Analysis of SI-Related BoGAPDH Family Genes and Response of BoGAPC to SI Signal in Brassica oleracea L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Conserved Domain Analysis of GAPDH Family Genes in B. oleracea and A. thaliana
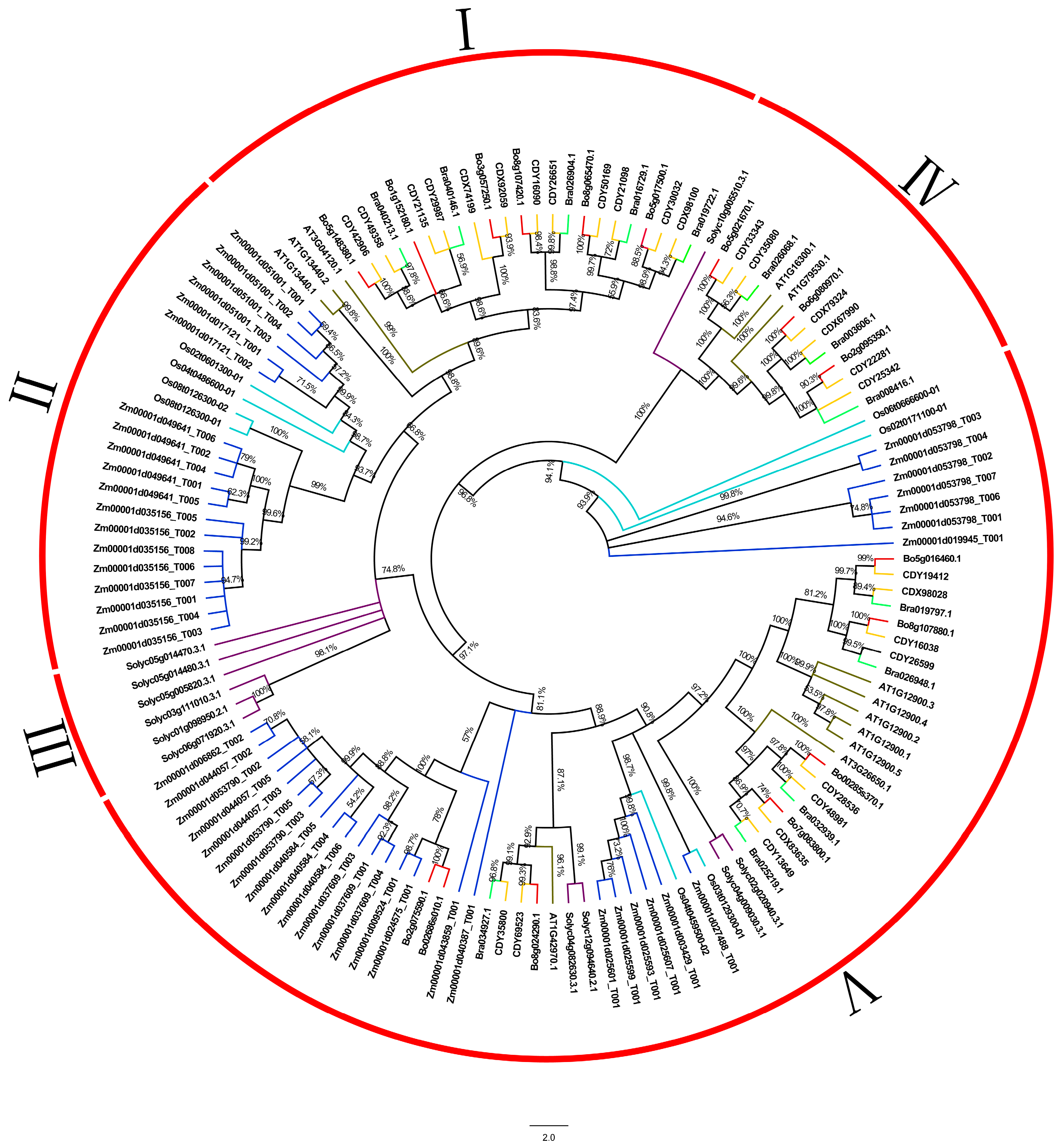
2.2. Evolutionary Analysis of Members of GAPDH Gene Family in B. oleracea and Related Species
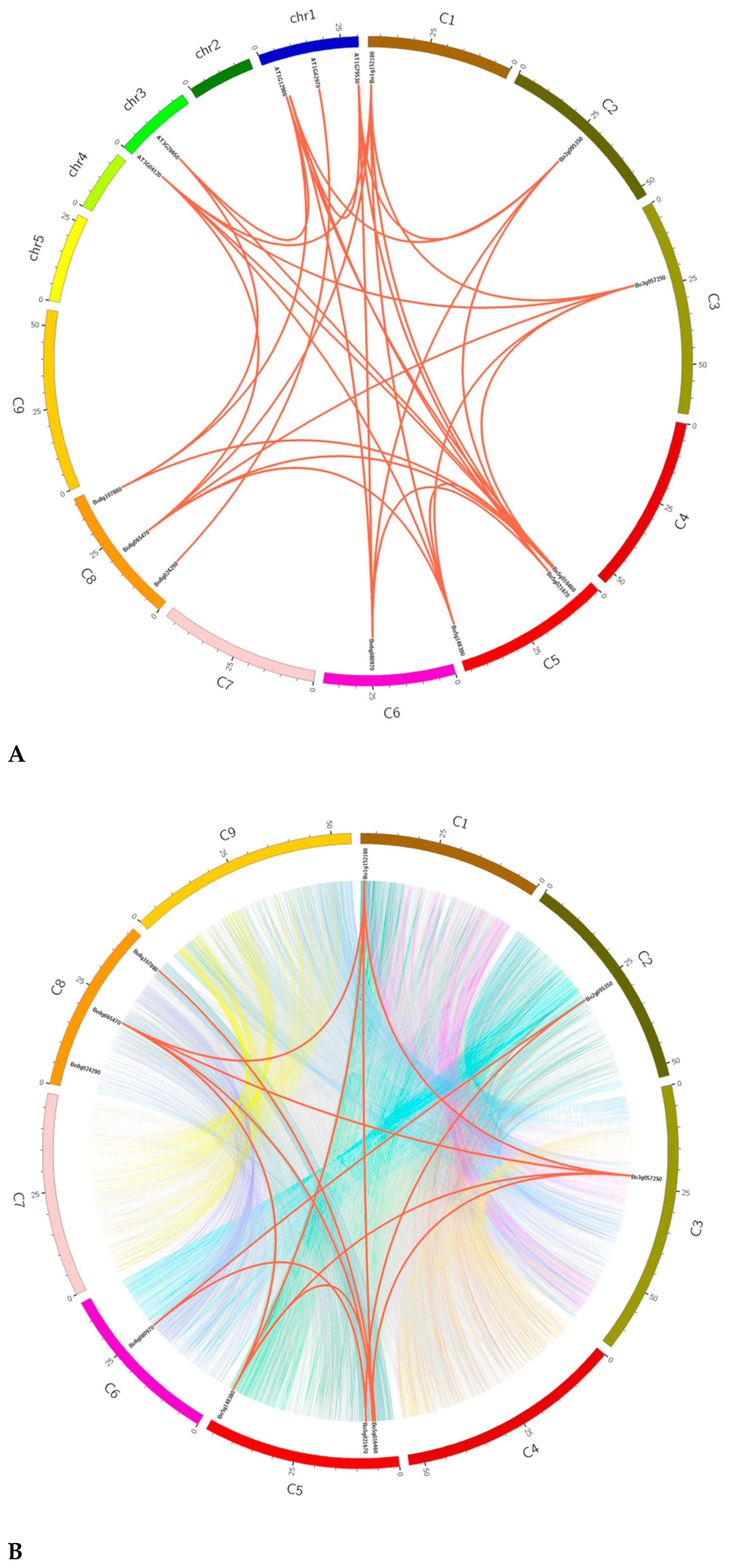
2.3. Chromosome Mapping of GAPDH Family Genes in B. oleracea
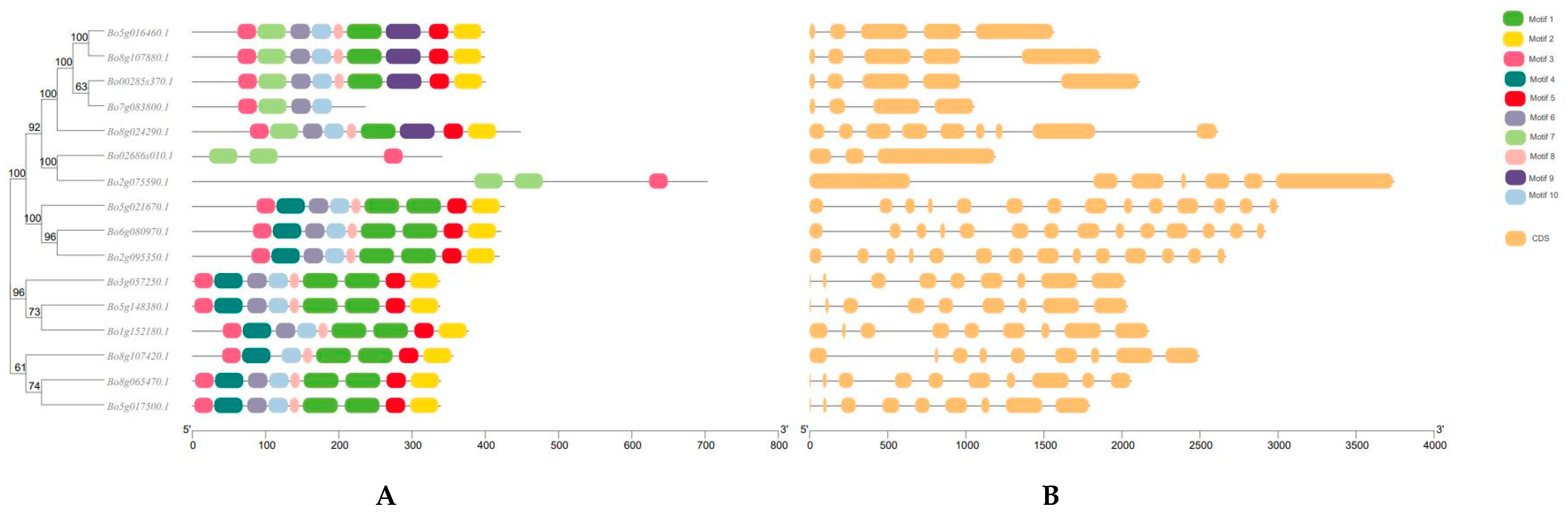
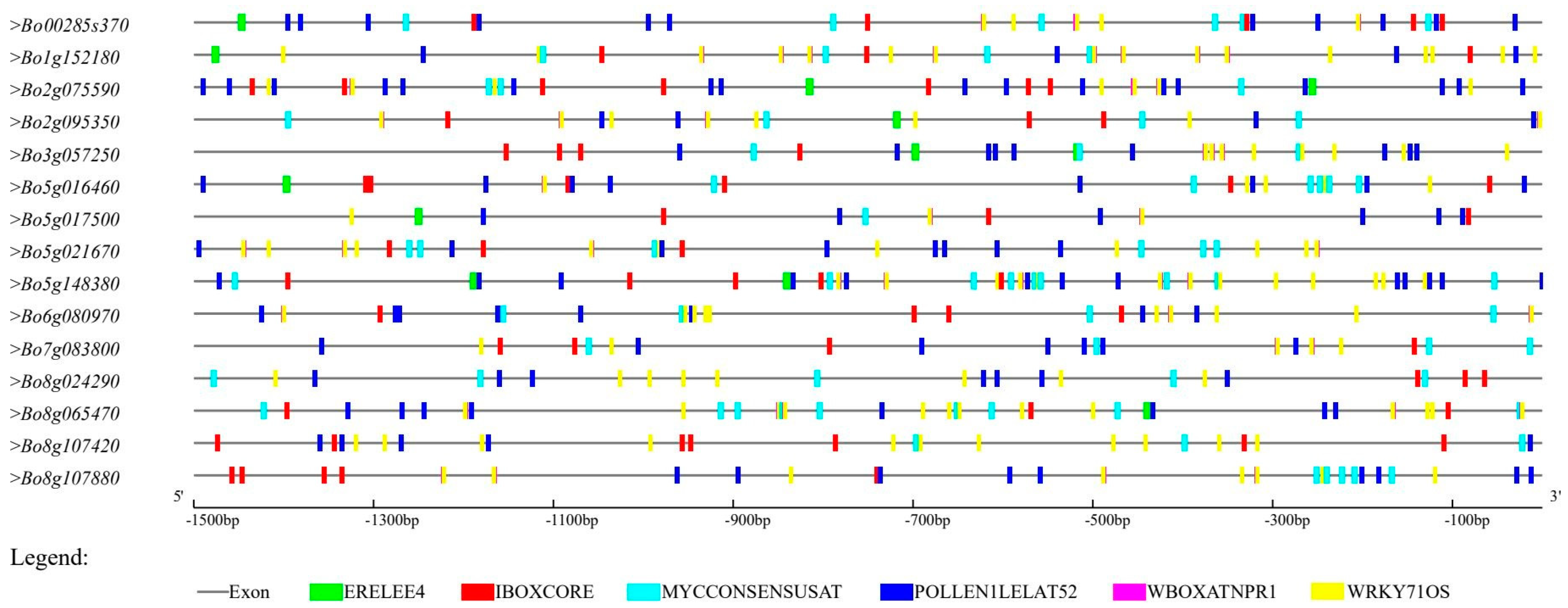
2.4. Transcript Structure of GAPDH Family Genes in B. oleracea and Analysis of Cis-Acting Regulatory Elements in Promoters
2.5. Expression Analysis of BoGAPDH Gene Family
2.6. Experimental Materials
2.7. Cloning of Target Genes
2.8. Bioinformatics Analysis
2.9. Analysis of BoGAPC Expression
2.10. Promoter Activity Analysis
2.11. Subcellular Localization
2.12. Prokaryotic Expression
2.13. Yeast Two-Hybrid Interaction Identification and Protein Interaction Detection by GST Pull-Down
3. Results
3.1. Evolutionary Analysis of GAPDH Gene in B. oleracea L.
3.2. Structure and Analysis of GAPDH Gene Family Members in B. oleracea L.
3.3. Chromosome Mapping of GAPDH Gene in B. oleracea
3.4. Cis-Acting Regulatory Elements of B. oleracea GAPDH Gene
3.5. Tandem Repeat Gene Pair and Collinearity Analysis of GAPDH Family Genes in B. oleracea and A. thaliana
3.6. Expression Analysis of GAPDH Family Genes in B. oleracea
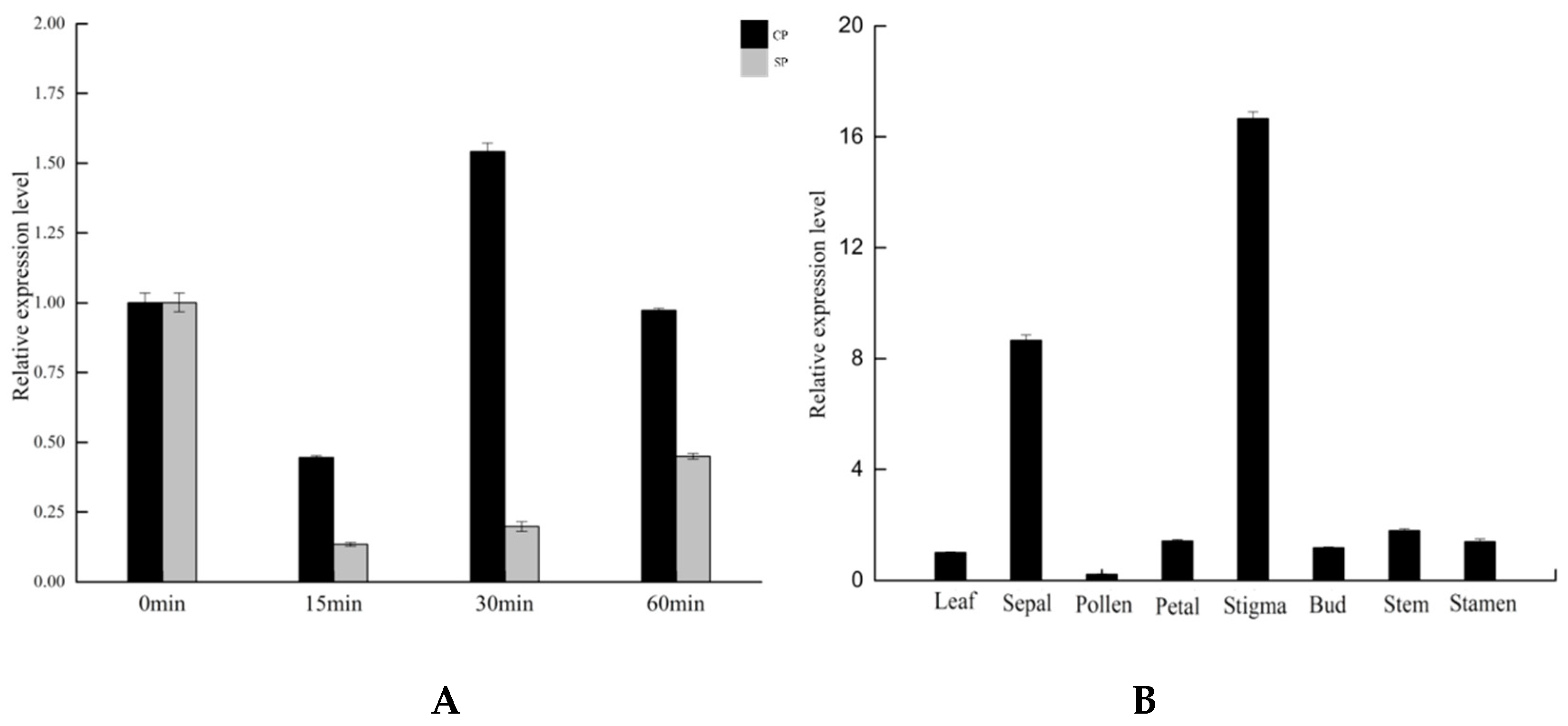
3.6.1. Expression Analysis of GAPDH Family Genes in B. oleracea during Self-Pollination and Cross-Pollination
3.6.2. Tissue Expression Analysis of GAPDH Gene in B. oleracea
3.6.3. Expression Analysis of BoGAPDH Gene in B. oleracea Transcriptome
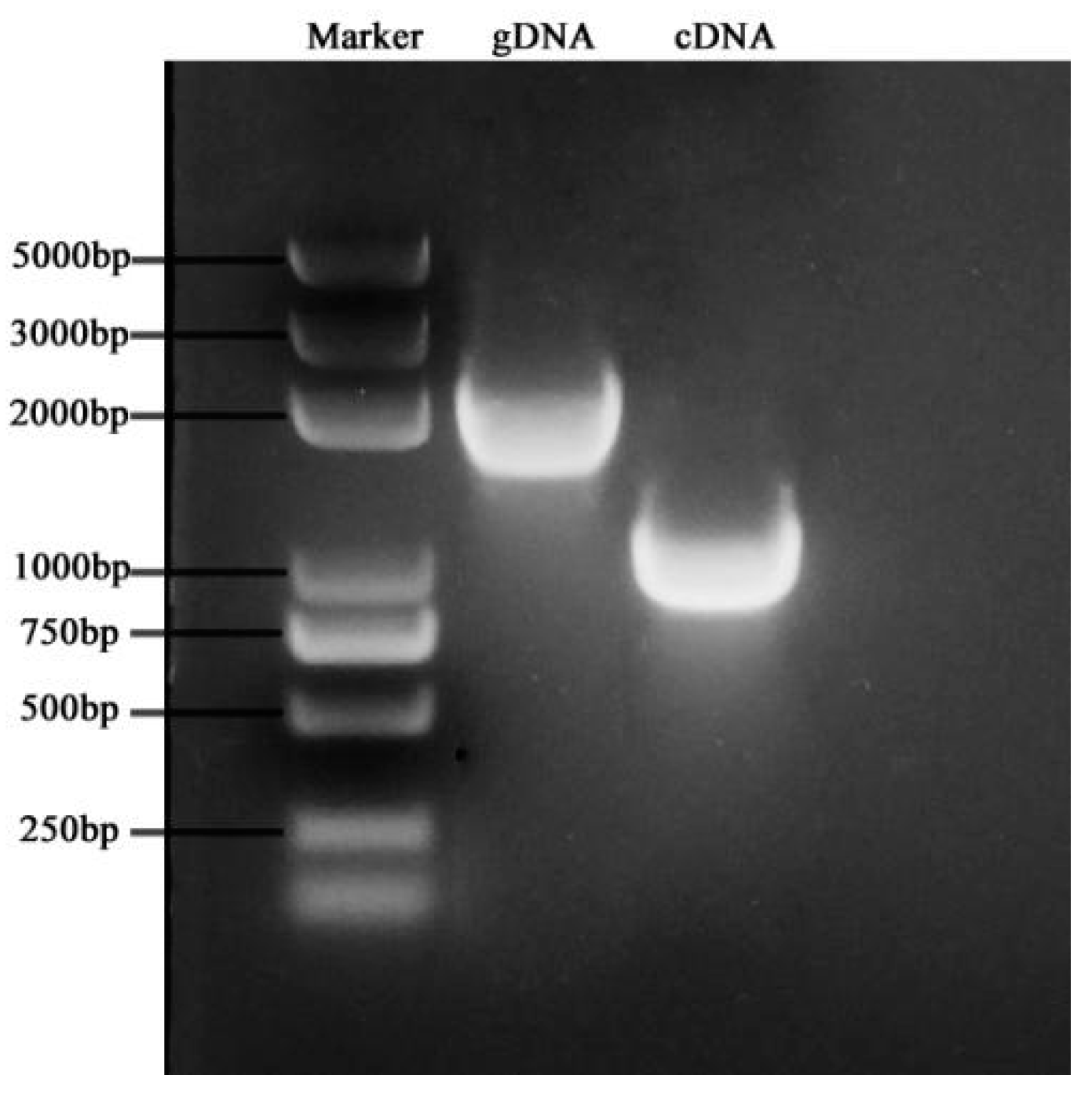
3.6.4. Full-Length Cloning of BoGAPC Gene
3.6.5. Protein Structure Analysis of BoGAPC
3.6.6. Protein Evolution Analysis of BoGAPC
3.6.7. Expression Analysis of BoGAPC Gene
3.6.8. Analysis of the Specificity of the Promoter of the BoGAPC Gene
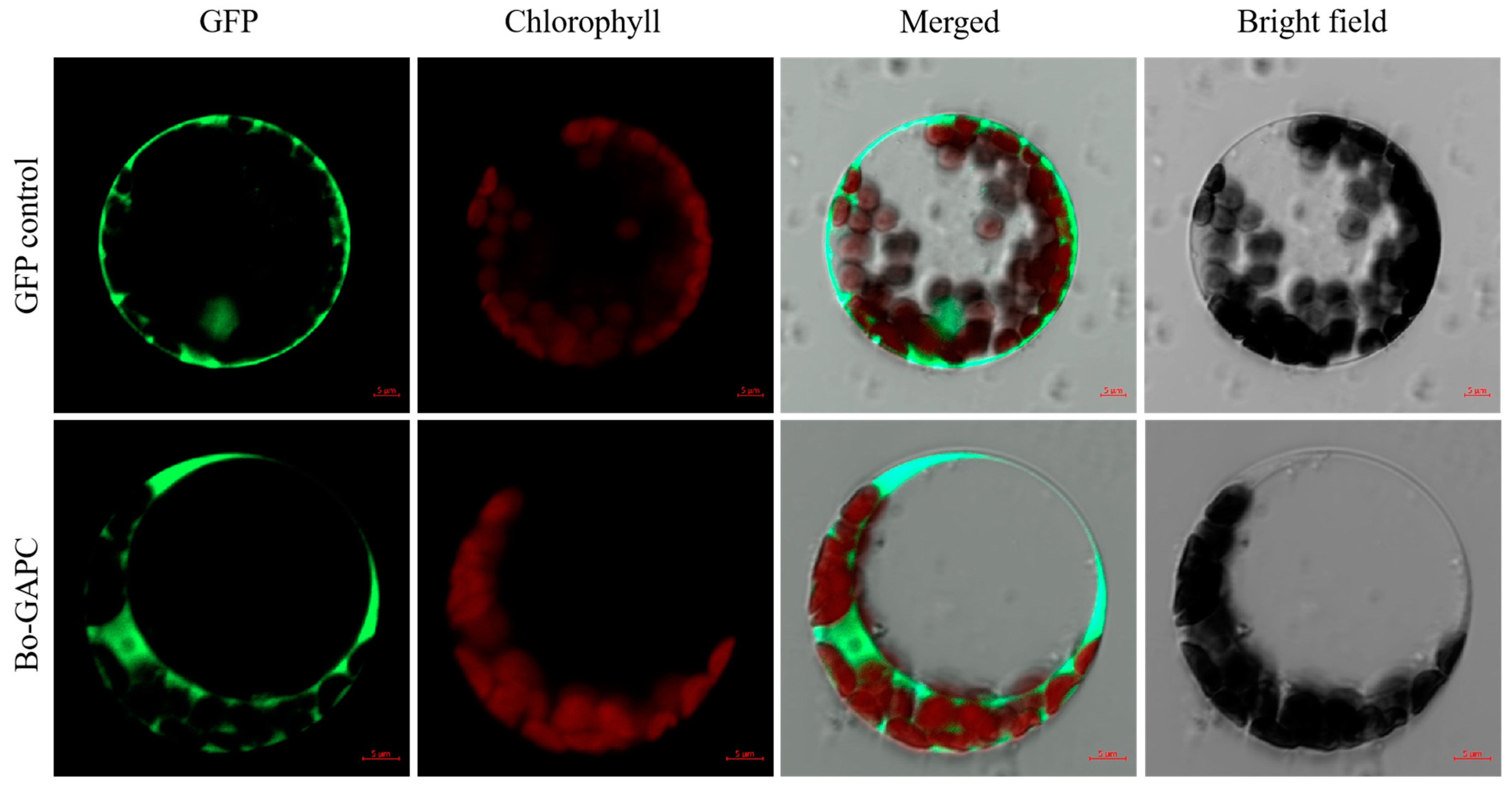
3.6.9. Subcellular Localization of BoGAPC
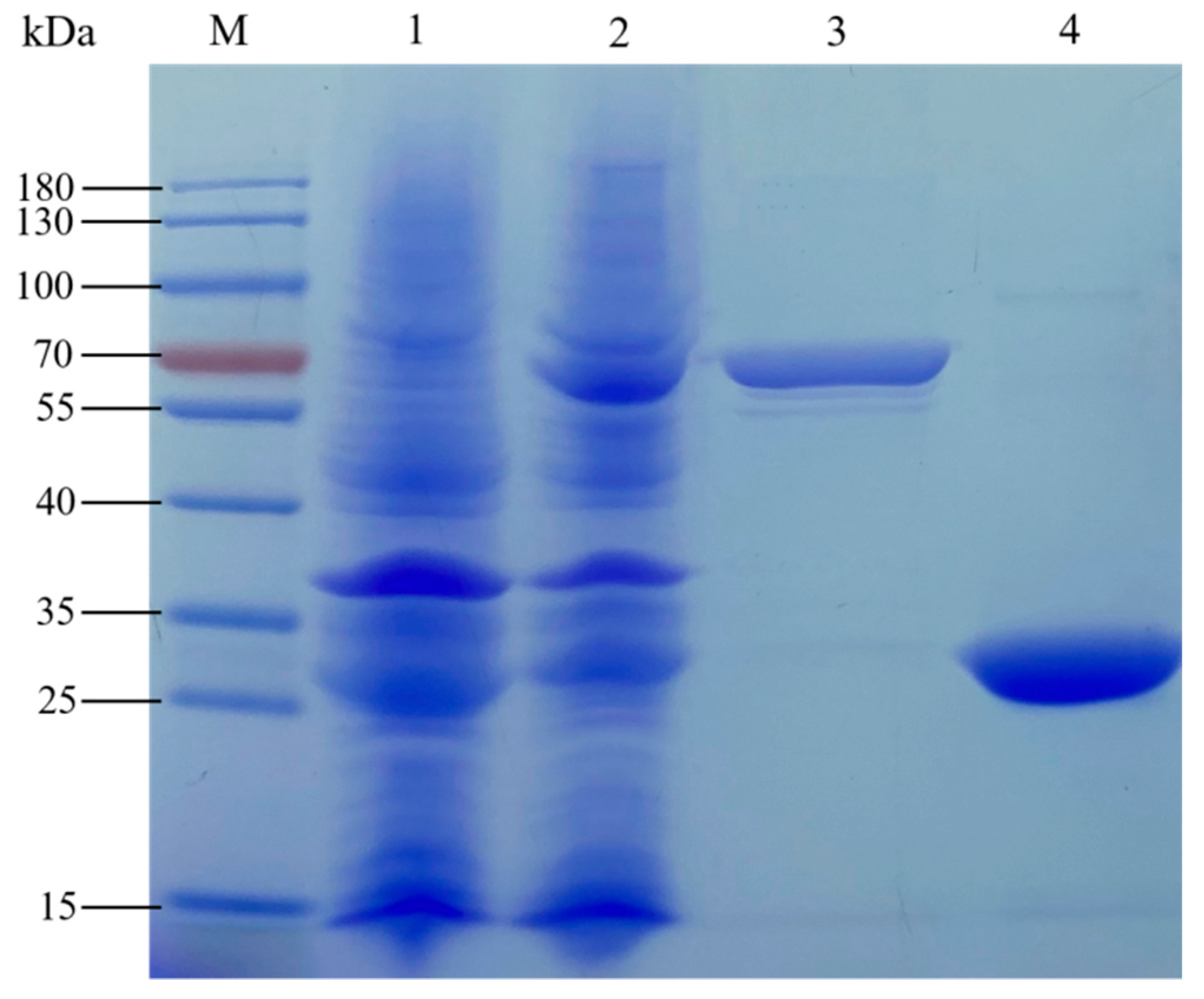
3.6.10. Prokaryotic Expression of BoGAPC
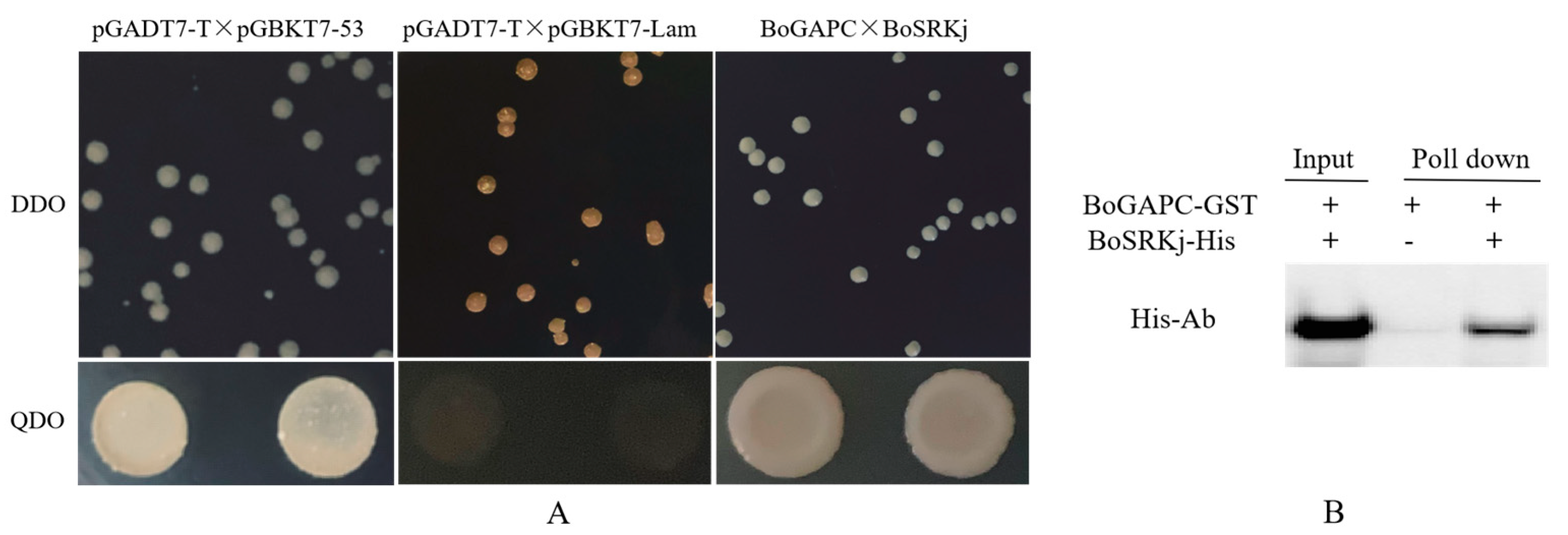
3.6.11. Interaction between BoGAPC and BoSRKj
4. Discussion
4.1. Members of GAPDH Family Participate in Plant Stress Resistance
4.2. Replication and Evolution of GAPDH Gene in B. oleracea
4.3. BoGAPC May Be Involved in Self-Incompatibility Signal Transduction in B. oleracea
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fujii, S.; Kubo, K.; Takayama, S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2016, 2, 16130. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Brinkmann, H.; Cerff, R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J. Mol. Evol. 2003, 57, 16–26. [Google Scholar] [CrossRef]
- Pelah, D.; Shoseyov, O.; Altman, A.; Bartels, D. Water-stress response in Aspen (Populus tremula): Differential accumulation of dehydrin, sucrose synthase, GAPDH homologues, and soluble sugars. J. Plant Physiol. 1997, 151, 96–100. [Google Scholar] [CrossRef]
- Yang, Y.; Kwon, H.B.; Peng, H.P.; Shih, M.C. Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol. 1993, 101, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Devaiah, S.P.; Narasimhan, R.; Pan, X.; Zhang, Y.; Zhang, W.; Wang, X. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Ddelta to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 2012, 24, 2200–2212. [Google Scholar] [CrossRef]
- Guo, L.; Ma, F.; Wei, F.; Fanella, B.; Allen, D.K.; Wang, X. Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 2014, 26, 3023–3035. [Google Scholar] [CrossRef]
- Kim, S.C.; Guo, L.; Wang, X. Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J. Biol. Chem. 2013, 288, 11834–11844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Rao, X.L.; Shi, H.T.; Li, R.J.; Lu, Y.T. Overexpression of a cytosolic glyceraldehyde-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tiss. Org. 2011, 107, 1–11. [Google Scholar] [CrossRef]
- Wawer, I. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 2010, 429, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, S.; Sachs, M.M. Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol. Biol. 1997, 33, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Deng, R.; Guo, Z.; Yang, S.; Deng, X. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genom. 2016, 17, 240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z.; Ji, H.; Zhou, Y.; Yang, S. TaWRKY40 transcription factor positively regulate the expression of TaGAPC1 to enhance drought tolerance. BMC Genom. 2019, 20, 795. [Google Scholar] [CrossRef] [PubMed]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez-Casati, D.F. Characterization of an A. thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol. Biol. 2006, 61, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bertomeu, J.; Bermudez, M.A.; Segura, J.; Ros, R. Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: Interaction between ABA and primary metabolism. J. Exp. Bot. 2011, 62, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen Acceptance or Rejection: A Tale of Two Pathways. Trends Plant Sci. 2016, 21, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H. Dry Stigmas, Water and Self-Incompatibility in Brassica. Sex. Plant Reprod. 1995, 8, 1–10. [Google Scholar] [CrossRef]
- Haque, T.; Eaves, D.J.; Lin, Z.; Zampronio, C.G.; Cooper, H.J.; Bosch, M.; Smirnoff, N.; Franklin-Tong, V.E. Self-incompatibility triggers irreversible oxidative modification of proteins in incompatible pollen. Plant Physiol. 2020, 183, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Xie, F.; Trivino, M.; Karimi, M.; Bosch, M.; Franklin-Tong, V.E.; Nowack, M.K. Ectopic expression of a self-incompatibility module triggers growth arrest and cell death in vegetative cells. Plant Physiol. 2020, 183, 1765–1779. [Google Scholar] [CrossRef]
- Scheibe, R.; Baalmann, E.; Backhausen, J.E.; Rak, C.; Vetter, S. C-terminal truncation of spinach chloroplast NAD(P)-dependent glyceraldehyde-3-phosphate dehydrogenase prevents inactivation and reaggregation. Biochim. Biophys. Acta 1996, 1296, 228–234. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Lu, R.; Liu, D.; Ge, F.; Chen, C. Transient expression analysis of squalene epoxidase gene (SE) promoter of Panax notoginseng. Acta Physiol. Sin. 2015, 51, 495–500. [Google Scholar]
- Itzhaki, H.; Maxson, J.M.; Woodson, W.R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 1994, 91, 8925–8929. [Google Scholar] [CrossRef]
- Montgomery, J.; Goldman, S.; Deikman, J.; Margossian, L.; Fischer, R.L. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc. Natl. Acad. Sci. USA 1993, 90, 5939–5943. [Google Scholar] [CrossRef]
- Donald, R.G.; Cashmore, A.R. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990, 9, 1717–1726. [Google Scholar] [CrossRef]
- Rose, A.; Meier, I.; Wienand, U. The tomato I-box binding factor LeMYBI is a member of a novel class of myb-like proteins. Plant J. 1999, 20, 641–652. [Google Scholar] [CrossRef]
- Giuliano, G.; Pichersky, E.; Malik, V.S.; Timko, M.P.; Scolnik, P.A.; Cashmore, A.R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 1988, 85, 7089–7093. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef] [PubMed]
- Bate, N.; Twell, D. Functional architecture of a late pollen promoter: Pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 1998, 37, 859–869. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Leonard, J.M.; Monteros, A.; Liu, P.P.; Nonogaki, H. A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 2004, 134, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002, 129, 706–716. [Google Scholar] [CrossRef]
- Chen, W. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.H.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez-Casati, D.F. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 2008, 148, 1655–1667. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, C.; Liu, X.; Qin, R.; Shi, L.; Liu, S. The expression differences of GAPC and AT2G36580 in different tissues and organs of Brassica napus and their relationship with male sterility. Chin. J. Oil Crop. 2008, 2, 157–161. [Google Scholar]
- Hu, G.; Fearon, E.R. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell Biol. 1999, 19, 724–732. [Google Scholar] [CrossRef]
- Peralta, D.A.; Araya, A.; Busi, M.V.; Gomez-Casati, D.F. The E3 ubiquitin-ligase SEVEN IN ABSENTIA like 7 mono-ubiquitinates glyceraldehyde-3-phosphate dehydrogenase 1 isoform in vitro and is required for its nuclear localization in A. thaliana. Int. J. Biochem. Cell Biol. 2016, 70, 48–56. [Google Scholar] [CrossRef]
- Peralta, D.A.; Araya, A.; Gomez-Casati, D.F.; Busi, M.V. Over-expression of SINAL7 increases biomass and drought tolerance, and also delays senescence in Arabidopsis. J. Biotechnol. 2018, 283, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.K.; Reed, S.I. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 2013, 82, 387–414. [Google Scholar] [CrossRef]
- Lu, B.; He, W.; Shen, Y.; Jiang, J.; Chen, X.; Chen, W.; Lu, L. CDNA cloning and sequence analysis of glyceraldehyde 3-phosphate dehydrogenase gene from longan cotyledon embryo. J. Fujian Agric. For. Univ. 2009, 38, 507–511. [Google Scholar]
- Liu, Y.; Pan, T.; Tang, Y.; Zhuang, Y.; Liu, Z.; Li, P.; Li, H.; Huang, W.; Tu, S.; Ren, G.; et al. Proteomic Analysis of Rice Subjected to Low Light Stress and Overexpression of OsGAPB Increases the Stress Tolerance. Rice 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Ryazanova, L.P.; Ledova, L.A.; Andreeva, N.A.; Zvonarev, A.N.; Eldarov, M.A.; Kulakovskaya, T.V. Inorganic Polyphosphate and Physiological Properties of Saccharomyces cerevisiae Yeast Overexpressing Ppn2. Biochemistry 2020, 85, 516–522. [Google Scholar] [CrossRef]
- Zhang, L.; Lei, D.; Deng, X.; Li, F.; Ji, H.; Yang, S. Cytosolic glyceraldehyde-3-phosphate dehydrogenase 2/5/6 increase drought tolerance via stomatal movement and reactive oxygen species scavenging in wheat. Plant Cell Environ. 2020, 43, 836–853. [Google Scholar] [CrossRef]
- Zhang, L.; Song, Z.; Li, F.; Li, X.; Ji, H.; Yang, S. The specific MYB binding sites bound by TaMYB in the GAPCp2/3 promoters are involved in the drought stress response in wheat. BMC Plant Biol. 2019, 19, 366. [Google Scholar] [CrossRef]
- Miao, L.; Chen, C.; Yao, L.; Tran, J.; Zhang, H. Genome-wide identification, characterization, interaction network and expression profile of GAPDH gene family in sweet orange (Citrus sinensis). PeerJ 2019, 7, e7934. [Google Scholar] [CrossRef]
- Singh, P.; Kaloudas, D.; Raines, C.A. Expression analysis of the Arabidopsis CP12 gene family suggests novel roles for these proteins in roots and floral tissues. J. Exp. Bot. 2008, 59, 3975–3985. [Google Scholar] [CrossRef]
- Zeng, H.; Xie, Y.; Liu, G.; Lin, D.; He, C.; Shi, H. Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Mol. Biol. 2018, 97, 201–214. [Google Scholar] [CrossRef]
- Hu, D.; Xie, Q.; Liu, Q.; Zuo, T.; Zhang, H.; Zhang, Y.; Lian, X.; Zhu, L. Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L. Genes 2019, 10, 1000. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Zeng, J.; Zhang, H.; Converse, R.; Wang, Y.; Bai, X.; Zhu, L. PUB7, a pollen expression gene induced by self-pollination, negatively regulates pollen germination. Acta Biochim. Biophys. Sin. 2019, 51, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Gao, Q.; Zuo, T.; Lei, Z.; Pu, Q.; Wang, Y.; Liu, G.; He, X.; Ren, X.; Zhu, L. Identification and characterization of BoPUB3: A novel interaction protein with S-locus receptor kinase in Brassica oleracea L. Acta Biochim. Biophys. Sin. 2019, 51, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, J.B.; Nasrallah, M.E. Robust self-incompatibility in the absence of a functional ARC1 gene in Arabidopsis thaliana. Plant Cell 2014, 26, 3838–3841. [Google Scholar] [CrossRef] [PubMed][Green Version]



| Gene ID | Chr | Gene Length (bp) | CDS (bp) | No. of AA | PI | Molecular Weight (D) |
|---|---|---|---|---|---|---|
| Bo00285s370.1 | Scaffold00285 | 2112 | 1203 | 400 | 5.03 | 99,916.96 |
| Bo02686s010.1 | Scaffold02686 | 1190 | 1026 | 341 | 5.08 | 82,080.15 |
| Bo1g152180.1 | C1 | 2172 | 1134 | 377 | 5.02 | 94,630.74 |
| Bo2g075590.1 | C2 | 3743 | 2112 | 703 | 4.92 | 171,516.98 |
| Bo2g095350.1 | C2 | 2666 | 1260 | 419 | 5.02 | 104,769.31 |
| Bo3g057250.1 | C3 | 2022 | 1017 | 356 | 5.07 | 83,516.28 |
| Bo5g016460.1 | C5 | 1565 | 1200 | 399 | 5.02 | 99,854.00 |
| Bo5g017500.1 | C5 | 1794 | 1020 | 339 | 5.06 | 83,749.58 |
| Bo5g021670.1 | C5 | 3001 | 1281 | 426 | 5.02 | 106,195.90 |
| Bo5g148380.1 | C5 | 2037 | 1017 | 338 | 5.05 | 83,667.04 |
| Bo6g080970.1 | C6 | 2919 | 1266 | 421 | 5.02 | 105,067.33 |
| Bo7g083800.1 | C7 | 1055 | 711 | 236 | 5.13 | 59,271.32 |
| Bo8g024290.1 | C8 | 2614 | 1347 | 448 | 4.99 | 110,675.48 |
| Bo8g065470.1 | C8 | 2061 | 1020 | 339 | 5.07 | 83,721.50 |
| Bo8g107420.1 | C8 | 2495 | 1071 | 356 | 5.05 | 88,265.82 |
| Bo8g107880.1 | C8 | 1862 | 1200 | 399 | 5.02 | 100,014.22 |
| AT1G12900.1 | Chr1 | 1950 | 1200 | 399 | 5.04 | 99,919.44 |
| AT1G12900.2 | Chr1 | 1914 | 954 | 317 | 5.09 | 79,272.45 |
| AT1G12900.3 | Chr1 | 1857 | 1053 | 350 | 5.08 | 86,837.59 |
| AT1G12900.4 | Chr1 | 1784 | 1053 | 350 | 5.08 | 86,837.59 |
| AT1G12900.5 | Chr1 | 2016 | 1326 | 441 | 5.01 | 111,216.46 |
| AT1G13440.1 | Chr1 | 2801 | 1017 | 338 | 5.08 | 83,708.07 |
| AT1G13440.2 | Chr1 | 2451 | 933 | 310 | 5.10 | 76,429.79 |
| AT1G16300.1 | Chr1 | 3365 | 1263 | 420 | 5.02 | 104,385.82 |
| AT1G42970.1 | Chr1 | 2542 | 1344 | 447 | 5.01 | 110,193.00 |
| AT1G79530.1 | Chr1 | 3506 | 1269 | 442 | 5.02 | 105,332.66 |
| AT3G04120.1 | Chr3 | 2748 | 1017 | 338 | 5.06 | 83,798.81 |
| AT3G26650.1 | Chr3 | 2183 | 1191 | 396 | 5.04 | 99,013.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Q.; Zhang, H.; Hu, D.; Liu, Q.; Zuo, T.; Zhang, Y.; Liu, Y.; Zhou, S.; Zhu, L. Analysis of SI-Related BoGAPDH Family Genes and Response of BoGAPC to SI Signal in Brassica oleracea L. Genes 2021, 12, 1719. https://doi.org/10.3390/genes12111719
Xie Q, Zhang H, Hu D, Liu Q, Zuo T, Zhang Y, Liu Y, Zhou S, Zhu L. Analysis of SI-Related BoGAPDH Family Genes and Response of BoGAPC to SI Signal in Brassica oleracea L. Genes. 2021; 12(11):1719. https://doi.org/10.3390/genes12111719
Chicago/Turabian StyleXie, Qinqin, Hecui Zhang, Dengke Hu, Qianying Liu, Tonghong Zuo, Yizhong Zhang, Yimei Liu, Siru Zhou, and Liquan Zhu. 2021. "Analysis of SI-Related BoGAPDH Family Genes and Response of BoGAPC to SI Signal in Brassica oleracea L." Genes 12, no. 11: 1719. https://doi.org/10.3390/genes12111719
APA StyleXie, Q., Zhang, H., Hu, D., Liu, Q., Zuo, T., Zhang, Y., Liu, Y., Zhou, S., & Zhu, L. (2021). Analysis of SI-Related BoGAPDH Family Genes and Response of BoGAPC to SI Signal in Brassica oleracea L. Genes, 12(11), 1719. https://doi.org/10.3390/genes12111719





