Effect of High Hydrostatic Pressure on Stress-Related dnaK, hrcA, and ctsR Expression Patterns in Selected Lactobacilli Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. LAB Strains and Culture Conditions
2.2. High Hydrostatic Pressure Application
2.3. Plate Count Analytical Methods
2.4. Bacterial RNA Extraction
2.5. Real-Time PCR (RT-qPCR)
2.6. Statistical Data Analysis
3. Results
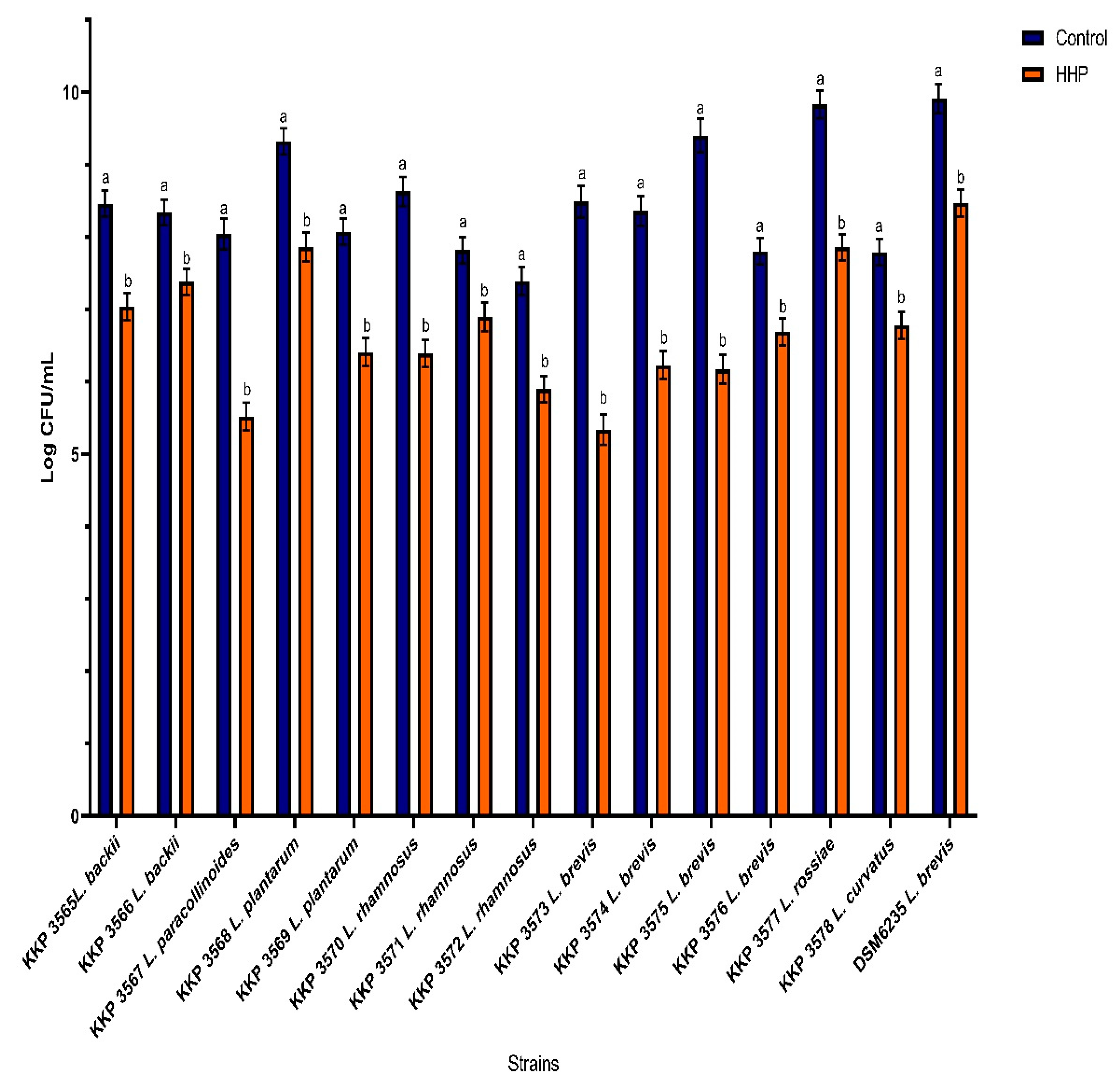
3.1. The Effect of the HHP on the Bacterial Cells
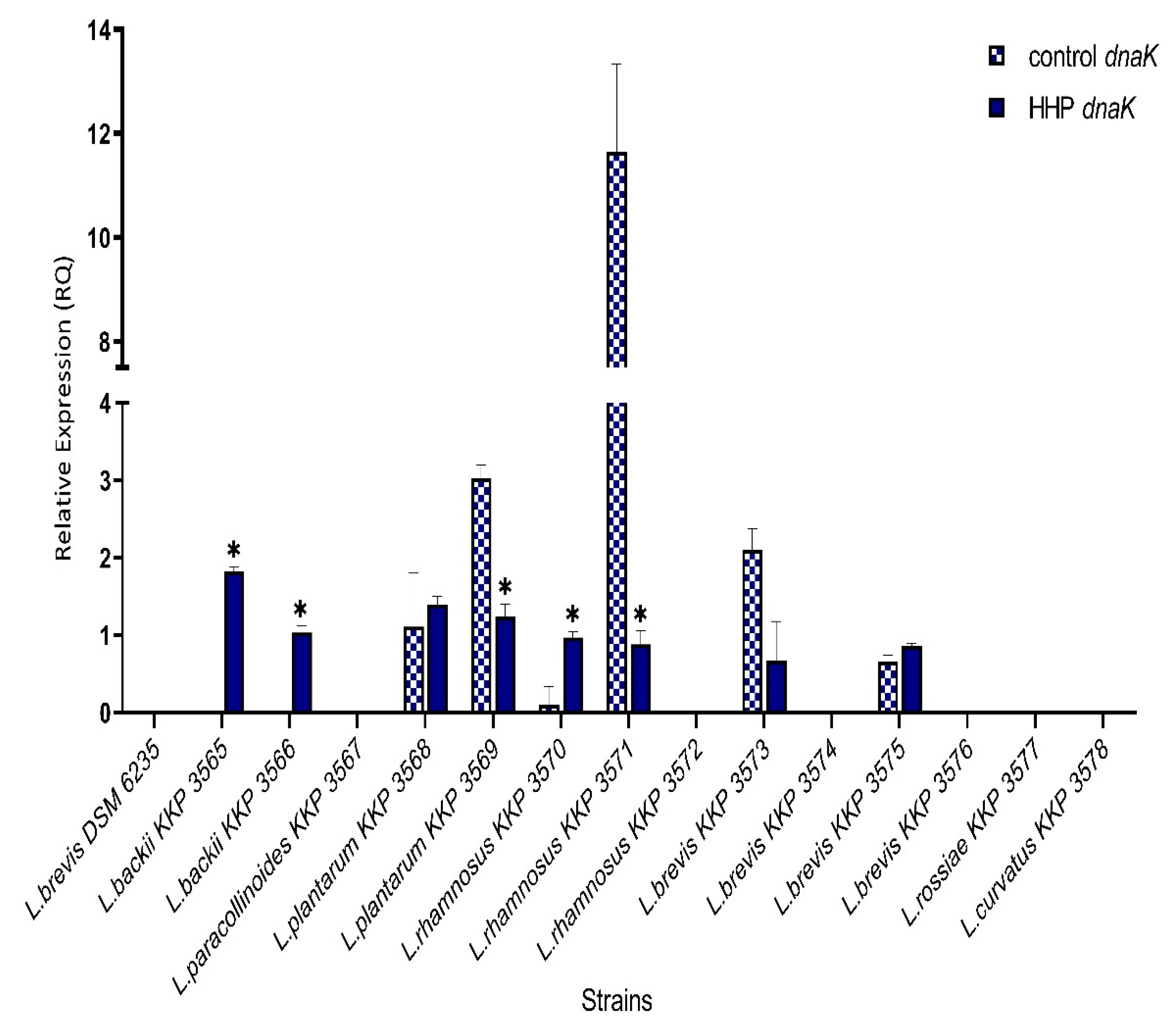
3.2. DnaK Gene
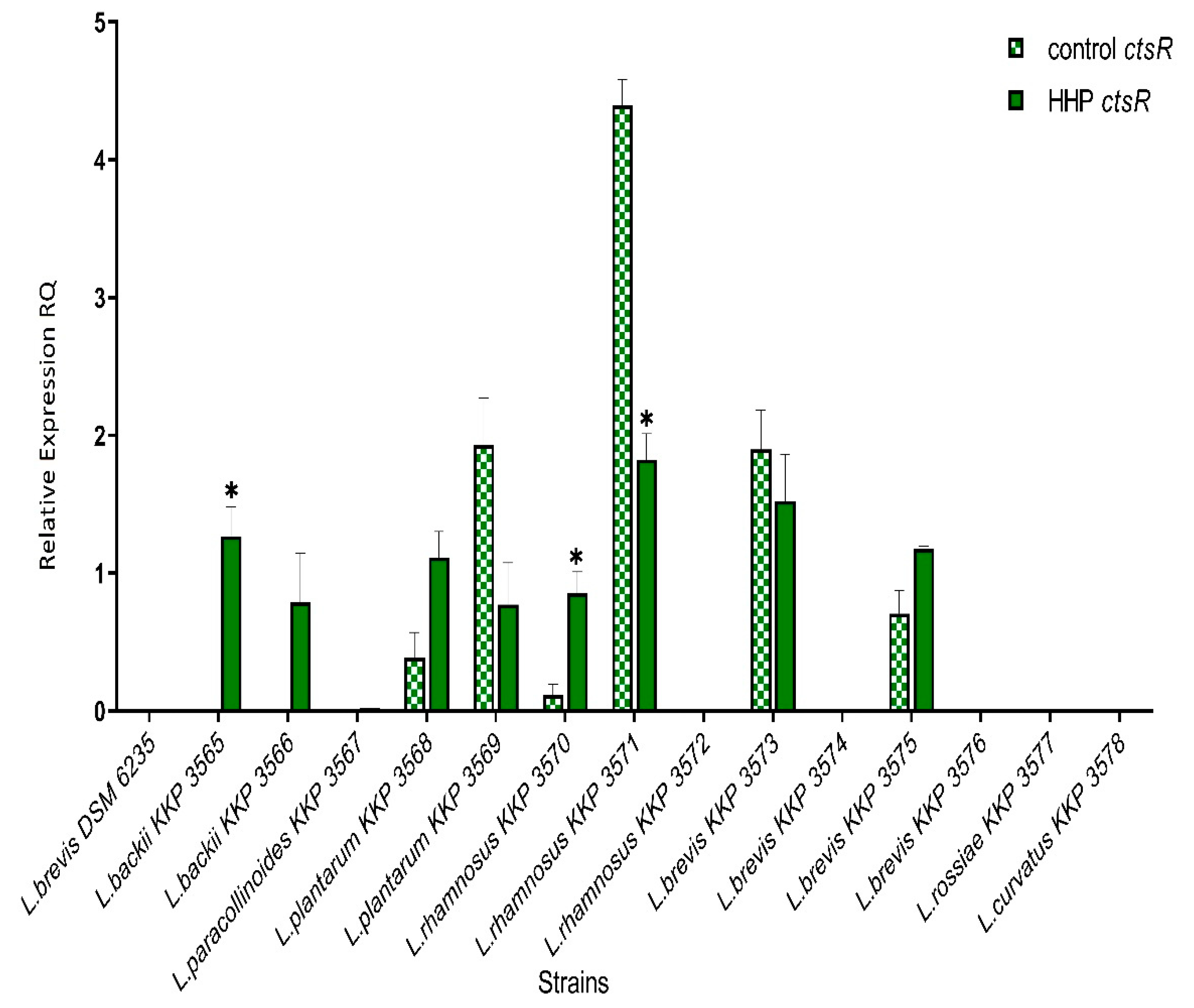
3.3. ctsR Gene
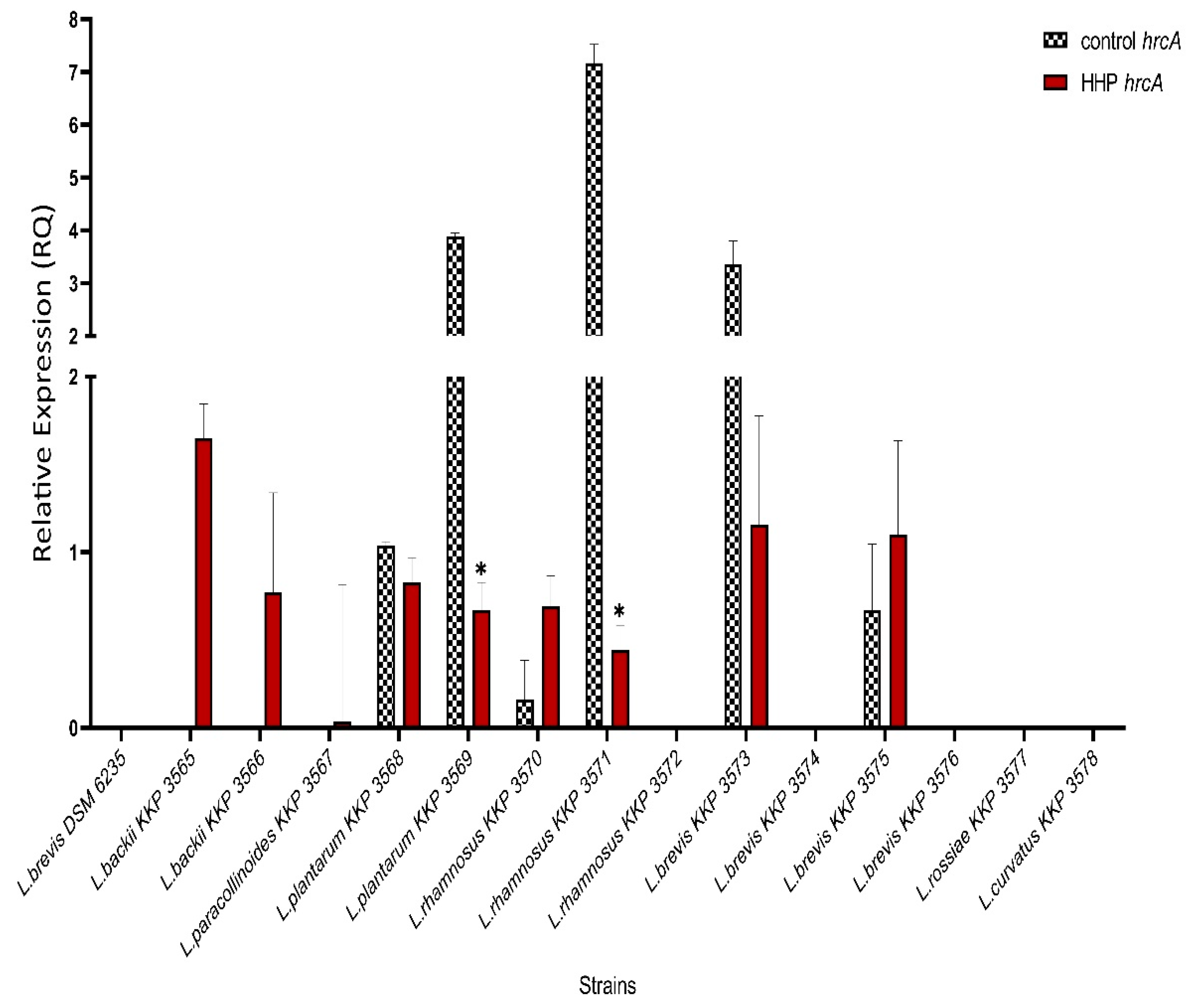
3.4. hrcA Gene
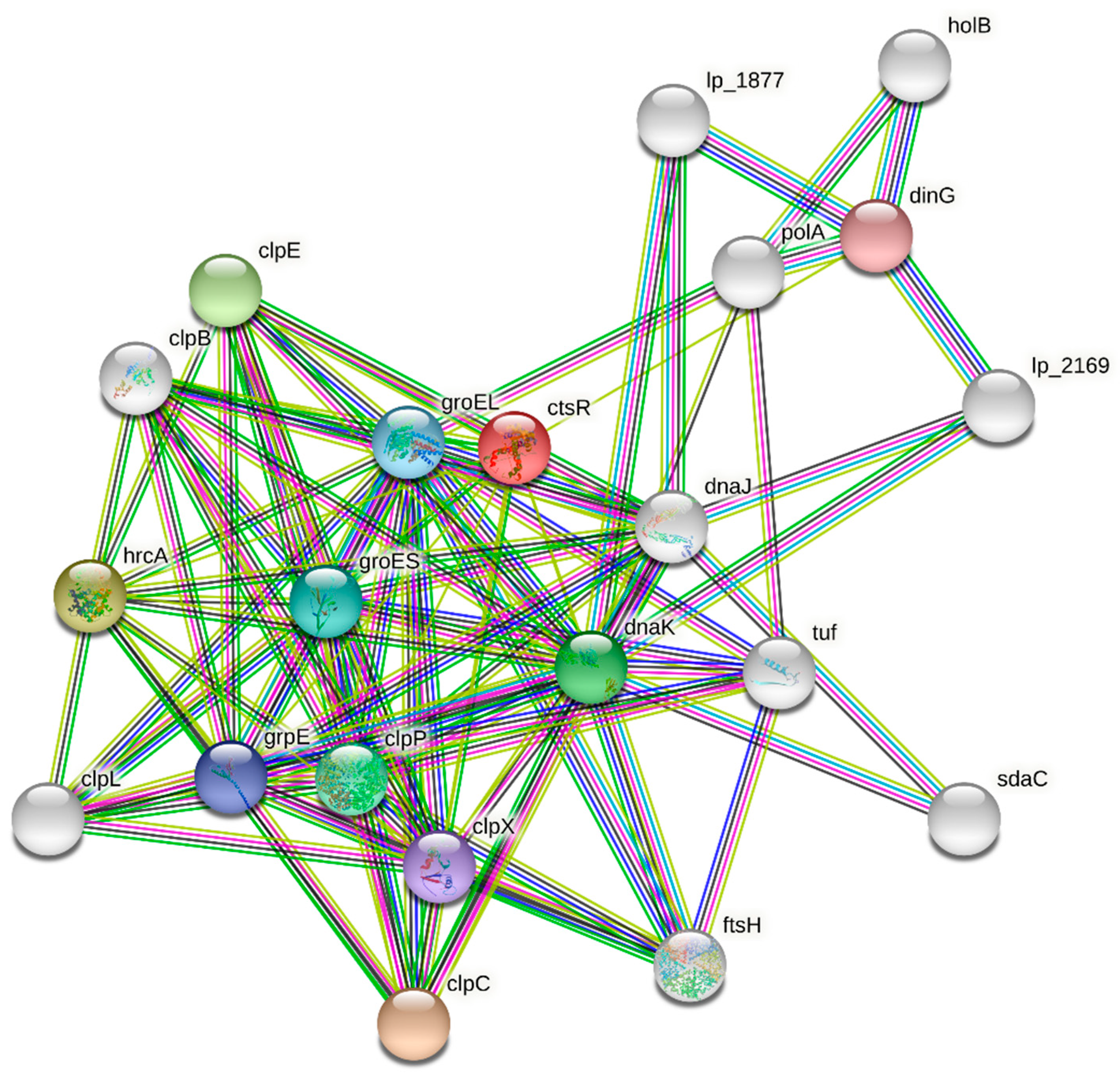
3.5. Protein–Protein Interaction (PPI) Network Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bucka-Kolendo, J.; Sokołowska, B. Lactic acid bacteria stress response to preservation processes in the beverage and juice industry. Acta Biochim. Pol. 2017, 64, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, D.; Scarlato, V. Regulation of heat-shock genes in bacteria: From signal sensing to gene expression output. FEMS Microbiol. Rev. 2017, 41, 549–574. [Google Scholar] [CrossRef]
- Mbye, M.; Baig, M.A.; AbuQamar, S.F.; El-Tarabily, K.A.; Obaid, R.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Turner, M.S.; Nagendra, P.S.; Ayyash, M.M. Updates on understanding of probiotic lactic acid bacteria responses to environmental stresses and highlights on proteomic analyses. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Pepper, S. Characterisation of Stress Responses in Lactobacillus Paracasei and Bifidobacterium Animalis (syn. lactis). Ph.D Thesis, Victoria University, Wellington, Australia, 2004. [Google Scholar]
- Sokołowska, B.; Nasiłowska, J. Controlling spoilage and pathogenic microorganisms in beetroot (Beta vulgaris) juice by high hydrostatic pressure. In Safety Issues in Beverage Production; Grumezescu, A.M., Holban, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 18, pp. 79–104. [Google Scholar] [CrossRef]
- Sokołowska, B.; Skąpska, S.; Fonberg-Broczek, M.; Niezgoda, J.; Rutkowska, M.; Dobros, N.; Rzoska, J.S. The impact of high hydrostatic pressure (HHP) on native microflora and the colour of beetroot juice—A preliminary shelf-life study. In Industrial, Medical and Environmental Applications of Microorganisms: Current Status and Trends; Mendez-Vilas, A., Ed.; Wageningen Academic Publisher: Wageningen, The Netherlands, 2014; pp. 380–384. [Google Scholar] [CrossRef]
- Darans, N.; Badosa, E.; Frances, J.; Montesinos, E.; Bonaterra, A. Enhancing water stress tolerance improves fitness in biological control strains of Lactobacillus plantarum in plant environments. PLoS ONE 2018, 13, e0190931. [Google Scholar] [CrossRef] [PubMed]
- Van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Koch, B.; Kilstrup, M.; Vogensen, F.V.; Hammer, K. Induced levels of heat shock proteins in a dnaK mutant of Lactococcus lactis. J. Bacteriol. 1998, 180, 3873–3881. [Google Scholar] [CrossRef]
- Palud, A.; Scornec, H.; Calvin, J.-F.; Licandro, H. New genes involved in mild stress response identified by transposon mutagenesis in Lactobacillus paracasei. Front. Microbiol. 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Alegria, A.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 3, 837–890. [Google Scholar] [CrossRef]
- Guidone, A.; Parente, E.; Zotta, T.; Guinane, C.M.; Rea, M.C.; Stanton, C.; Ross, R.P.; Ricciardi, A. Polymorphisms in stress response genes in Lactobacillus plantarum: Implications for classification and heat stress response. Ann. Microbiol. 2015, 65, 297–305. [Google Scholar] [CrossRef]
- Capozzi, V.; Arena, P.A.; Crisetti, E.; Spano, G.; Fiocco, D. The hsp16 gene of the probiotic Lactobacillus acidophilus is differently regulated by salt, high temperature and acidic stresses, as revealed by Reverse Transcription Quantitative PCR (qRT-PCR) Analysis. Int. J. Mol. Sci. 2011, 12, 5390–5405. [Google Scholar] [CrossRef]
- Van Bokhorst-van de Veen, H.; Bongers, R.S.; Wels, M.; Bron, P.A.; Kleerebezern, M. Transcriptome signatures of class I and III stress response deregulation in Lactobacillus plantarum reveal pleiotropic adaptation. Microb. Cell Factories 2013, 12, 112. Available online: http://www.microbialcellfactories.com/content/12/1/112 (accessed on 15 September 2021). [CrossRef]
- Drews, O.; Weiss, W.; Reil, G.; Parlar, H.; Wait, R.; Görg, A. High pressure effects step-wise altered protein expression in Lactobacillus sanfranciscensis. Proteomics 2002, 2, 765–774. [Google Scholar] [CrossRef]
- Vogel, R.F.; Pavlovic, M.; Hörmann, S.; Ehrmann, M.A. High pressure-sensitive expression in Lactobacillus sanfranciscensis. Braz. J. Med. Biol Res. 2005, 38, 1247–1252. [Google Scholar] [CrossRef]
- Bucka-Kolendo, J.; Sokołowska, B.; Winiarczyk, S. Influence of High Hydrostatic Pressure on the Identification of Lactobacillus by MALDI-TOF MS-Preliminary Study. Microorganisms 2020, 8, 813. [Google Scholar] [CrossRef]
- Zheng, Z.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Sliver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2(-Delta Delta C(T)) method. Methods 2001, 25, 4, 402–408. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Schumann, W. The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 2003, 8, 207–217. [Google Scholar] [CrossRef]
- Ricciardi, A.; Parente, E.; Guidone, A.; Ianniello, R.G.; Zotta, T.; Abu Sayem, S.M.; Varcamonti, M. Genotypic diversity of stress response in Lactobacillus plantarum, Lactobacillus paraplantalum and Lactobacillus pentosus. Int. Food Microbiol. 2012, 157, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Darsonval, M.; Julliat, F.; Msadek, T.; Alexandre, H.; Grandvalet, C. CtsR, the Master Regulator of Stress-Response in Oenococcus oeni, Is a Heat Sensor Interacting With ClpL1. Front. Microbiol. 2018, 9, 3135. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Bi, Y.T.; Dong, C.; Yang, J.F.; Liang, W.D. Transcriptome analysis of adaptive heat shock response of Streptococcus thermophilus. PLoS ONE 2011, 6, e25777. [Google Scholar] [CrossRef]
- Russo, P.; De la Luz Mohedano, M.; Capozzi, V.; De Palencia, P.F.; Lopez, P.; Spano, G.; Fiocco, D. Comparative Proteomic Analysis of Lactobacillus plantarum WCFS1 and ΔctsR Mutant Strains Under Physiological and Heat Stress Conditions. Int. J. Mol. Sci. 2012, 13, 10680–10696. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Ciocia, F.; Ricciardi, A.; Zotta, T.; Felis, G.E.; Torriani, S. Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: A multivariate screening study. Int. J. Food Microbiol. 2010, 144, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, C.; Siciliano, R.A.; Muscariello, L.; Marasco, R.; Sacco, M. CcpA affects expression of the groESL and dnaK operons in Lactobacillus plantarum. Microb. Cell Factories 2006, 5, 35. [Google Scholar] [CrossRef] [PubMed]
| Strain | Source | 16S rDNA Identification | New Nomenclature |
|---|---|---|---|
| KKP 3565 | Beer | Lactobacillus backii | Loigolactobacillus backii |
| KKP 3566 | Beer | Lactobacillus backii | Loigolactobacillus backii |
| KKP 3567 | Beer | Lactobacillus paracollinoides | Secundilactobacillus paracollinoides |
| KKP 3568 | Bread | Lactobacillus plantarum | Lactiplantibacillus plantarum |
| KKP 3569 | Tomato juice | Lactobacillus plantarum | Lactiplantibacillus plantarum |
| KKP 3570 | Tomato juice | Lactobacillus rhamnosus | Lacticaseibacillus rhamnosus |
| KKP 3571 | Probiotic | Lactobacillus rhamnosus | Lacticaseibacillus rhamnosus |
| KKP 3572 | Ice Cream | Lactobacillus rhamnosus | Lacticaseibacillus rhamnosus |
| KKP 3573 | Beer | Lactobacillus brevis | Levitlactobacillus brevis |
| KKP 3574 | Beer | Lactobacillus brevis | Levitlactobacillus brevis |
| KKP 3575 | Beer | Lactobacillus brevis | Levitlactobacillus brevis |
| KKP 3576 | Beer | Lactobacillus brevis | Levitlactobacillus brevis |
| KKP 3577 | Beer | Lactobacillus rossiae | Furfurilactobacillus rossiae |
| KKP 3578 | Sauerkraut juice | Lactobacillus curvatus | Latilactobacillus curvatus |
| DSM6235 | Beer | Lactobacillus brevis | Levitlactobacillus brevis |
| Gene | Sequence—5′ to 3′ | Amplicon (bp) | TagMan Assay No. | GenBank |
|---|---|---|---|---|
| ctsR | F: TGGTCGATGATGCTGATGTG | 128 | AP47ZY7 | CP052869.1 |
| FAM-ACAACGAGATGCTTATGCGGTCGT-MGB | ||||
| R: TAAATCGTCGAGAAGCGCAA | ||||
| hrcA | F: TGCAAGATCCAGACGGATTC | 116 | APXGWAE | JX967738.1 |
| P: FAM-TTTGGCAGTGTGTTGTCCAAGGC-MGB | ||||
| R: ACTATACGGACCAGCAAAGC | ||||
| dnaK | F: TGTCGGTCTTATCCAAACC | 110 | APZTJE9 | CP053571.1 |
| P: FAM-CTGCCGCCGTTGGTTCGTTAATAA-MGB | ||||
| R: CTTTAGTTGCTTGCCGTTGG | ||||
| 16S | Assay by design TFS | Ba04230899_s1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bucka-Kolendo, J.; Juszczuk-Kubiak, E.; Sokołowska, B. Effect of High Hydrostatic Pressure on Stress-Related dnaK, hrcA, and ctsR Expression Patterns in Selected Lactobacilli Strains. Genes 2021, 12, 1720. https://doi.org/10.3390/genes12111720
Bucka-Kolendo J, Juszczuk-Kubiak E, Sokołowska B. Effect of High Hydrostatic Pressure on Stress-Related dnaK, hrcA, and ctsR Expression Patterns in Selected Lactobacilli Strains. Genes. 2021; 12(11):1720. https://doi.org/10.3390/genes12111720
Chicago/Turabian StyleBucka-Kolendo, Joanna, Edyta Juszczuk-Kubiak, and Barbara Sokołowska. 2021. "Effect of High Hydrostatic Pressure on Stress-Related dnaK, hrcA, and ctsR Expression Patterns in Selected Lactobacilli Strains" Genes 12, no. 11: 1720. https://doi.org/10.3390/genes12111720
APA StyleBucka-Kolendo, J., Juszczuk-Kubiak, E., & Sokołowska, B. (2021). Effect of High Hydrostatic Pressure on Stress-Related dnaK, hrcA, and ctsR Expression Patterns in Selected Lactobacilli Strains. Genes, 12(11), 1720. https://doi.org/10.3390/genes12111720








