scholar] [CrossRef]
- Nabarro, L.E.; Nolder, D.; Broderick, C.; Nadjm, B.; Smith, V.; Blaze, M.; Checkley, A.M.; Chiodini, P.L.; Sutherland, C.J.; Whitty, C.J.M. Geographical and temporal trends and seasonal relapse in Plasmodium ovale spp. and Plasmodium mlariae infections imported to the UK between 1987 and 2015. BMC Med. 2018, 16, 218. [Google Scholar] [CrossRef] [PubMed]
- Nolder, D.; Oguike, M.C.; Maxwell-Scott, H.; Niyazi, H.A.; Smith, V.; Chiodini, P.; Sutherland, C.J. An observational study of malaria in British travellers: Plasmodium ovale wallikeri and Plasmodium ovale curtisi differ significantly in the duration of latency. BMJ Open 2013, 3, e002711. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Marcos, G.; Rubio-Muñoz, J.M.; Angheben, A.; Jaureguiberry, S.; García-Bujalance, S.; Tomasoni, L.R.; Rodríguez-Valero, N.; Ruiz-Giardín, J.M.; Salas-Coronas, J.; Cuadros-González, J.; et al. Prospective comparative multi-centre study on imported Plasmodium ovale wallikeri and Plasmodium ovale curtisi infections. Malar. J. 2018, 17, 399. [Google Scholar] [CrossRef]
- Neafsey, D.E.; Taylor, A.R.; MacInnis, B.L. Advances and opportunities in malaria population genomics. Nat. Rev. Genet. 2021, 1–16. [Google Scholar] [CrossRef]
- Malaria, G.E.N. Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. eLife 2016, 5. [Google Scholar] [CrossRef]
- Pearson, R.D.; Ahouidi, A.; Ali, M.; Almagro-Garcia, J.; Amambua-Ngwa, A.; Amaratunga, C.; Amato, R.; Amenga-Etego, L.; Andagalu, B.; Anderson, T.J.C.; et al. An open dataset of Plasmodium falciparum genome variation in 7000 worldwide samples. Wellcome Open Res. 2021, 6, 42. [Google Scholar] [CrossRef]
- Noviyanti, R.; Miotto, O.; Barry, A.; Marfurt, J.; Siegel, S.; Thuy-Nhien, N.; Quang, H.H.; Anggraeni, N.D.; Laihad, F.; Liu, Y.; et al. Implementing parasite genotyping into national surveillance frameworks: Feedback from control programmes and researchers in the Asia–Pacific region. Malar. J. 2020, 19, 1–20. [Google Scholar] [CrossRef]
- Tessema, S.K.; Raman, J.; Duffy, C.W.; Ishengoma, D.S.; Amambua-Ngwa, A.; Greenhouse, B. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar. J. 2019, 18, 268. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Learn, G.H.; Rudicell, R.S.; Robertson, J.D.; Keele, B.F.; Ndjango, J.-B.N.; Sanz, C.M.; Morgan, D.B.; Locatelli, S.; et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 2010, 467, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Plenderleith, L.J.; Hahn, B.H. Ape Origins of Human Malaria. Annu. Rev. Microbiol. 2020, 74, 39–63. [Google Scholar] [CrossRef]
- Rich, S.M.; Licht, M.C.; Hudson, R.R.; Ayala, F.J. Malaria’s Eve: Evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 1998, 95, 4425–4430. [Google Scholar] [CrossRef] [PubMed]
- Neafsey, D.E.; Galinsky, K.; Jiang, R.H.; Young, L.; Sykes, S.M.; Saif, S.; Gujja, S.; Goldberg, J.M.; Young, S.; Zeng, Q.; et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat. Genet. 2012, 44, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.J.; Haubold, B.; Williams, J.T.; Estrada-Franco, J.G.; Richardson, L.; Mollinedo, R.; Bockarie, M.; Mokili, J.; Mharakurwa, S.; French, N.; et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 2000, 17, 1467–1482. [Google Scholar] [CrossRef]
- Manske, M.; Miotto, O.; Campino, S.; Auburn, S.; Almagro-Garcia, J.; Maslen, G.; O’Brien, J.; Djimde, A.; Doumbo, O.; Zongo, I.; et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 2012, 487, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Amambua-Ngwa, A.; Amenga-Etego, L.; Kamau, E.; Amato, R.; Ghansah, A.; Golassa, L.; Randrianarivelojosia, M.; Ishengoma, D.; Apinjoh, T.; Maïga-Ascofaré, O.; et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science 2019, 365, 813–816. [Google Scholar] [CrossRef]
- Baniecki, M.L.; Faust, A.L.; Schaffner, S.F.; Park, D.J.; Galinsky, K.; Daniels, R.F.; Hamilton, E.; Ferreira, M.U.; Karunaweera, N.D.; Serre, D.; et al. Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLoS Negl. Trop. Dis. 2015, 9, e0003539. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R.; Volkman, S.K.; Milner, D.A.; Mahesh, N.; Neafsey, D.E.; Park, D.J.; Rosen, D.; Angelino, E.; Sabeti, P.C.; Wirth, D.F.; et al. A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar. J. 2008, 7, 223. [Google Scholar] [CrossRef]
- Preston, M.D.; Campino, S.; Assefa, S.A.; Echeverry, D.F.; Ocholla, H.; Amambua-Ngwa, A.; Stewart, L.B.; Conway, D.; Borrmann, S.; Michon, P.; et al. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat. Commun. 2014, 5, 4052. [Google Scholar] [CrossRef]
- Albsheer, M.M.A.; Gebremeskel, E.I.; Kepple, D.; Lo, E.; Rougeron, V.; Ibrahim, M.E.; Hamid, M.M.A. Extensive genetic diversity in Plasmodium vivax from Sudan and its genetic relationships with other geographical isolates. BioRxiv 2020. [Google Scholar] [CrossRef]
- Bâ, H.; Duffy, C.W.; Ahouidi, A.D.; Deh, Y.B.; Diallo, M.Y.; Tandia, A.; Conway, D.J. Widespread distribution of Plasmodium vivax malaria in Mauritania on the interface of the Maghreb and West Africa. Malar. J. 2016, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Ba, H.; Auburn, S.; Jacob, C.G.; Goncalves, S.; Duffy, C.W.; Stewart, L.B.; Price, R.N.; Deh, Y.B.; Diallo, M.Y.; Tandia, A.; et al. Multi-locus genotyping reveals established endemicity of a geographically distinct Plasmodium vivax population in Mauritania, West Africa. PLoS Negl. Trop. Dis. 2020, 14, e0008945. [Google Scholar] [CrossRef] [PubMed]
- Dewasurendra, R.L.; Baniecki, M.L.; Schaffner, S.; Siriwardena, Y.; Moon, J.; Doshi, R.; Gunawardena, S.; Daniels, R.F.; Neafsey, D.; Volkman, S.; et al. Use of a Plasmodium vivax genetic barcode for genomic surveillance and parasite tracking in Sri Lanka. Malar. J. 2020, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Diez Benavente, E.; Campos, M.; Phelan, J.; Nolder, D.; Dombrowski, J.G.; Marinho, C.R.F.; Sriprawat, K.; Taylor, A.R.; Watson, J.; Roper, C.; et al. A molecular barcode to inform the geographical origin and transmission dynamics of Plasmodium vivax malaria. PLoS Genet. 2020, 16, e1008576. [Google Scholar] [CrossRef]
- Fola, A.A.; Kattenberg, E.; Razook, Z.; Lautu-Gumal, D.; Lee, S.; Mehra, S.; Bahlo, M.; Kazura, J.; Robinson, L.J.; Laman, M.; et al. SNP barcodes provide higher resolution than microsatellite markers to measure Plasmodium vivax population genetics. Malar. J. 2020, 19, 1–15. [Google Scholar] [CrossRef]
- Rougeron, V.; Elguero, E.; Arnathau, C.; Hidalgo, B.A.; Durand, P.; Houze, S.; Berry, A.; Zakeri, S.; Haque, R.; Alam, M.S.; et al. Human Plasmodium vivax diversity, population structure and evolutionary origin. PLOS Negl. Trop. Dis. 2020, 14, e0008072. [Google Scholar] [CrossRef]
- Price, R.N.; Commons, R.J.; Battle, K.E.; Thriemer, K.; Mendis, K. Plasmodium vivax in the Era of the Shrinking P. falciparum Map. Trends Parasitol. 2020, 36, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Jennison, C.; Arnott, A.; Tessier, N.; Tavul, L.; Koepfli, C.; Felger, I.; Siba, P.M.; Reeder, o.C.; Bahlo, M.; Mueller, I.; et al. Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl. Trop. Dis. 2015, 9, e0003634. [Google Scholar] [CrossRef] [PubMed]
- Orjuela-Sánchez, P.; Sá, J.M.; Brandi, M.C.; Rodrigues, P.T.; Bastos, M.S.; Amaratunga, C.; Duong, S.; Fairhurst, R.M.; Ferreira, M.U. Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp. Parasitol. 2013, 134, 318–326. [Google Scholar] [CrossRef]
- Parobek, C.M.; Lin, J.T.; Saunders, D.L.; Barnett, E.J.; Lon, C.; Lanteri, C.A.; Balasubramanian, S.; Brazeau, N.; DeConti, D.K.; Garba, D.L.; et al. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc. Natl. Acad. Sci. USA 2016, 113, E8096–E8105. [Google Scholar] [CrossRef]
- Ferreira, M.U.; Karunaweera, N.D.; da Silva-Nunes, M.; Da Silva, N.S.; Wirth, D.F.; Hartl, D.L. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J. Infect. Dis. 2007, 195, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Imwong, M.; Nair, S.; Pukrittayakamee, S.; Sudimack, D.; Williams, J.T.; Mayxay, M.; Newton, P.N.; Kim, J.R.; Nandy, A.; Osorio, L.; et al. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int. J. Parasitol. 2007, 37, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.E.; Pacheco, M.A.; Bacon, D.J.; Beg, M.A.; Machado, R.L.; Fairhurst, R.M.; Herrera, S.; Kim, J.; Menard, D.; Póvoa, M.M.; et al. The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: Parasite genetic diversity in the Americas. Mol. Biol. Evol. 2013, 30, 2050–2064. [Google Scholar] [CrossRef]
- Eede, P.V.D.; Van Der Auwera, G.; Delgado, C.; Huyse, T.; Soto-Calle, V.E.; Gamboa, D.; Grande, T.; Rodriguez, H.; Llanos, A.; Anné, J.; et al. Multilocus genotyping reveals high heterogeneity and strong local population structure of the Plasmodium vivax population in the Peruvian Amazon. Malar. J. 2010, 9, 151. [Google Scholar] [CrossRef][Green Version]
- Gunawardena, S.; Wirth, D.F.; Konradsen, F.; Ferreira, M.U.; Abeyasinghe, R.R.; Alifrangis, M.; Schousboe, M.L.; Amerasinghe, P.H.; Hartl, D.L.; Phone-Kyaw, M.; et al. Geographic Structure of Plasmodium vivax: Microsatellite Analysis of Parasite Populations from Sri Lanka, Myanmar, and Ethiopia. Am. J. Trop. Med. Hyg. 2010, 82, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.-A.; Dowd, S.; Bain, L.; Bobogare, A.; Wini, L.; Shanks, G.D.; Cheng, Q. Population genetics of Plasmodium falciparum and Plasmodium vivax and asymptomatic malaria in Temotu Province, Solomon Islands. Malar. J. 2013, 12, 429. [Google Scholar] [CrossRef]
- Koepfli, C.; Timinao, L.; Antao, T.; Barry, A.E.; Siba, P.; Mueller, I.; Felger, I. A Large Plasmodium vivax Reservoir and Little Population Structure in the South Pacific. PLoS ONE 2013, 8, e66041. [Google Scholar] [CrossRef]
- Twohig, K.A.; Pfeffer, D.A.; Baird, J.K.; Price, R.N.; Zimmerman, P.A.; Hay, S.I.; Gething, P.W.; Battle, K.E.; Howes, R.E. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLOS Neglected Trop. Dis. 2019, 13, e0007140. [Google Scholar] [CrossRef]
- Gunalan, K.; Niangaly, A.; Thera, M.A.; Doumbo, O.K.; Miller, L.H. Plasmodium vivax Infections of Duffy-Negative Erythrocytes: Historically Undetected or a Recent Adaptation? Trends Parasitol. 2018, 34, 420–429. [Google Scholar] [CrossRef]
- Battle, K.E.; Lucas, T.C.D.; Nguyen, M.; Howes, R.E.; Nandi, A.K.; Twohig, K.A.; Pfeffer, D.A.; Cameron, E.; Rao, P.C.; Casey, D.; et al. Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000–2017: A spatial and temporal modelling study. Lancet 2019, 394, 332–343. [Google Scholar] [CrossRef]
- De Oliveira, T.C.; Corder, R.M.; Early, A.; Rodrigues, P.T.; Ladeia-Andrade, S.; Alves, J.M.P.; Neafsey, D.E.; Ferreira, M.U. Population genomics reveals the expansion of highly inbred Plasmodium vivax lineages in the main malaria hotspot of Brazil. PLOS Negl. Trop. Dis. 2020, 14, e0008808. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, S.; Ferreira, M.U.; Kapilananda GM, G.; Wirth, D.F.; Karunaweera, N.D. The Sri Lankan paradox: High genetic diversity in Plasmodium vivax populations despite de-creasing levels of malaria transmission. Parasitology 2014, 141, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Divis, P.C.; Singh, B.; Anderios, F.; Hisam, S.; Matusop, A.; Kocken, C.H.; Assefa, S.A.; Duffy, C.W.; Conway, D.J. Admixture in Humans of Two Divergent Plasmodium knowlesi Populations Associated with Different Macaque Host Species. PLoS Pathog. 2015, 11, e1004888. [Google Scholar] [CrossRef]
- Benavente, E.D.; Gomes, A.R.; De Silva, J.R.; Grigg, M.; Walker, H.; Barber, B.E.; William, T.; Yeo, T.W.; De Sessions, P.F.; Ramaprasad, A.; et al. Whole genome sequencing of amplified Plasmodium knowlesi DNA from unprocessed blood reveals genetic exchange events between Malaysian Peninsular and Borneo subpopulations. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Benavente, E.D.; de Sessions, P.F.; Moon, R.W.; Holder, A.A.; Blackman, M.J.; Roper, C.; Drakeley, C.J.; Pain, A.; Sutherland, C.J.; Hibberd, M.L.; et al. Analysis of nuclear and organellar genomes of Plasmodium knowlesi in humans reveals ancient population structure and recent recombination among host-specific subpopulations. PLoS Genet. 2017, 13, e1007008. [Google Scholar] [CrossRef]
- Divis, P.C.S.; Duffy, C.W.; Kadir, K.A.; Singh, B.; Conway, D.J. Genome-wide mosaicism in divergence between zoonotic malaria parasite subpopulations with separate sympatric transmission cycles. Mol. Ecol. 2018, 27, 860–870. [Google Scholar] [CrossRef]
- Pinheiro, M.M.; Ahmed, M.A.; Millar, S.B.; Sanderson, T.; Otto, T.D.; Lu, W.C.; Krishna, S.; Rayner, J.C.; Cox-Singh, J. Plasmodium knowlesi genome sequences from clinical isolates reveal extensive genomic dimorphism. PLoS ONE 2015, 10, e0121303. [Google Scholar] [CrossRef]
- Divis, P.C.; Lin, L.C.; Rovie-Ryan, J.J.; Kadir, K.A.; Anderios, F.; Hisam, S.; Sharma, R.S.; Singh, B.; Conway, D.J. Three Divergent Subpopulations of the Malaria Parasite Plasmodium knowlesi. Emerg. Infect. Dis. 2017, 23, 616–624. [Google Scholar] [CrossRef]
- Hocking, S.E.; Divis, P.C.; Kadir, K.A.; Singh, B.; Conway, D.J. Population Genomic Structure and Recent Evolution of Plasmodium knowlesi, Peninsular Malaysia. Emerg. Infect. Dis. 2020, 26, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Yusof, R.; Ahmed, A.; Jelip, J.; Ngian, H.U.; Mustakim, S.; Hussin, H.M.; Fong, M.Y.; Mahmud, R.; Sitam, F.A.T.; Japning, J.R.-R.; et al. Phylogeographic Evidence for 2 Genetically Distinct Zoonotic Plasmodium knowlesi Parasites, Malaysia. Emerg. Infect. Dis. 2016, 22, 1371–1380. [Google Scholar] [CrossRef]
- Weedall, G.D.; Conway, D.J. Detecting signatures of balancing selection to identify targets of anti-parasite im-munity. Trends Parasitol. 2010, 26, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Nwakanma, D.C.; Duffy, C.W.; Amambua-Ngwa, A.; Oriero, E.C.; Bojang, K.A.; Pinder, M.; Drakeley, C.J.; Sutherland, C.J.; Milligan, P.J.; MacInnis, B.; et al. Changes in Malaria Parasite Drug Resistance in an Endemic Population Over a 25-Year Period With Resulting Genomic Evidence of Selection. J. Infect. Dis. 2013, 209, 1126–1135. [Google Scholar] [CrossRef]
- Ravenhall, M.; Benavente, E.D.; Mipando, M.; Jensen, A.T.; Sutherland, C.J.; Roper, C.; Sepúlveda, N.; Kwiatkowski, D.P.; Montgomery, J.; Phiri, K.S.; et al. Characterizing the impact of sustained sulfadoxine/pyrimethamine use upon the Plasmodium falciparum population in Malawi. Malar. J. 2016, 15, 575. [Google Scholar] [CrossRef]
- Hupalo, D.N.; Luo, Z.; Melnikov, A.; Sutton, P.L.; Rogov, P.; Escalante, A.; Vallejo, A.F.; Herrera, S.; Arévalo-Herrera, M.; Fan, Q.; et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat. Genet. 2016, 48, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.D.; Amato, R.; Auburn, S.; Miotto, O.; Almagro-Garcia, J.; Amaratunga, C.; Suon, S.; Mao, S.; Noviyanti, R.; Trimarsanto, H.; et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet. 2016, 48, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.J.; Pacheco, M.A.; Vallejo, A.F.; Schwartz, R.S.; Arevalo-Herrera, M.; Herrera, S.; Cartwright, R.A.; Escalante, A.A. Whole Genome Sequencing of Field Isolates Reveals Extensive Genetic Diversity in Plasmodium vivax from Colombia. PLoS Negl. Trop. Dis. 2015, 9, e0004252. [Google Scholar] [CrossRef]
- Costa, G.L.; Amaral, L.C.; Fontes, C.J.F.; Carvalho, L.H.; De Brito, C.F.A.; De Sousa, T.N. Assessment of copy number variation in genes related to drug resistance in Plasmodium vivax and Plasmodium falciparum isolates from the Brazilian Amazon and a systematic review of the literature. Malar. J. 2017, 16, 152. [Google Scholar] [CrossRef]
- Lin, J.T.; Patel, J.C.; Kharabora, O.; Sattabongkot, J.; Muth, S.; Ubalee, R.; Schuster, A.L.; Rogers, W.O.; Wongsrichanalai, C.; Juliano, J.J. Plasmodium vivax Isolates from Cambodia and Thailand Show High Genetic Complexity and Distinct Patterns of P. vivax Multidrug Resistance Gene 1 (pvmdr1) Polymorphisms. Am. J. Trop. Med. Hyg. 2013, 88, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Auburn, S.; Getachew, S.; Pearson, R.D.; Amato, R.; Miotto, O.; Trimarsanto, H.; Zhu, S.J.; Rumaseb, A.; Marfurt, J.; Noviyanti, R.; et al. Genomic Analysis of Plasmodium vivax in Southern Ethiopia Reveals Selective Pressures in Multiple Parasite Mechanisms. J. Infect. Dis. 2019, 220, 1738–1749. [Google Scholar] [CrossRef]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of Artemisinin Resistance in Plasmodium falciparum Malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Dondorp, A.M.; Nosten, F.; Yi, P.; Das, D.; Phyo, A.P.; Tarning, J.; Lwin, K.M.; Ariey, F.; Hanpithakpong, W.; Lee, S.J.; et al. Artemisinin Resistance in Plasmodium falciparum Malaria. N. Engl. J. Med. 2009, 361, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Noedl, H.; Se, Y.; Schaecher, K.; Smith, B.L.; Socheat, D.; Fukuda, M.M. Evidence of Artemisinin-Resistant Malaria in Western Cambodia. N. Engl. J. Med. 2008, 359, 2619–2620. [Google Scholar] [CrossRef]
- Ghorbal, M.; Gorman, M.; Macpherson, C.R.; Martins, R.M.; Scherf, A.; Lopez-Rubio, J.J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature Biotechnol. 2014, 32, 819–821. [Google Scholar] [CrossRef] [PubMed]
- Miotto, O.; Amato, R.; Ashley, E.A.; MacInnis, B.; Almagro-Garcia, J.; Amaratunga, C.; Lim, P.; Mead, D.; Oyola, S.O.; Dhorda, M.; et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015, 47, 226–234. [Google Scholar] [CrossRef]
- Straimer, J.; Gnädig, N.F.; Witkowski, B.; Amaratunga, C.; Duru, V.; Ramadani, A.P.; Dacheux, M.; Khim, N.; Zhang, L.; Lam, S.; et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 2015, 347, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Takala-Harrison, S.; Jacob, C.G.; Arze, C.; Cummings, M.P.; Silva, J.C.; Dondorp, A.M.; Fukuda, M.M.; Hien, T.T.; Mayxay, M.; Noedl, H.; et al. Independent Emergence of Artemisinin Resistance Mutations Among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015, 211, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Ménard, D.; Khim, N.; Beghain, J.; Adegnika, A.A.; Shafiul-Alam, M.; Amodu, O.; Rahim-Awab, G.; Barnadas, C.; Berry, A.; Boum, Y.; et al. A Worldwide Map of Plasmodium falciparum K13-Propeller Polymorphisms. N. Engl. J. Med. 2016, 374, 2453–2464. [Google Scholar] [CrossRef]
- Muwanguzi, J.; Henriques, G.; Sawa, P.; Bousema, T.; Sutherland, C.J.; Beshir, K.B. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar. J. 2016, 15, 1–6. [Google Scholar] [CrossRef]
- Dogovski, C.; Xie, S.C.; Burgio, G.; Bridgford, J.; Mok, S.; McCaw, J.; Chotivanich, K.; Kenny, S.; Gnädig, N.; Straimer, J.; et al. Targeting the Cell Stress Response of Plasmodium falciparum to Overcome Artemisinin Resistance. PLoS Biol. 2015, 13, e1002132. [Google Scholar] [CrossRef]
- Mok, S.; Ashley, E.A.; Ferreira, P.E.; Zhu, L.; Lin, Z.; Yeo, T.; Chotivanich, K.; Imwong, M.; Pukrittayakamee, S.; Dhorda, M.; et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of arte-misinin resistance. Science 2015, 347, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Mok, S.; Stokes, B.H.; Gnädig, N.F.; Ross, L.S.; Yeo, T.; Amaratunga, C.; Allman, E.; Solyakov, L.; Bottrill, A.R.; Tripathi, J.; et al. Artemisinin-resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic pro-gram to enhance survival. Nat. Commun. 2021, 12, 530. [Google Scholar] [CrossRef] [PubMed]
- Rocamora, F.; Zhu, L.; Liong, K.Y.; Dondorp, A.; Miotto, O.; Mok, S.; Bozdech, Z. Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLOS Pathog. 2018, 14, e1006930. [Google Scholar] [CrossRef] [PubMed]
- Duffy, C.W.; Amambua-Ngwa, A.; Ahouidi, A.D.; Diakite, M.; Awandare, G.A.; Ba, H.; Tarr, S.J.; Murray, L.; Stewart, L.B.; D’Alessandro, U.; et al. Multi-population genomic analysis of malaria parasites indicates local selection and differentiation at the gdv1 locus regulating sexual development. Sci. Rep. 2018, 8, 15763. [Google Scholar] [CrossRef] [PubMed]
- Mobegi, V.A.; Duffy, C.W.; Amambua-Ngwa, A.; Loua, K.M.; Laman, E.; Nwakanma, D.; MacInnis, B.; Aspeling-Jones, H.; Murray, L.; Clark, T.; et al. Genome-Wide Analysis of Selection on the Malaria Parasite Plasmodium falciparum in West African Populations of Differing Infection Endemicity. Mol. Biol. Evol. 2014, 31, 1490–1499. [Google Scholar] [CrossRef] [PubMed]
- Tagliamonte, M.S.; Yowell, C.A.; ElBadry, M.A.; Boncy, J.; Raccurt, C.P.; Okech, B.A.; Goss, E.M.; Salemi, M.; Dame, J.B. Genetic Markers of Adaptation of Plasmodium falciparum to Transmission by American Vectors Identified in the Genomes of Parasites from Haiti and South America. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Bozdech, Z.; Llinás, M.; Pulliam, B.L.; Wong, E.D.; Zhu, J.; DeRisi, J.L. The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum. PLoS Biol. 2003, 1, e5. [Google Scholar] [CrossRef] [PubMed]
- Chappell, L.; Ross, P.; Orchard, L.; Russell, T.J.; Otto, T.D.; Berriman, M.; Rayner, J.C.; Llinás, M. Refining the transcriptome of the human malaria parasite Plasmodium falciparum using amplifica-tion-free RNA-seq. BMC Genom. 2020, 21, 395. [Google Scholar] [CrossRef]
- Le Roch, K.G.; Zhou, Y.; Blair, P.L.; Grainger, M.; Moch, J.K.; Haynes, J.D.; De La Vega, P.; Holder, A.; Batalov, S.; Carucci, D.J.; et al. Discovery of Gene Function by Expression Profiling of the Malaria Parasite Life Cycle. Science 2003, 301, 1503–1508. [Google Scholar] [CrossRef]
- Llinás, M.; Bozdech, Z.; Wong, E.D.; Adai, A.T.; DeRisi, J.L. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006, 34, 1166–1173. [Google Scholar] [CrossRef]
- Otto, T.D.; Wilinski, D.; Assefa, S.; Keane, T.M.; Sarry, L.R.; Böhme, U.; Lemieux, J.; Barrell, B.; Pain, A.; Berriman, M.; et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010, 76, 12–24. [Google Scholar] [CrossRef]
- Painter, H.J.; Chung, N.C.; Sebastian, A.; Albert, I.; Storey, J.; Llinás, M. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Bechtsi, D.P.; Waters, A.P. Genomics and epigenetics of sexual commitment in Plasmodium. Int. J. Parasitol. 2017, 47, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.; Deitsch, K.W. Malaria Epigenetics. Cold Spring Harb. Perspect. Med. 2017, 7, a025528. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.P.; Bozdech, Z. Epigenetic landscapes underlining global patterns of gene expression in the human malaria parasite, Plasmodium falciparum. Int. J. Parasitol. 2017, 47, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Hammam, E.; Ananda, G.; Sinha, A.; Scheidig-Benatar, C.; Bohec, M.; Preiser, P.R.; Dedon, P.C.; Scherf, A.; Vembar, S.S. Discovery of a new predominant cytosine DNA modification that is linked to gene expression in malaria parasites. Nucleic Acids Res. 2020, 48, 184–199. [Google Scholar] [CrossRef]
- McInroy, G.R.; Beraldi, D.; Raiber, E.-A.; Modrzynska, K.; Van Delft, P.; Billker, O.; Balasubramanian, S. Enhanced Methylation Analysis by Recovery of Unsequenceable Fragments. PLoS ONE 2016, 11, e0152322. [Google Scholar] [CrossRef]
- Ponts, N.; Fu, L.; Harris, E.Y.; Zhang, J.; Chung, D.-W.D.; Cervantes, M.C.; Prudhomme, J.; Atanasova-Penichon, V.; Zehraoui, E.; Bunnik, E.; et al. Genome-wide Mapping of DNA Methylation in the Human Malaria Parasite Plasmodium falciparum. Cell Host Microbe 2013, 14, 696–706. [Google Scholar] [CrossRef]
- Lopez-Rubio, J.-J.; Mancio-Silva, L.; Scherf, A. Genome-wide analysis of heterochromatin associates clonally var-iant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 2009, 5, 179–190. [Google Scholar] [CrossRef]
- Salcedo-Amaya, A.M.; van Driel, M.A.; Alako, B.T.; Trelle, M.B.; van den Elzen, A.M.; Cohen, A.M.; Janssen-Megens, E.M.; van de Vegte-Bolmer, M.; Selzer, R.R.; Iniguez, A.L.; et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2009, 106, 9655–9660. [Google Scholar] [CrossRef]
- Baum, J.; Papenfuss, A.T.; Mair, G.R.; Janse, C.J.; Vlachou, D.; Waters, A.P.; Cowman, A.F.; Crabb, B.S.; De Koning-Ward, T.F. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009, 37, 3788–3798. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Li, S.; Chavez, V.; Lanting, L.; Natarajan, R. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol. Endocrinol. 2006, 20, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Deobagkar, D. Epigenetics with special reference to the human X chromosome inactivation and the enigma of Drosophila DNA methylation. J. Genet. 2018, 97, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Albalat, R.; Martí-Solans, J.; Cañestro, C. DNA methylation in amphioxus: From ancestral functions to new roles in vertebrates. Briefings Funct. Genom. 2012, 11, 142–155. [Google Scholar] [CrossRef] [PubMed]
- De Mendoza, A.; Lister, R.; Bogdanovic, O. Evolution of DNA Methylome Diversity in Eukaryotes. J. Mol. Biol. 2020, 432, 1687–1705. [Google Scholar] [CrossRef] [PubMed]
- Borodinova, A.A.; Balaban, P.M. Epigenetic Regulation as a Basis for Long-Term Changes in the Nervous Sys-tem: In Search of Specificity Mechanisms. Biochem. Biokhimiia 2020, 85, 994–1010. [Google Scholar] [CrossRef]
- Nikolic, D.; Jankovic, M.; Petrovic, B.; Novakovic, I. Genetic Aspects of Inflammation and Immune Response in Stroke. Int. J. Mol. Sci. 2020, 21, 7409. [Google Scholar] [CrossRef]
- Rossnerova, A.; Izzotti, A.; Pulliero, A.; Bast, A.; Rattan, S.; Rossner, P. The Molecular Mechanisms of Adaptive Response Related to Environmental Stress. Int. J. Mol. Sci. 2020, 21, 7053. [Google Scholar] [CrossRef]
- Thamban, T.; Agarwaal, V.; Khosla, S. Role of genomic imprinting in mammalian development. J. Biosci. 2020, 45, 1–21. [Google Scholar] [CrossRef]
- Choi, S.-W.; Keyes, M.K.; Horrocks, P. LC/ESI-MS demonstrates the absence of 5-methyl-2′-deoxycytosine in Plasmodium falciparum genomic DNA. Mol. Biochem. Parasitol. 2006, 150, 350–352. [Google Scholar] [CrossRef]
- Gissot, M.; Choi, S.-W.; Thompson, R.F.; Greally, J.M.; Kim, K. Toxoplasma gondii and Cryptosporidium parvum Lack Detectable DNA Cytosine Methylation. Eukaryot. Cell 2008, 7, 537–540. [Google Scholar] [CrossRef][Green Version]
- Pollack, Y.; Kogan, N.; Golenser, J. Plasmodium falciparum: Evidence for a DNA methylation pattern. Exp. Parasitol. 1991, 72, 339–344. [Google Scholar] [CrossRef]
- Krueger, F.; Kreck, B.; Franke, A.; Andrews, S.R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 2012, 9, 145–151. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Pastor, W.A.; Aravind, L.; Rao, A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013, 14, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Hastert, F.D.; Cardoso, M.C. DNA Modification Readers and Writers and Their Interplay. J. Mol. Biol. 2020, 432, 1731–1746. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Xia, B.; Yi, C. Single-base resolution analysis of DNA epigenome via high-throughput sequencing. Sci. China Life Sci. 2016, 59, 219–226. [Google Scholar] [CrossRef]
- Tost, J. Current and Emerging Technologies for the Analysis of the Genome-Wide and Locus-Specific DNA Methylation Patterns. Adv. Exp. Med. Biol. 2016, 945, 343–430. [Google Scholar] [PubMed]
- Williams, K.; Christensen, J.; Pedersen, M.T.; Johansen, J.V.; Cloos, P.; Rappsilber, J.; Helin, K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011, 473, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, F.; Tan, L.; Kong, L.; Xiong, L.; Deng, J.; Barbera, A.J.; Zheng, L.; Zhang, H.; Huang, S.; et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol. Cell 2011, 42, 451–464. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Sung, W.-K.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2006, 39, 61–69. [Google Scholar] [CrossRef]
- Gaiti, F.; Jindrich, K.; Fernandez-Valverde, S.L.; Roper, K.E.; Degnan, B.M.; Tanurdžić, M. Landscape of histone modifications in a sponge reveals the origin of animal cis-regulatory complexity. eLife 2017, 6. [Google Scholar] [CrossRef]
- Guo, X.; Wang, L.; Li, J.; Ding, Z.; Xiao, J.; Yin, X.; He, S.; Shi, P.; Dong, L.; Li, G.; et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 2015, 517, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, G.; Guzzardo, P.M.; Olova, N.; Fantappie, M.R.; Rampp, M.; Schaefer, M.; Reik, W.; Hannon, G.J.; Lyko, F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. USA 2013, 110, 8627–8631. [Google Scholar] [CrossRef]
- Govindaraju, G.; Jabeena, C.; Sethumadhavan, D.V.; Rajaram, N.; Rajavelu, A. DNA methyltransferase homologue TRDMT1 in Plasmodium falciparum specifically methylates endogenous aspartic acid tRNA. Biochim. Biophys. Acta (BBA)—Bioenerg. 2017, 1860, 1047–1057. [Google Scholar] [CrossRef]
- Bewick, A.J.; Hofmeister, B.T.; Powers, R.A.; Mondo, S.J.; Grigoriev, I.V.; James, T.Y.; Stajich, J.E.; Schmitz, R.J. Diversity of cytosine methylation across the fungal tree of life. Nat. Ecol. Evol. 2019, 3, 479–490. [Google Scholar] [CrossRef]
- Huff, J.T.; Zilberman, D. Dnmt1-Independent CG Methylation Contributes to Nucleosome Positioning in Diverse Eukaryotes. Cell 2014, 156, 1286–1297. [Google Scholar] [CrossRef]
- Yaari, R.; Katz, A.; Domb, K.; Harris, K.D.; Zemach, A.; Ohad, N. RdDM-independent de novo and heterochromatin DNA methylation by plant CMT and DNMT3 orthologs. Nat. Commun. 2019, 10, 1613. [Google Scholar] [CrossRef]
- Gabel, H.W.; Kinde, B.Z.; Stroud, H.; Gilbert, C.S.; Harmin, D.A.; Kastan, N.R.; Hemberg, M.; Ebert, D.H.; Greenberg, M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522, 89–93. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Song, H. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Liu, K.; Xu, C.; Lei, M.; Yang, A.; Loppnau, P.; Hughes, T.R.; Min, J. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J. Biol. Chem. 2018, 293, 7344–7354. [Google Scholar] [CrossRef] [PubMed]
- Sperlazza, M.J.; Bilinovich, S.M.; Sinanan, L.M.; Javier, F.R.; Williams, D.C., Jr. Structural Basis of MeCP2 Distribution on Non-CpG Methylated and Hydroxymethylated DNA. J. Mol. Biol. 2017, 429, 1581–1594. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.K.; Zhang, P.; Cardoso, M.C. Modifiers and Readers of DNA Modifications and Their Impact on Genome Structure, Expression, and Stability in Disease. Front. Genet. 2016, 7, 115. [Google Scholar] [CrossRef]
- Hashimoto, H.; Zhang, X.; Cheng, X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glyco-sylase domain: Structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 8276–8284. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.J.; Branco, M.R.; Ficz, G.; Oxley, D.; Krueger, F.; Reik, W.; Balasubramanian, S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012, 336, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Branco, M.R.; Seisenberger, S.; Santos, F.; Krueger, F.; Hore, T.; Marques, C.J.; Andrews, S.; Reik, W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011, 473, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.-G.; Wu, X.; Li, A.X.; Pfeifer, G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011, 39, 5015–5024. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Pape, U.J.; Huang, Y.; Henderson, H.R.; Lister, R.; Ko, M.; McLoughlin, E.M.; Brudno, Y.; Mahapatra, S.; Kapranov, P.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011, 473, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Incarnato, D.; Krepelova, A.; Rapelli, S.; Anselmi, F.; Parlato, C.; Medana, C.; Bello, F.; Oliviero, S. Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep. 2015, 10, 674–683. [Google Scholar] [CrossRef]
- Raiber, E.A.; Beraldi, D.; Ficz, G.; Burgess, H.E.; Branco, M.R.; Murat, P.; Oxley, D.; Booth, M.J.; Reik, W.; Balasubramanian, S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcrip-tion and depends on thymine DNA glycosylase. Genome Biol. 2012, 13, R69. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-X.; Szulwach, K.E.; Dai, Q.; Fu, Y.; Mao, S.-Q.; Lin, L.; Street, C.; Li, Y.; Poidevin, M.; Wu, H.; et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell 2013, 153, 678–691. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).