The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity
Abstract
:1. Introduction
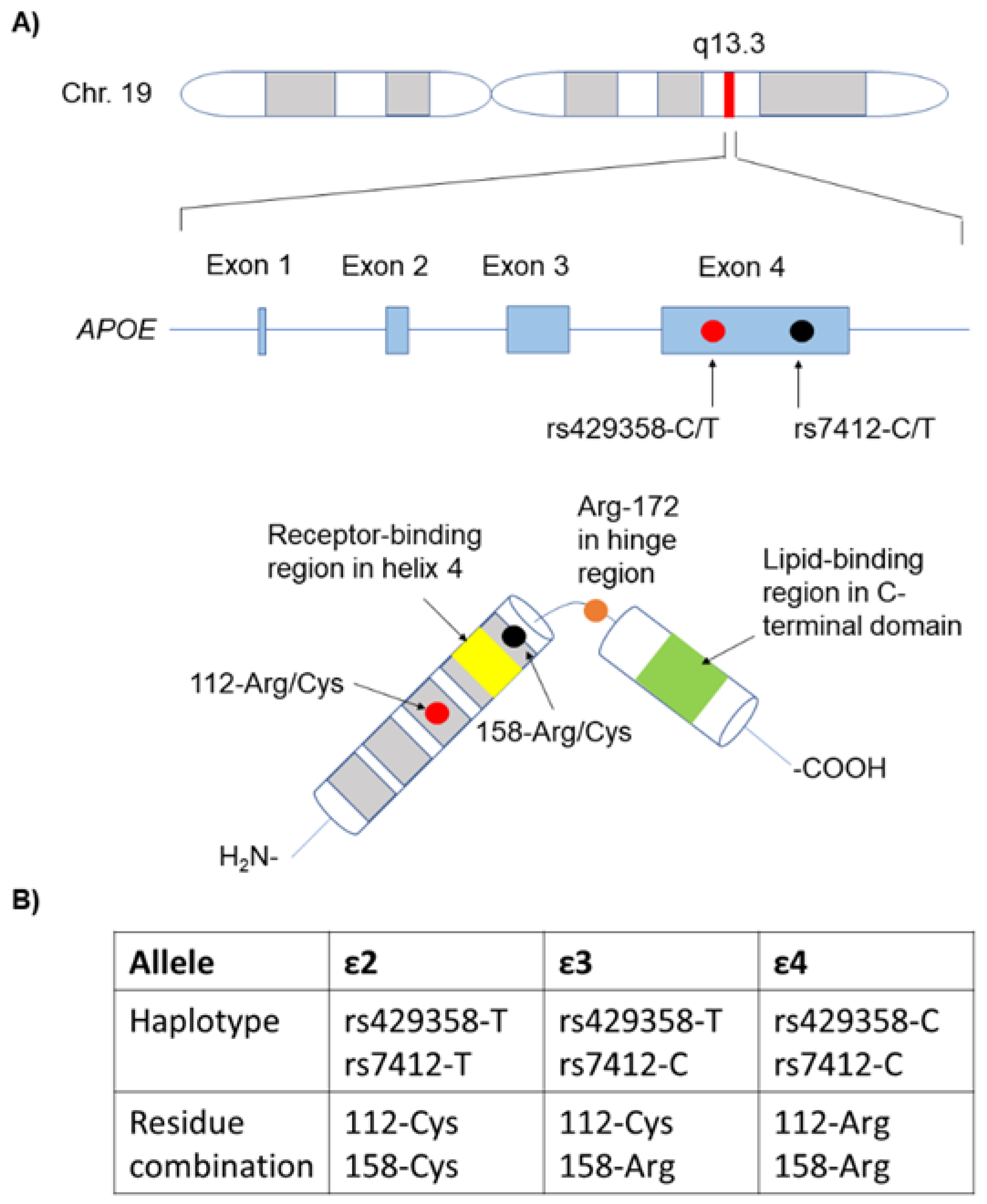
- APOE gene and protein structure and function, including the latest theoretical models describing its mechanism of action
- The role of APOE in human longevity, its physiological functions, and the involvement in pathological traits in modern populations
- APOE evolution and variability among human populations, including a novel analysis of modern and ancient data
- The evolutionary mechanisms that maintained APOE deleterious variants in modern human populations.
2. APOE Structure and Models
3. APOE Function and Pathology
4. APOE and Human Longevity
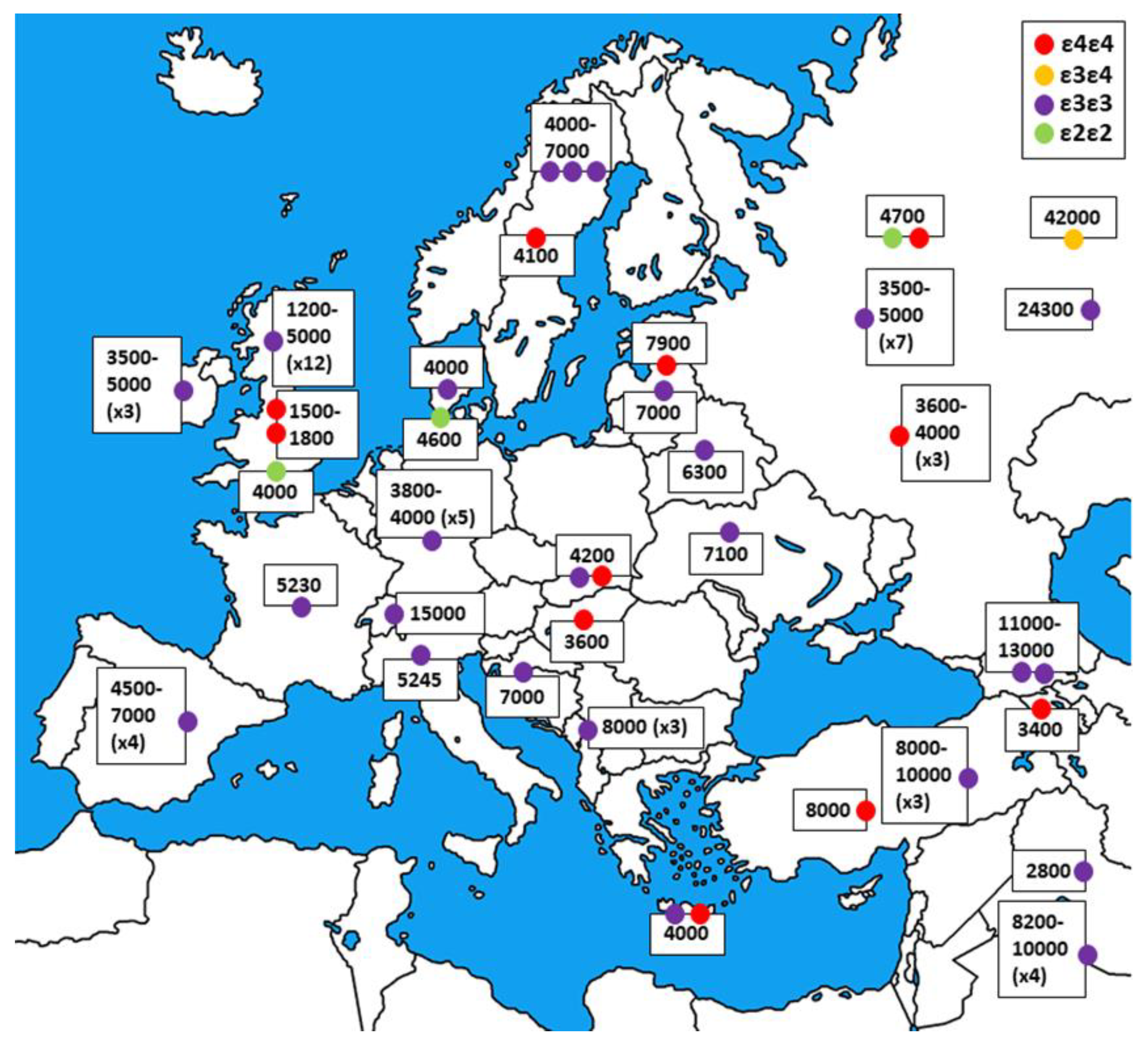
5. APOE Evolution and Variability among Human Populations
6. APOE Trade-Offs
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Blue, M.L.; Williams, D.L.; Zucker, S.; Khan, S.A.; Blum, C.B. Apolipoprotein E synthesis in human kidney, adrenal gland, and liver. Proc. Natl. Acad. Sci. USA 1983, 80, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Kockx, M.; Traini, M.; Kritharides, L. Cell-specific production, secretion, and function of apolipoprotein E. J. Mol. Med. 2018, 96, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Tedla, N.; Glaros, E.N.; Brunk, U.T.; Jessup, W.; Garner, B. Heterogeneous expression of apolipoprotein-E by human macrophages. Immunology 2004, 113, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Boyles, J.K.; Pitas, R.E.; Wilson, E.; Mahley, R.W.; Taylor, J.M. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J. Clin. Investig. 1985, 76, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Wetterau, J.R.; Aggerbeck, L.P.; Rall, S.C.; Weisgraber, K.H. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988, 263, 6240–6248. [Google Scholar] [PubMed]
- Wilson, C.; Wardell, M.R.; Weisgraber, K.H.; Mahley, R.W.; Agard, D.A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 1991, 252, 1817–1822. [Google Scholar] [CrossRef]
- Mahley, R.W.; Innerarity, T.L.; Rall, S.C.; Weisgraber, K.H. Plasma lipoproteins: Apolipoprotein structure and function. J. Lipid Res. 1984, 25, 1277–1294. [Google Scholar]
- Morrow, J.A.; Arnold, K.S.; Dong, J.; Balestra, M.E.; Innerarity, T.L.; Weisgraber, K.H. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J. Biol. Chem. 2000, 275, 2576–2580. [Google Scholar] [CrossRef]
- Huang, R.Y.-C.; Garai, K.; Frieden, C.; Gross, M.L. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry 2011, 50, 9273–9282. [Google Scholar] [CrossRef]
- Chou, C.-Y.; Lin, Y.-L.; Huang, Y.-C.; Sheu, S.-Y.; Lin, T.-H.; Tsay, H.-J.; Chang, G.-G.; Shiao, M.-S. Structural variation in human apolipoprotein E3 and E4: Secondary structure, tertiary structure, and size distribution. Biophys. J. 2005, 88, 455–466. [Google Scholar] [CrossRef]
- Subramanian, S.; Gottschalk, W.K.; Kim, S.Y.; Roses, A.D.; Chiba-Falek, O. The effects of PPARγ on the regulation of the TOMM40-APOE-C1 genes cluster. Biochim. Biophys. Acta 2017, 1863, 810–816. [Google Scholar] [CrossRef]
- Roses, A.; Sundseth, S.; Saunders, A.; Gottschalk, W.; Burns, D.; Lutz, M. Understanding the genetics of APOE and TOMM40 and role of mitochondrial structure and function in clinical pharmacology of Alzheimer’s disease. Alzheimers Dement. 2016, 12, 687–694. [Google Scholar] [CrossRef]
- Cervantes, S.; Samaranch, L.; Vidal-Taboada, J.M.; Lamet, I.; Bullido, M.J.; Frank-García, A.; Coria, F.; Lleó, A.; Clarimón, J.; Lorenzo, E.; et al. Genetic variation in APOE cluster region and Alzheimer’s disease risk. Neurobiol. Aging 2011, 32, 2107.e7–2107.e17. [Google Scholar] [CrossRef]
- Papaioannou, I.; Simons, J.P.; Owen, J.S. Targeted in situ gene correction of dysfunctional APOE alleles to produce atheroprotective plasma ApoE3 protein. Cardiol. Res. Pract. 2012, 2012, 148796. [Google Scholar] [CrossRef]
- Kulminski, A.M.; Huang, J.; Wang, J.; He, L.; Loika, Y.; Culminskaya, I. Apolipoprotein E region molecular signatures of Alzheimer’s disease. Aging Cell 2018, 17, e12779. [Google Scholar] [CrossRef]
- Weisgraber, K.H.; Rall, S.C.; Mahley, R.W. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem. 1981, 256, 9077–9083. [Google Scholar]
- Weisgraber, K.H. Apolipoprotein E distribution among human plasma lipoproteins: Role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990, 31, 1503–1511. [Google Scholar]
- Chetty, P.S.; Mayne, L.; Lund-Katz, S.; Englander, S.W.; Phillips, M.C. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA 2017, 114, 968–973. [Google Scholar] [CrossRef]
- Matsunaga, A.; Saito, T. Apolipoprotein E mutations: A comparison between lipoprotein glomerulopathy and type III hyperlipoproteinemia. Clin. Exp. Nephrol. 2014, 18, 220–224. [Google Scholar] [CrossRef]
- Frieden, C.; Garai, K. Concerning the structure of apoE: Structure of apoE. Protein Sci. 2013, 22, 1820–1825. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72, 3–12. [Google Scholar] [CrossRef]
- Mahley, R.W.; Weisgraber, K.H.; Huang, Y. Apolipoprotein E: Structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 2009, 50, S183–S188. [Google Scholar] [CrossRef]
- Nguyen, D.; Dhanasekaran, P.; Nickel, M.; Mizuguchi, C.; Watanabe, M.; Saito, H.; Phillips, M.C.; Lund-Katz, S. Influence of domain stability on the properties of human apolipoprotein E3 and E4 and mouse Apolipoprotein E. Biochemistry 2014, 53, 4025–4033. [Google Scholar] [CrossRef]
- Henry, N.; Krammer, E.-M.; Stengel, F.; Adams, Q.; Van Liefferinge, F.; Hubin, E.; Chaves, R.; Efremov, R.; Aebersold, R.; Vandenbussche, G.; et al. Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach. PLoS Comput. Biol. 2018, 14, e1006165. [Google Scholar] [CrossRef]
- Nguyen, D.; Dhanasekaran, P.; Nickel, M.; Nakatani, R.; Saito, H.; Phillips, M.C.; Lund-Katz, S. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry 2010, 49, 10881–10889. [Google Scholar] [CrossRef]
- Nguyen, D.; Dhanasekaran, P.; Phillips, M.C.; Lund-Katz, S. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry 2009, 48, 3025–3032. [Google Scholar] [CrossRef]
- Weisgraber, K.H.; Innerarity, T.L.; Mahley, R.W. Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. J. Biol. Chem. 1982, 257, 2518–2521. [Google Scholar] [PubMed]
- Dong, L.-M.; Parkin, S.; Trakhanov, S.D.; Rupp, B.; Simmons, T.; Arnold, K.S.; Newhouse, Y.M.; Innerarity, T.L.; Weisgraber, K.H. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat. Struct. Mol. Biol. 1996, 3, 718–722. [Google Scholar] [CrossRef]
- Dong, L.-M.; Weisgraber, K.H. Human Apolipoprotein E4 Domain Interaction arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996, 271, 19053–19057. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.M.; Wilson, C.; Wardell, M.R.; Simmons, T.; Mahley, R.W.; Weisgraber, K.H.; Agard, D.A. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 1994, 269, 22358–22365. [Google Scholar] [PubMed]
- Hatters, D.M.; Peters-Libeu, C.A.; Weisgraber, K.H. Engineering conformational destabilization into mouse apolipoprotein E A model for a unique property of human apolipoprotein E4. J. Biol. Chem. 2005, 280, 26477–26482. [Google Scholar] [CrossRef]
- Hatters, D.M.; Budamagunta, M.S.; Voss, J.C.; Weisgraber, K.H. Modulation of apolipoprotein E structure by domain interaction differences in lipid-bound and lipid-free forms. J. Biol. Chem. 2005, 280, 34288–34295. [Google Scholar] [CrossRef]
- Kara, E.; Marks, J.D.; Fan, Z.; Klickstein, J.A.; Roe, A.D.; Krogh, K.A.; Wegmann, S.; Maesako, M.; Luo, C.C.; Mylvaganam, R.; et al. Isoform- and cell type-specific structure of apolipoprotein E lipoparticles as revealed by a novel Forster resonance energy transfer assay. J. Biol. Chem. 2017, 292, 14720–14729. [Google Scholar] [CrossRef]
- Xu, Q.; Brecht, W.J.; Weisgraber, K.H.; Mahley, R.W.; Huang, Y. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J. Biol. Chem. 2004, 279, 25511–25516. [Google Scholar] [CrossRef]
- Morrow, J.A.; Segall, M.L.; Lund-Katz, S.; Phillips, M.C.; Knapp, M.; Rupp, B.; Weisgraber, K.H. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 2000, 39, 11657–11666. [Google Scholar] [CrossRef]
- Morrow, J.A.; Hatters, D.M.; Lu, B.; Höchtl, P.; Oberg, K.A.; Rupp, B.; Weisgraber, K.H. Apolipoprotein E4 forms a Molten Globule A potential basis for its association with disease. J. Biol. Chem. 2002, 277, 50380–50385. [Google Scholar] [CrossRef]
- Acharya, P.; Segall, M.L.; Zaiou, M.; Morrow, J.; Weisgraber, K.H.; Phillips, M.C.; Lund-Katz, S.; Snow, J. Comparison of the stabilities and unfolding pathways of human apolipoprotein E isoforms by differential scanning calorimetry and circular dichroism. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2002, 1584, 9–19. [Google Scholar] [CrossRef]
- Bychkova, V.E.; Ptitsyn, O.B. Folding intermediates are involved in genetic diseases? FEBS Lett. 1995, 359, 6–8. [Google Scholar] [CrossRef]
- Ptitsyn, O.B.; Bychkova, V.E.; Uversky, V.N. Kinetic and equilibrium folding intermediates. Philos. Trans. R. Soc. Lond. B 1995, 348, 35–41. [Google Scholar]
- Gursky, O.; Atkinson, D. Thermal unfolding of human high-density apolipoprotein A-1: Implications for a lipid-free molten globular state. Proc. Natl. Acad. Sci. USA 1996, 93, 2991–2995. [Google Scholar] [CrossRef]
- Gursky, O.; Atkinson, D. High- and low-temperature unfolding of human high-density apolipoprotein A-2. Protein Sci. Publ. Protein Soc. 1996, 5, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Haddy, N.; Bacquer, D.D.; Chemaly, M.M.; Maurice, M.; Ehnholm, C.; Evans, A.; Sans, S.; do Martins, M.C.; Backer, G.D.; Siest, G.; et al. The importance of plasma apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: Results from Apo Europe. Eur. J. Hum. Genet. 2002, 10, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Girod, M.; Fonbonne, C.; Salvador, A.; Clément, Y.; Lantéri, P.; Amouyel, P.; Lambert, J.C.; Lemoine, J. Total ApoE and ApoE4 isoform assays in an Alzheimer’s disease case-control study by targeted mass spectrometry (n = 669): A pilot assay for methionine-containing proteotypic peptides. Mol. Cell. Proteom. 2012, 11, 1389–1403. [Google Scholar] [CrossRef]
- Martínez-Morillo, E.; Hansson, O.; Atagi, Y.; Bu, G.; Minthon, L.; Diamandis, E.P.; Nielsen, H.M. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 2014, 127, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Rezeli, M.; Zetterberg, H.; Blennow, K.; Brinkmalm, A.; Laurell, T.; Hansson, O.; Marko-Varga, G. Quantification of total apolipoprotein E and its specific isoforms in cerebrospinal fluid and blood in Alzheimer’s disease and other neurodegenerative diseases. EuPA Open Proteomics 2015, 8, 137–143. [Google Scholar] [CrossRef]
- Brecht, W.J.; Harris, F.M.; Chang, S.; Tesseur, I.; Yu, G.-Q.; Xu, Q.; Fish, J.D.; Wyss-Coray, T.; Buttini, M.; Mucke, L.; et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increased Tau phosphorylation in brains of transgenic mice. J. Neurosci. 2004, 24, 2527–2534. [Google Scholar] [CrossRef]
- Riddell, D.R.; Zhou, H.; Atchison, K.; Warwick, H.K.; Atkinson, P.J.; Jefferson, J.; Xu, L.; Aschmies, S.; Kirksey, Y.; Hu, Y.; et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J. Neurosci. 2008, 28, 11445–11453. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.A.; Tsoi, K.; Holinkova, S.; Chan, S.L.; Kim, W.S.; Halliday, G.M.; Rye, K.-A.; Garner, B. Isoform-specific proteolysis of apolipoprotein-E in the brain. Neurobiol. Aging 2011, 32, 257–271. [Google Scholar] [CrossRef]
- Williams II, B.; Convertino, M.; Das, J.; Dokholyan, N.V. ApoE4-specific misfolded intermediate identified by molecular dynamics simulations. PLoS Comput. Biol. 2015, 11, e1004359. [Google Scholar] [CrossRef]
- Love, J.E.; Day, R.J.; Gause, J.W.; Brown, R.J.; Pu, X.; Theis, D.I.; Caraway, C.A.; Poon, W.W.; Rahman, A.A.; Morrison, B.E.; et al. Nuclear uptake of an amino-terminal fragment of apolipoprotein E4 promotes cell death and localizes within microglia of the Alzheimer’s disease brain. Int. J. Physiol. Pathophysiol. Pharmacol. 2017, 9, 40–57. [Google Scholar] [PubMed]
- Yeh, Y.-Q.; Liao, K.-F.; Shih, O.; Shiu, Y.-J.; Wu, W.-R.; Su, C.-J.; Lin, P.-C.; Jeng, U.-S. Probing the acid-induced packing structure changes of the molten globule domains of a protein near equilibrium unfolding. J. Phys. Chem. Lett. 2017, 8, 470–477. [Google Scholar] [CrossRef]
- Fisher, C.A.; Narayanaswami, V.; Ryan, R.O. The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E. J. Biol. Chem. 2000, 275, 33601–33606. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswami, V.; Maiorano, J.N.; Dhanasekaran, P.; Ryan, R.O.; Phillips, M.C.; Lund-Katz, S.; Davidson, W.S. Helix orientation of the functional domains in apolipoprotein E in discoidal high density lipoprotein particles. J. Biol. Chem. 2004, 279, 14273–14279. [Google Scholar] [CrossRef]
- Newhouse, Y.; Peters-Libeu, C.; Weisgraber, K.H. Crystallization and preliminary X-ray diffraction analysis of apolipoprotein E-containing lipoprotein particles. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 981–984. [Google Scholar] [CrossRef]
- Drury, J.; Narayanaswami, V. Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence. J. Biol. Chem. 2005, 280, 14605–14610. [Google Scholar] [CrossRef]
- Krul, E.S.; Tikkanen, M.J.; Schonfeld, G. Heterogeneity of apolipoprotein E epitope expression on human lipoproteins: Importance for apolipoprotein E function. J. Lipid Res. 1988, 29, 1309–1325. [Google Scholar]
- Saito, H.; Dhanasekaran, P.; Baldwin, F.; Weisgraber, K.H.; Lund-Katz, S.; Phillips, M.C. Lipid binding-induced conformational change in human apolipoprotein E evidence for two lipid-bound states on spherical particles. J. Biol. Chem. 2001, 276, 40949–40954. [Google Scholar] [CrossRef] [PubMed]
- Raussens, V.; Drury, J.; Forte, T.M.; Choy, N.; Goormaghtigh, E.; Ruysschaert, J.-M.; Narayanaswami, V. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. Biochem. J. 2005, 387, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Frieden, C.; Wang, H.; Ho, C.M.W. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain–domain interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 6292–6297. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, F.; Wang, N.; Chen, W.; Jiang, X.-C.; Tall, A.R. HDL from CETP-deficient subjects shows enhanced ability to promote cholesterol efflux from macrophages in an apoE- and ABCG1-dependent pathway. J. Clin. Investig. 2006, 116, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Huang, Y.; Weisgraber, K.H. Putting cholesterol in its place: apoE and reverse cholesterol transport. J. Clin. Investig. 2006, 116, 1226–1229. [Google Scholar] [CrossRef]
- Ang, L.S.; Cruz, R.P.; Hendel, A.; Granville, D.J. Apolipoprotein E, an important player in longevity and age-related diseases. Exp. Gerontol. 2008, 43, 615–622. [Google Scholar] [CrossRef]
- Ilaria, Z.; Matteo, P.; Francesco, P.; Grazia, S.; Monica, G.; Laura, C.; Franco, B. Macrophage, but not systemic, apolipoprotein E is necessary for macrophage reverse cholesterol transport in vivo. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 74–80. [Google Scholar]
- Spinney, L. Alzheimer’s disease: The forgetting gene. Nat. News 2014, 510, 26. [Google Scholar] [CrossRef]
- Raber, J.; Huang, Y.; Ashford, J.W. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol. Aging 2004, 25, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.; George-Hyslop, P.H.; Pericak-Vance, M.A.; Joo, S.H.; Rosi, B.L.; Gusella, J.F.; Crapper-MacLachlan, D.R.; Alberts, M.J. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 1993, 43, 1467–1472. [Google Scholar] [CrossRef]
- Hyman, B.T.; Gomez-Isla, T.; West, H.; Briggs, M.; Chung, H.; Growdon, J.H.; Rebeck, G.W. Clinical and neuropathological correlates of apolipoprotein E genotype in Alzheimer’s disease. Window on molecular epidemiology. Ann. N. Y. Acad. Sci. 1996, 777, 158–165. [Google Scholar] [CrossRef]
- van Duijn, C.M.; de Knijff, P.; Cruts, M.; Wehnert, A.; Havekes, L.M.; Hofman, A.; Broeckhoven, C.V. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer’s disease. Nat. Genet. 1994, 7, 74. [Google Scholar] [CrossRef]
- Piedrahita, J.A.; Zhang, S.H.; Hagaman, J.R.; Oliver, P.M.; Maeda, N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc. Natl. Acad. Sci. USA 1992, 89, 4471–4475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.H.; Reddick, R.L.; Piedrahita, J.A.; Maeda, N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 1992, 258, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Hayek, T.; Oiknine, J.; Brook, J.G.; Aviram, M. Increased plasma and lipoprotein lipid peroxidation in apo E-deficient mice. Biochem. Biophys. Res. Commun. 1994, 201, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Roselaar, S.E.; Daugherty, A. Apolipoprotein E-deficient mice have impaired innate immune responses to Listeria monocytogenes in vivo. J. Lipid Res. 1998, 39, 1740–1743. [Google Scholar] [PubMed]
- de Bont, N.; Netea, M.G.; Demacker, P.N.M.; Verschueren, I.; Kullberg, B.J.; van Dijk, K.W.; van der Meer, J.W.M.; Stalenhoef, A.F.H. Apolipoprotein E knock-out mice are highly susceptible to endotoxemia and Klebsiella pneumoniae infection. J. Lipid Res. 1999, 40, 680–685. [Google Scholar] [CrossRef]
- Robertson, T.A.; Dutton, N.S.; Martins, R.N.; Taddei, K.; Papadimitriou, J.M. Comparison of astrocytic and myocytic metabolic dysregulation in apolipoprotein E deficient and human apolipoprotein E transgenic mice. Neuroscience 2000, 98, 353–359. [Google Scholar] [CrossRef]
- Moghadasian, M.H.; Mcmanus, B.M.; Nguyen, L.B.; Shefer, S.; Nadji, M.; Godin, D.V.; Green, T.J.; Hill, J.; Yang, Y.; Scudamore, C.H.; et al. Pathophysiology of apolipoprotein E deficiency in mice: Relevance to apo E-related disorders in humans. FASEB J. 2001, 15, 2623–2630. [Google Scholar] [CrossRef] [PubMed]
- Kulminski, A.M.; Loika, Y.; Culminskaya, I.; Huang, J.; Arbeev, K.G.; Bagley, O.; Feitosa, M.F.; Zmuda, J.M.; Christensen, K.; Yashin, A.I. Independent associations of TOMM40 and APOE variants with body mass index. Aging Cell 2019, 18, e12869. [Google Scholar] [CrossRef]
- Harman, D. Aging: Overview. Ann. N. Y. Acad. Sci. 2001, 928, 1–21. [Google Scholar] [CrossRef]
- Vasto, S.; Candore, G.; Balistreri, C.R.; Caruso, M.; Colonna-Romano, G.; Grimaldi, M.P.; Listi, F.; Nuzzo, D.; Lio, D.; Caruso, C. Inflammatory networks in ageing, age-related diseases and longevity. Mech. Ageing Dev. 2007, 128, 83–91. [Google Scholar] [CrossRef]
- Kregel, K.C.; Zhang, H.J. An integrated view of oxidative stress in aging: Basic mechanisms, functional effects, and pathological considerations. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2007, 292, R18–R36. [Google Scholar] [CrossRef]
- Broer, L.; Buchman, A.S.; Deelen, J.; Evans, D.S.; Faul, J.D.; Lunetta, K.L.; Sebastiani, P.; Smith, J.A.; Smith, A.V.; Tanaka, T.; et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J. Gerontol. A. Biol. Sci. Med. Sci. 2015, 70, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Iurescia, S.; Fioretti, D.; Mangialasche, F.; Rinaldi, M. The pathological cross talk between apolipoprotein E and amyloid-β peptide in Alzheimer’s disease: Emerging gene-based therapeutic approaches. J. Alzheimers Dis. 2010, 21, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Dafnis, I.; Stratikos, E.; Tzinia, A.; Tsilibary, E.C.; Zannis, V.I.; Chroni, A. An apolipoprotein E4 fragment can promote intracellular accumulation of amyloid peptide β42. J. Neurochem. 2010, 115, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Argyri, L.; Dafnis, I.; Theodossiou, T.A.; Gantz, D.; Stratikos, E.; Chroni, A. Molecular basis for increased risk for late-onset Alzheimer disease due to the naturally occurring L28P mutation in apolipoprotein E4. J. Biol. Chem. 2014, 289, 12931–12945. [Google Scholar] [CrossRef]
- Dafnis, I.; Argyri, L.; Sagnou, M.; Tzinia, A.; Tsilibary, E.C.; Stratikos, E.; Chroni, A. The ability of apolipoprotein E fragments to promote intraneuronal accumulation of amyloid β peptide 42 is both isoform and size-specific. Sci. Rep. 2016, 6, 30654. [Google Scholar] [CrossRef]
- Ji, Z.-S.; Miranda, R.D.; Newhouse, Y.M.; Weisgraber, K.H.; Huang, Y.; Mahley, R.W. Apolipoprotein E4 potentiates amyloid β peptide-induced lysosomal leakage and apoptosis in neuronal cells. J. Biol. Chem. 2002, 277, 21821–21828. [Google Scholar] [CrossRef]
- Ji, Z.-S.; Müllendorff, K.; Cheng, I.H.; Miranda, R.D.; Huang, Y.; Mahley, R.W. Reactivity of apolipoprotein E4 and amyloid β peptide lysosomal stability and neurodegeneration. J. Biol. Chem. 2006, 281, 2683–2692. [Google Scholar] [CrossRef]
- Buttini, M.; Orth, M.; Bellosta, S.; Akeefe, H.; Pitas, R.E.; Wyss-Coray, T.; Mucke, L.; Mahley, R.W. Expression of human apolipoprotein E3 or E4 in the brains ofApoE−/− mice: Isoform-specific effects on neurodegeneration. J. Neurosci. 1999, 19, 4867–4880. [Google Scholar] [CrossRef] [PubMed]
- Hartman, R.E.; Wozniak, D.F.; Nardi, A.; Olney, J.W.; Sartorius, L.; Holtzman, D.M. Behavioral phenotyping of GFAP-ApoE3 and -ApoE4 transgenic mice: ApoE4 mice show profound working memory impairments in the absence of Alzheimer’s-like neuropathology. Exp. Neurol. 2001, 170, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Bour, A.; Grootendorst, J.; Vogel, E.; Kelche, C.; Dodart, J.-C.; Bales, K.; Moreau, P.-H.; Sullivan, P.M.; Mathis, C. Middle-aged human apoE4 targeted-replacement mice show retention deficits on a wide range of spatial memory tasks. Behav. Brain Res. 2008, 193, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Raber, J.; Wong, D.; Buttini, M.; Orth, M.; Bellosta, S.; Pitas, R.E.; Mahley, R.W.; Mucke, L. Isoform-specific effects of human apolipoprotein E on brain function revealed in ApoE knockout mice: Increased susceptibility of females. Proc. Natl. Acad. Sci. USA 1998, 95, 10914–10919. [Google Scholar] [CrossRef]
- Nathan, B.P.; Bellosta, S.; Sanan, D.A.; Weisgraber, K.H.; Mahley, R.W.; Pitas, R.E. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science 1994, 264, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Nathan, B.P.; Chang, K.-C.; Bellosta, S.; Brisch, E.; Ge, N.; Mahley, R.W.; Pitas, R.E. The inhibitory effect of apolipoprotein E4 on neurite outgrowth is associated with microtubule depolymerization. J. Biol. Chem. 1995, 270, 19791–19799. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, D.M.; Pitas, R.E.; Kilbridge, J.; Nathan, B.; Mahley, R.W.; Bu, G.; Schwartz, A.L. Low density lipoprotein receptor-related protein mediates apolipoprotein E-dependent neurite outgrowth in a central nervous system-derived neuronal cell line. Proc. Natl. Acad. Sci. USA 1995, 92, 9480–9484. [Google Scholar] [CrossRef]
- Li, G.; Bien-Ly, N.; Andrews-Zwilling, Y.; Xu, Q.; Bernardo, A.; Ring, K.; Halabisky, B.; Deng, C.; Mahley, R.W.; Huang, Y. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell 2009, 5, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Reiman, E.M.; Chen, K.; Alexander, G.E.; Caselli, R.J.; Bandy, D.; Osborne, D.; Saunders, A.M.; Hardy, J. Correlations between apolipoprotein E ε4 gene dose and brain-imaging measurements of regional hypometabolism. Proc. Natl. Acad. Sci. USA 2005, 102, 8299–8302. [Google Scholar] [CrossRef]
- Chang, S.; Ran Ma, T.; Miranda, R.D.; Balestra, M.E.; Mahley, R.W.; Huang, Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl. Acad. Sci. USA 2005, 102, 18694–18699. [Google Scholar] [CrossRef]
- Scarmeas, N.; Habeck, C.; Hilton, J.; Anderson, K.; Flynn, J.; Park, A.; Stern, Y. APOE related alterations in cerebral activation even at college age. J. Neurol. Neurosurg. Psychiatry 2005, 76, 1440–1444. [Google Scholar] [CrossRef]
- Nakamura, T.; Watanabe, A.; Fujino, T.; Hosono, T.; Michikawa, M. Apolipoprotein E4 (1–272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells. Mol. Neurodegener. 2009, 4, 35. [Google Scholar] [CrossRef]
- Tanaka, M.; Vedhachalam, C.; Sakamoto, T.; Dhanasekaran, P.; Phillips, M.C.; Lund-Katz, S.; Saito, H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry 2006, 45, 4240–4247. [Google Scholar] [CrossRef]
- Chou, C.-Y.; Jen, W.-P.; Hsieh, Y.-H.; Shiao, M.-S.; Chang, G.-G. Structural and functional variations in human apolipoprotein E3 and E4. J. Biol. Chem. 2006, 281, 13333–13344. [Google Scholar] [CrossRef]
- Tambini, M.D.; Pera, M.; Kanter, E.; Yang, H.; Guardia-Laguarta, C.; Holtzman, D.; Sulzer, D.; Area-Gomez, E.; Schon, E.A. ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep. 2016, 17, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bravo, R.; Gutierrez, T.; Paredes, F.; Gatica, D.; Rodriguez, A.E.; Pedrozo, Z.; Chiong, M.; Parra, V.; Quest, A.F.G.; Rothermel, B.A.; et al. Endoplasmic reticulum: ER stress regulates mitochondrial bioenergetics. Int. J. Biochem. Cell Biol. 2012, 44, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.S.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 2014, 21, 396–413. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; Lefebvre d’Hellencourt, C.; Ravanan, P. A Molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell. Neurosci. 2014, 8. [Google Scholar] [CrossRef]
- Miyata, M.; Smith, J.D. Apolipoprotein E allele–specific antioxidant activity and effects on cytotoxicity by oxidative insults and β–amyloid peptides. Nat. Genet. 1996, 14, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Jofre-Monseny, L.; Loboda, A.; Wagner, A.E.; Huebbe, P.; Boesch-Saadatmandi, C.; Jozkowicz, A.; Minihane, A.-M.; Dulak, J.; Rimbach, G. Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem. Biophys. Res. Commun. 2007, 357, 319–324. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Herpes and Alzheimer’s disease: Subversion in the central nervous system and how it might be halted. J. Alzheimers Dis. JAD 2016, 54, 1273–1281. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Herpes simplex virus type 1 and Alzheimer’s disease: Possible mechanisms and signposts. FASEB J. 2017, 31, 3216–3226. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, R.F. Corroboration of a major role for Herpes simplex virus type 1 in Alzheimer’s disease. Front. Aging Neurosci. 2018, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Cun, W.; Jiang, J.; Luo, G. The C-terminal α-helix domain of apolipoprotein E Is required for interaction with nonstructural protein 5A and assembly of Hepatitis C virus. J. Virol. 2010, 84, 11532–11541. [Google Scholar] [CrossRef] [PubMed]
- Chiba-Falek, O.; Linnertz, C.; Guyton, J.; Gardner, S.D.; Roses, A.D.; McCarthy, J.J.; Patel, K. Pleiotropy and allelic heterogeneity in the TOMM40-APOE genomic region related to clinical and metabolic features of hepatitis C infection. Hum. Genet. 2012, 131, 1911–1920. [Google Scholar] [CrossRef]
- Bankwitz, D.; Doepke, M.; Hueging, K.; Weller, R.; Bruening, J.; Behrendt, P.; Lee, J.-Y.; Vondran, F.W.R.; Manns, M.P.; Bartenschlager, R.; et al. Maturation of secreted HCV particles by incorporation of secreted ApoE protects from antibodies by enhancing infectivity. J. Hepatol. 2017, 67, 480–489. [Google Scholar] [CrossRef]
- Weller, R.; Hueging, K.; Brown, R.J.P.; Todt, D.; Joecks, S.; Vondran, F.W.R.; Pietschmann, T. Hepatitis C virus strain-dependent usage of apolipoprotein E modulates assembly efficiency and specific infectivity of secreted virions. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Gondar, V.; Molina-Jiménez, F.; Hishiki, T.; García-Buey, L.; Koutsoudakis, G.; Shimotohno, K.; Benedicto, I.; Majano, P.L. Apolipoprotein E, but not apolipoprotein B, Is essential for efficient cell-to-cell transmission of Hepatitis C virus. J. Virol. 2015, 89, 9962–9973. [Google Scholar] [CrossRef]
- Popescu, C.-I.; Dubuisson, J. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol. Cell 2010, 102, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Andres, M.; Sadino, J.; Jiang, C.S.; Nakama, H.; Miller, E.; Ernst, T. Impact of apolipoprotein E ε4 and HIV on cognition and brain atrophy: antagonistic pleiotropy and premature brain aging. NeuroImage 2011, 58, 1017–1027. [Google Scholar] [CrossRef]
- Chang, L.; Jiang, C.; Cunningham, E.; Buchthal, S.; Douet, V.; Andres, M.; Ernst, T. Effects of APOE ε4, age, and HIV on glial metabolites and cognitive deficits. Neurology 2014, 82, 2213–2222. [Google Scholar] [CrossRef]
- Wendelken, L.A.; Jahanshad, N.; Rosen, H.J.; Busovaca, E.; Allen, I.; Coppola, G.; Adams, C.; Rankin, K.P.; Milanini, B.; Clifford, K.; et al. ApoE ε4 is associated with cognition, brain integrity and atrophy in HIV over age 60. J. Acquir. Immune Defic. Syndr. 1999 2016, 73, 426–432. [Google Scholar] [CrossRef]
- Suwalak, T.; Srisawasdi, P.; Puangpetch, A.; Santon, S.; Koomdee, N.; Chamnanphon, M.; Charoenyingwattana, A.; Chantratita, W.; Sukasem, C. Polymorphisms of the ApoE (Apolipoprotein E) gene and their influence on dyslipidemia in hiv-1-infected individuals. Jpn. J. Infect. Dis. 2015, 68, 5–12. [Google Scholar] [CrossRef]
- Cooley, S.A.; Paul, R.H.; Fennema-Notestine, C.; Morgan, E.E.; Vaida, F.; Deng, Q.; Chen, J.A.; Letendre, S.; Ellis, R.; Clifford, D.B.; et al. Apolipoprotein E ε4 genotype status is not associated with neuroimaging outcomes in a large cohort of HIV+ individuals. J. Neurovirol. 2016, 22, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Geffin, R.; McCarthy, M. Aging and apolipoprotein E in HIV infection. J. Neurovirol. 2018, 24, 529–548. [Google Scholar] [CrossRef] [PubMed]
- Stengard, J.H.; Weiss, K.M.; Sing, C.F. An ecological study of association between coronary heart disease mortality rates in men and the relative frequencies of common allelic variations in the gene coding for apolipoprotein E. Hum. Genet. 1998, 103, 234–241. [Google Scholar] [CrossRef]
- Gerdes, L.U.; Gerdes, C.; Kervinen, K.; Savolainen, M.; Klausen, I.C.; Hansen, P.S.; Kesäniemi, Y.A.; Faergeman, O. The apolipoprotein ε4 allele determines prognosis and the effect on prognosis of simvastatin in survivors of myocardial infarction: A substudy of the Scandinavian simvastatin survival study. Circulation 2000, 101, 1366–1371. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.T.; Liestøl, K.; Løberg, E.M.; Reims, H.M.; Brorson, S.-H.; Mæhlen, J. The apolipoprotein E polymorphism and cardiovascular diseases—An autopsy study. Cardiovasc. Pathol. 2012, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Satizabal Claudia, L.; Samieri, C.; Davis-Plourde, K.L.; Voetsch, B.; Aparicio, H.J.; Pase, M.P.; Romero, J.R.; Helmer, C.; Vasan, R.S.; Kase, C.S.; et al. APOE and the association of fatty acids with the risk of stroke, coronary heart disease, and mortality. Stroke 2018, 49, 2822–2829. [Google Scholar] [CrossRef] [PubMed]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.-E. The APOE gene is differentially methylated in Alzheimer’s disease. J. Alzheimers Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, W.; Ware, E.B.; Turner, S.T.; Mosley, T.H.; Smith, J.A. DNA methylation in the APOE genomic region is associated with cognitive function in African Americans. BMC Med. Genomics 2018, 11. [Google Scholar] [CrossRef]
- Giuliani, C.; Sazzini, M.; Pirazzini, C.; Bacalini, M.G.; Marasco, E.; Ruscone, G.A.G.; Fang, F.; Sarno, S.; Gentilini, D.; Di Blasio, A.M.; et al. Impact of demography and population dynamics on the genetic architecture of human longevity. Aging 2018, 10, 1947–1963. [Google Scholar] [CrossRef]
- Sebastiani, P.; Solovieff, N.; DeWan, A.T.; Walsh, K.M.; Puca, A.; Hartley, S.W.; Melista, E.; Andersen, S.; Dworkis, D.A.; Wilk, J.B.; et al. Genetic Signatures of exceptional longevity in humans. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Revelas, M.; Thalamuthu, A.; Oldmeadow, C.; Evans, T.-J.; Armstrong, N.J.; Kwok, J.B.; Brodaty, H.; Schofield, P.R.; Scott, R.J.; Sachdev, P.S.; et al. Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech. Ageing Dev. 2018, 175, 24–34. [Google Scholar] [CrossRef]
- Pilling, L.C.; Atkins, J.L.; Bowman, K.; Jones, S.E.; Tyrrell, J.; Beaumont, R.N.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; Wood, A.R.; et al. Human longevity is influenced by many genetic variants: Evidence from 75,000 UK Biobank participants. Aging 2016, 8, 547–560. [Google Scholar] [CrossRef]
- Pilling, L.C.; Kuo, C.-L.; Sicinski, K.; Tamosauskaite, J.; Kuchel, G.A.; Harries, L.W.; Herd, P.; Wallace, R.; Ferrucci, L.; Melzer, D. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging 2017, 9, 2504–2520. [Google Scholar] [CrossRef]
- Newman, A.B.; Walter, S.; Lunetta, K.L.; Garcia, M.E.; Slagboom, P.E.; Christensen, K.; Arnold, A.M.; Aspelund, T.; Aulchenko, Y.S.; Benjamin, E.J.; et al. A meta-analysis of four genome-wide association studies of survival to age 90 Years or older: The cohorts for heart and aging research in Genomic Epidemiology Consortium. J. Gerontol. A Biol. Sci. Med. Sci. 2010, 65A, 478–487. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, G.; Franceschi, C. The unusual genetics of human longevity. Sci. Aging Knowl. Environ. 2006, 2006, pe20. [Google Scholar] [CrossRef]
- Zeng, Y.; Nie, C.; Min, J.; Liu, X.; Li, M.; Chen, H.; Xu, H.; Wang, M.; Ni, T.; Li, Y.; et al. Novel loci and pathways significantly associated with longevity. Sci. Rep. 2016, 6, 21243. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Garagnani, P.; Franceschi, C. Genetics of Human longevity within an eco-evolutionary nature-nurture framework. Circ. Res. 2018, 123, 745–772. [Google Scholar] [CrossRef] [PubMed]
- Fuku, N.; Díaz-Peña, R.; Arai, Y.; Abe, Y.; Zempo, H.; Naito, H.; Murakami, H.; Miyachi, M.; Spuch, C.; Serra-Rexach, J.A.; et al. Epistasis, physical capacity-related genes and exceptional longevity: FNDC5 gene interactions with candidate genes FOXOA3 and APOE. BMC Genomics 2017, 18, 803. [Google Scholar] [CrossRef]
- Fortney, K.; Dobriban, E.; Garagnani, P.; Pirazzini, C.; Monti, D.; Mari, D.; Atzmon, G.; Barzilai, N.; Franceschi, C.; Owen, A.B.; et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, L.; Zou, Y.; Tang, J.; Cai, J.; Wei, Y.; Qin, J.; Zhang, Z. Positive association of familial longevity with the moderate-high HDL-C concentration in Bama aging Study. Aging 2018, 10, 3528–3540. [Google Scholar] [CrossRef]
- Silva-Sena, G.G.; Camporez, D.; dos Santos, L.R.; da Silva, A.S.; Sagrillo Pimassoni, L.H.; Tieppo, A.; do Pimentel Batitucci, M.C.; Morelato, R.L.; de Paula, F. An association study of FOXO3 variant and longevity. Genet. Mol. Biol. 2018, 41, 386–396. [Google Scholar] [CrossRef]
- Garatachea, N.; Emanuele, E.; Calero, M.; Fuku, N.; Arai, Y.; Abe, Y.; Murakami, H.; Miyachi, M.; Yvert, T.; Verde, Z.; et al. ApoE gene and exceptional longevity: Insights from three independent cohorts. Exp. Gerontol. 2014, 53, 16–23. [Google Scholar] [CrossRef]
- Ryu, S.; Atzmon, G.; Barzilai, N.; Raghavachari, N.; Suh, Y. Genetic landscape of APOE in human longevity revealed by high-throughput sequencing. Mech. Ageing Dev. 2016, 155, 7–9. [Google Scholar] [CrossRef]
- Louhija, J.; Miettinen, H.E.; Kontula, K.; Tikkanen, M.J.; Miettinen, T.A.; Tilvis, R.S. Aging and genetic variation of plasma apolipoproteins. Relative loss of the apolipoprotein E4 phenotype in centenarians. Arterioscler. Thromb. J. Vasc. Biol. 1994, 14, 1084–1089. [Google Scholar] [CrossRef]
- Schächter, F.; Faure-Delanef, L.; Guénot, F.; Rouger, H.; Froguel, P.; Lesueur-Ginot, L.; Cohen, D. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 1994, 6, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Asada, T.; Kariya, T.; Yamagata, Z.; Kinoshita, T.; Asaka, A. Apolipoprotein E allele in centenarians. Neurology 1996, 46, 1484. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Z.; Asada, T.; Kinoshita, A.; Zhang, Y.; Asaka, A. Distribution of apolipoprotein E gene polymorphisms in Japanese patients with Alzheimer’s disease and in Japanese centenarians. Hum. Hered. 1997, 47, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Ogburn, C.E.; Hunt, K.E.; Tilvis, R.; Louhija, J.; Penttinen, R.; Erkkola, R.; Panduro, A.; Riestra, R.; Piussan, C.; et al. Polymorphisms at the Werner locus: I. Newly identified polymorphisms, ethnic variability of 1367Cy/Arg, and its stability in a population of Finnish centenarians. Am. J. Med. Genet. 1999, 82, 399–403. [Google Scholar] [CrossRef]
- Gerdes, L.U.; Jeune, B.; Ranberg, K.A.; Nybo, H.; Vaupel, J.W. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: Apolipoprotein E gene is a “frailty gene,” not a “longevity gene. ” Genet. Epidemiol. 2000, 19, 202–210. [Google Scholar] [CrossRef]
- Blanché, H.; Cabanne, L.; Sahbatou, M.; Thomas, G. A study of French centenarians: Are ACE and APOE associated with longevity? Comptes Rendus Académie Sci. Ser. III Sci. Vie 2001, 324, 129–135. [Google Scholar] [CrossRef]
- Arai, Y.; Hirose, N.; Nakazawa, S.; Yamamura, K.; Shimizu, K.; Takayama, M.; Ebihara, Y.; Osono, Y.; Homma, S. Lipoprotein metabolism in Japanese centenarians: Effects of apolipoprotein E polymorphism and nutritional status. J. Am. Geriatr. Soc. 2001, 49, 1434–1441. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-H.; Kim, J.-H.; Kim, D.K.; Kim, J.-W.; Kim, D.-K.; Lee, M.S.; Kim, C.H.; Park, S.C. Distributions of ACE and APOE polymorphisms and their relations with dementia status in Korean centenarians. J. Gerontol. Ser. A 2003, 58, M227–M231. [Google Scholar] [CrossRef]
- Panza, F.; Solfrizzi, V.; Colacicco, A.M.; Basile, A.M.; D’Introno, A.; Capurso, C.; Sabba, M.; Capurso, S.; Capurso, A. Apolipoprotein E (APOE) polymorphism influences serum APOE levels in Alzheimer’s disease patients and centenarians. NeuroReport 2003, 14, 605. [Google Scholar] [CrossRef]
- Capurso, C.; Solfrizzi, V.; D’Introno, A.; Colacicco, A.M.; Capurso, S.A.; Semeraro, C.; Capurso, A.; Panza, F. Interleukin 6−174 G/C promoter gene polymorphism in centenarians: No evidence of association with human longevity or interaction with apolipoprotein E alleles. Exp. Gerontol. 2004, 39, 1109–1114. [Google Scholar] [CrossRef]
- Garatachea, N.; Marín, P.J.; Santos-Lozano, A.; Sanchis-Gomar, F.; Emanuele, E.; Lucia, A. The ApoE gene is related with exceptional longevity: A systematic review and meta-analysis. Rejuvenation Res. 2014, 18, 3–13. [Google Scholar] [CrossRef]
- Rea, I.M.; Mc Dowell, I.; McMaster, D.; Smye, M.; Stout, R.; Evans, A. Apolipoprotein E alleles in nonagenarian subjects in the Belfast Elderly Longitudinal Free-living Ageing Study (BELFAST). Mech. Ageing Dev. 2001, 122, 1367–1372. [Google Scholar] [CrossRef]
- Zubenko, G.S.; Stiffler, J.S.; Hughes, H.B.; Fatigati, M.J.; Zubenko, W.N. Genome survey for loci that influence successful aging: sample characterization, method validation, and initial results for the Y chromosome. Am. J. Geriatr. Psychiatry 2002, 10, 619–630. [Google Scholar] [CrossRef]
- Geesaman, B.J.; Benson, E.; Brewster, S.J.; Kunkel, L.M.; Blanché, H.; Thomas, G.; Perls, T.T.; Daly, M.J.; Puca, A.A. Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proc. Natl. Acad. Sci. USA 2003, 100, 14115–14120. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xiang, L.; Wan, G.; Qi, K.; Sun, L.; Huang, Z.; Zheng, C.; Lv, Z.; Hu, C.; Yang, Z. Is APOE ε3 a favourable factor for the longevity: An association study in Chinese population. J. Genet. 2011, 90, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, P.; Gurinovich, A.; Nygaard, M.; Sasaki, T.; Sweigart, B.; Bae, H.; Andersen, S.L.; Villa, F.; Atzmon, G.; Christensen, K.; et al. APOE alleles and extreme human longevity. J. Gerontol. Ser. A 2019, 74, 44–51. [Google Scholar] [CrossRef]
- Mostafavi, H.; Berisa, T.; Day, F.R.; Perry, J.R.B.; Przeworski, M.; Pickrell, J.K. Identifying genetic variants that affect viability in large cohorts. PLoS Biol. 2017, 15, e2002458. [Google Scholar] [CrossRef] [PubMed]
- Christensen, K.; Johnson, T.E.; Vaupel, J.W. The quest for genetic determinants of human longevity: Challenges and insights. Nat. Rev. Genet. 2006, 7, 436–448. [Google Scholar] [CrossRef]
- Huebbe, P.; Rimbach, G. Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res. Rev. 2017, 37, 146–161. [Google Scholar] [CrossRef]
- Luo, C.-C.; Li, W.-H.; Moore, M.N.; Chan, L. Structure and evolution of the apolipoprotein multigene family. J. Mol. Biol. 1986, 187, 325–340. [Google Scholar] [CrossRef]
- Smith, A.F.; Owen, L.M.; Strobel, L.M.; Chen, H.; Kanost, M.R.; Hanneman, E.; Wells, M.A. Exchangeable apolipoproteins of insects share a common structural motif. J. Lipid Res. 1994, 35, 1976–1984. [Google Scholar]
- Peterson, K.J.; Lyons, J.B.; Nowak, K.S.; Takacs, C.M.; Wargo, M.J.; McPeek, M.A. Estimating metazoan divergence times with a molecular clock. Proc. Natl. Acad. Sci. USA 2004, 101, 6536–6541. [Google Scholar] [CrossRef]
- Reich, D.; Green, R.E.; Kircher, M.; Krause, J.; Patterson, N.; Durand, E.Y.; Viola, B.; Briggs, A.W.; Stenzel, U.; Johnson, P.L.F.; et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 2010, 468, 1053–1060. [Google Scholar] [CrossRef]
- McIntosh, A.M.; Bennett, C.; Dickson, D.; Anestis, S.F.; Watts, D.P.; Webster, T.H.; Fontenot, M.B.; Bradley, B.J. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes). PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Finch, C.E.; Stanford, C.B. Meat-Adaptive genes and the evolution of slower aging in humans. Q. Rev. Biol. 2004, 79, 3–50. [Google Scholar] [CrossRef]
- Raichlen, D.A.; Alexander, G.E. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014, 37, 247–255. [Google Scholar] [CrossRef]
- Fullerton, S.M.; Clark, A.G.; Weiss, K.M.; Nickerson, D.A.; Taylor, S.L.; Stengård, J.H.; Salomaa, V.; Vartiainen, E.; Perola, M.; Boerwinkle, E.; et al. Apolipoprotein E variation at the sequence haplotype level: Implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000, 67, 881–900. [Google Scholar] [CrossRef]
- Antón, S.C.; Leonard, W.R.; Robertson, M.L. An ecomorphological model of the initial hominid dispersal from Africa. J. Hum. Evol. 2002, 43, 773–785. [Google Scholar] [CrossRef]
- Bramble, D.M.; Lieberman, D.E. Endurance running and the evolution of Homo. Nature 2004, 432, 345–352. [Google Scholar] [CrossRef]
- Malina, R.M.; Little, B.B. Physical activity: The present in the context of the past. Am. J. Hum. Biol. Off. J. Hum. Biol. Counc. 2008, 20, 373–391. [Google Scholar] [CrossRef]
- Caspari, R.; Lee, S.-H. Older age becomes common late in human evolution. Proc. Natl. Acad. Sci. USA 2004, 101, 10895–10900. [Google Scholar] [CrossRef]
- Hawkes, K.; O’Connell, J.F.; Jones, N.G.B.; Alvarez, H.; Charnov, E.L. Grandmothering, menopause, and the evolution of human life histories. Proc. Natl. Acad. Sci. USA 1998, 95, 1336–1339. [Google Scholar] [CrossRef]
- Hawkes, K. Genomic evidence for the evolution of human postmenopausal longevity. Proc. Natl. Acad. Sci. USA 2016, 113, 17–18. [Google Scholar] [CrossRef]
- Ojeda-Granados, C.; Panduro, A.; Gonzalez-Aldaco, K.; Sepulveda-Villegas, M.; Rivera-Iñiguez, I.; Roman, S. Tailoring nutritional advice for Mexicans based on prevalence profiles of diet-related adaptive gene polymorphisms. J. Pers. Med. 2017, 7, 16. [Google Scholar] [CrossRef]
- Singh, P.P.; Singh, M.; Mastana, S.S. APOE distribution in world populations with new data from India and the UK. Ann. Hum. Biol. 2006, 33, 279–308. [Google Scholar] [CrossRef]
- Hu, P.; Qin, Y.H.; Jing, C.X.; Lu, L.; Hu, B.; Du, P.F. Does the geographical gradient of ApoE4 allele exist in China? A systemic comparison among multiple Chinese populations. Mol. Biol. Rep. 2011, 38, 489–494. [Google Scholar] [CrossRef]
- Zekraoui, L.; Lagarde, J.P.; Raisonnier, A.; Gérard, N.; Aouizérate, A.; Lucotte, G. High frequency of the apolipoprotein E *4 allele in African pygmies and most of the African populations in sub-Saharan Africa. Hum. Biol. 1997, 69, 575–581. [Google Scholar]
- Corbo, R.M.; Scacchi, R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE * 4 a ‘thrifty’ allele? Ann. Hum. Genet. 1999, 63, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Sazzini, M.; Gnecchi Ruscone, G.A.; Giuliani, C.; Sarno, S.; Quagliariello, A.; De Fanti, S.; Boattini, A.; Gentilini, D.; Fiorito, G.; Catanoso, M.; et al. Complex interplay between neutral and adaptive evolution shaped differential genomic background and disease susceptibility along the Italian peninsula. Sci. Rep. 2016, 6, 32513. [Google Scholar] [CrossRef] [PubMed]
- Boattini, A.; Martinez-Cruz, B.; Sarno, S.; Harmant, C.; Useli, A.; Sanz, P.; Yang-Yao, D.; Manry, J.; Ciani, G.; Luiselli, D.; et al. Uniparental markers in Italy reveal a sex-biased genetic structure and different historical strata. PLoS ONE 2013, 8, e65441. [Google Scholar] [CrossRef]
- Ye, K.; Gao, F.; Wang, D.; Bar-Yosef, O.; Keinan, A. Dietary adaptation of FADS genes in Europe varied across time and geography. Nat. Ecol. Evol. 2017, 1, 167. [Google Scholar] [CrossRef]
- Buckley, M.T.; Racimo, F.; Allentoft, M.E.; Jensen, M.K.; Jonsson, A.; Huang, H.; Hormozdiari, F.; Sikora, M.; Marnetto, D.; Eskin, E.; et al. Selection in Europeans on fatty acid desaturases associated with dietary changes. Mol. Biol. Evol. 2017, 34, 1307–1318. [Google Scholar] [CrossRef]
- Egert, S.; Rimbach, G.; Huebbe, P. ApoE genotype: From geographic distribution to function and responsiveness to dietary factors. Proc. Nutr. Soc. 2012, 71, 410–424. [Google Scholar] [CrossRef]
- Graeser, A.-C.; Boesch-Saadatmandi, C.; Lippmann, J.; Wagner, A.E.; Huebbe, P.; Storm, N.; Höppner, W.; Wiswedel, I.; Gardemann, A.; Minihane, A.M.; et al. Nrf2-dependent gene expression is affected by the proatherogenic apoE4 genotype—studies in targeted gene replacement mice. J. Mol. Med. 2011, 89, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Huebbe, P.; Nebel, A.; Siegert, S.; Moehring, J.; Boesch-Saadatmandi, C.; Most, E.; Pallauf, J.; Egert, S.; Müller, M.J.; Schreiber, S.; et al. APOE ε4 is associated with higher vitamin D levels in targeted replacement mice and humans. FASEB J. 2011, 25, 3262–3270. [Google Scholar] [CrossRef]
- Azevedo, O.G.R.; Bolick, D.T.; Roche, J.K.; Pinkerton, R.F.; Lima, A.A.M.; Vitek, M.P.; Warren, C.A.; Oriá, R.B.; Guerrant, R.L. Apolipoprotein E Plays a key role against cryptosporidial infection in transgenic undernourished mice. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Huebbe, P.; Dose, J.; Schloesser, A.; Campbell, G.; Glüer, C.-C.; Gupta, Y.; Ibrahim, S.; Minihane, A.-M.; Baines, J.F.; Nebel, A.; et al. Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization—Studies in gene-targeted replacement mice. Mol. Nutr. Food Res. 2015, 59, 334–343. [Google Scholar] [CrossRef]
- Huebbe, P.; Lange, J.; Lietz, G.; Rimbach, G. Dietary β-carotene and lutein metabolism is modulated by the APOE genotype. BioFactors 2016, 42, 388–396. [Google Scholar] [CrossRef]
- Luca, F.; Perry, G.H.; Di Rienzo, A. Evolutionary adaptations to dietary changes. Annu. Rev. Nutr. 2010, 30, 291–314. [Google Scholar] [CrossRef] [PubMed]
- Finch, C.E.; Sapolsky, R.M. The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol. Aging 1999, 20, 407–428. [Google Scholar] [CrossRef]
- Schwarz, F.; Springer, S.A.; Altheide, T.K.; Varki, N.M.; Gagneux, P.; Varki, A. Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc. Natl. Acad. Sci. 2016, 113, 74–79. [Google Scholar] [CrossRef] [PubMed]
- van Exel, E.; Koopman, J.J.E.; van Bodegom, D.; Meij, J.J.; de Knijff, P.; Ziem, J.B.; Finch, C.E.; Westendorp, R.G.J. Effect of APOE ε4 allele on survival and fertility in an adverse environment. PLoS ONE 2017, 12, e0179497. [Google Scholar] [CrossRef]
- Asgari, N.; Akbari, M.T.; Zare, S.; Babamohammadi, G. Positive association of apolipoprotein E4 polymorphism with recurrent pregnancy loss in Iranian patients. J. Assist. Reprod. Genet. 2013, 30, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.; Goodman, C.S.; Hur, J.; Jeyendran, R.S.; Coulam, C. The Association of apoprotien E polymorphisms with recurrent pregnancy loss. Am. J. Reprod. Immunol. 2009, 61, 34–38. [Google Scholar] [CrossRef]
- Yenicesu, G.I.; Cetin, M.; Ozdemir, O.; Cetin, A.; Ozen, F.; Yenicesu, C.; Yildiz, C.; Kocak, N. A Prospective case–control study analyzes 12 thrombophilic gene mutations in Turkish couples with recurrent pregnancy loss. Am. J. Reprod. Immunol. 2010, 63, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Ozornek, H.; Ergin, E.; Jeyendran, R.S.; Ozay, A.T.; Pillai, D.; Coulam, C. Is apolipoprotien E codon 112 polymorphisms associated with recurrent pregnancy loss? Am. J. Reprod. Immunol. 2010, 64, 87–92. [Google Scholar] [CrossRef]
- Ergin, E.; Jeyendran, R.S.; Özörnek, H.; Alev, Ö.; Pillai, M.D.; Coulam, C. Apolipoprotein E codon 112 polymorphisms is associated with recurrent pregnancy loss. Fertil. Steril. 2009, 92, S115. [Google Scholar] [CrossRef]
- Vasunilashorn, S.; Finch, C.E.; Crimmins, E.M.; Vikman, S.A.; Stieglitz, J.; Gurven, M.; Kaplan, H.; Allayee, H. Inflammatory gene variants in the Tsimane, an indigenous Bolivian population with a high infectious load. Biodemography Soc. Biol. 2011, 57, 33–52. [Google Scholar] [CrossRef]
- Trumble, B.C.; Stieglitz, J.; Blackwell, A.D.; Allayee, H.; Beheim, B.; Finch, C.E.; Gurven, M.; Kaplan, H. Apolipoprotein E4 is associated with improved cognitive function in Amazonian forager-horticulturalists with a high parasite burden. FASEB J. 2017, 31, 1508–1515. [Google Scholar] [CrossRef]
- Tzourio, C.; Arima, H.; Harrap, S.; Anderson, C.; Godin, O.; Woodward, M.; Neal, B.; Bousser, M.-G.; Chalmers, J.; Cambien, F.; et al. APOE genotype, ethnicity, and the risk of cerebral hemorrhage. Neurology 2008, 70, 1322–1328. [Google Scholar] [CrossRef]
- Zhang, M.; Gu, W.; Qiao, S.; Zhu, E.; Zhao, Q.; Lv, S. Apolipoprotein E gene polymorphism and risk for coronary heart disease in the Chinese population: A meta-analysis of 61 studies including 6634 cases and 6393 controls. PLoS ONE 2014, 9, e9546. [Google Scholar] [CrossRef]
- Tang, M.-X.; Stern, Y.; Marder, K.; Bell, K.; Gurland, B.; Lantigua, R.; Andrews, H.; Feng, L.; Tycko, B.; Mayeux, R. The APOE-∊4 allele and the risk of Alzheimer disease among African Americans, Whites, and Hispanics. JAMA 1998, 279, 751–755. [Google Scholar] [CrossRef]
- Wright, R.O.; Hu, H.; Silverman, E.K.; Tsaih, S.W.; Schwartz, J.; Bellinger, D.; Palazuelos, E.; Weiss, S.T.; Hernandez-Avila, M. Apolipoprotein E genotype predicts 24-Month Bayley scales infant development score. Pediatr. Res. 2003, 54, 819–825. [Google Scholar] [CrossRef]
- Becher, J.-C.; Bell, J.E.; McIntosh, N.; Keeling, J.W. Distribution of apolipoprotein E alleles in a Scottish healthy newborn population. Neonatology 2005, 88, 164–167. [Google Scholar] [CrossRef]
- Becher, J.; Keeling, J.W.; McIntosh, N.; Wyatt, B.; Bell, J. The distribution of apolipoprotein E alleles in Scottish perinatal deaths. J. Med. Genet. 2006, 43, 414–418. [Google Scholar] [CrossRef]
- Becher, J.-C.; Keeling, J.W.; Bell, J.; Wyatt, B.; McIntosh, N. Apolipoprotein E e4 and its prevalence in early childhood death due to sudden infant death syndrome or to recognised causes. Early Hum. Dev. 2008, 84, 549–554. [Google Scholar] [CrossRef]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: A meta-analysis. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Bennet, A.M.; Angelantonio, E.D.; Ye, Z.; Wensley, F.; Dahlin, A.; Ahlbom, A.; Keavney, B.; Collins, R.; Wiman, B.; de Faire, U.; et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 2007, 298, 1300–1311. [Google Scholar] [CrossRef]
- Rougeron, V.; Woods, C.M.; Tiedje, K.E.; Bodeau-Livinec, F.; Migot-Nabias, F.; Deloron, P.; Luty, A.J.F.; Fowkes, F.J.I.; Day, K.P. Epistatic interactions between apolipoprotein E and Hemoglobin S genes in regulation of Malaria Parasitemia. PLoS ONE 2013, 8, e76924. [Google Scholar] [CrossRef] [PubMed]
- Kulminski, A.M.; Raghavachari, N.; Arbeev, K.G.; Culminskaya, I.; Arbeeva, L.; Wu, D.; Ukraintseva, S.V.; Christensen, K.; Yashin, A.I. Protective role of the apolipoprotein E2 allele in age-related disease traits and survival: Evidence from the long life family study. Biogerontology 2016, 17, 893–905. [Google Scholar] [CrossRef]
- Konishi, K.; Bhat, V.; Banner, H.; Poirier, J.; Joober, R.; Bohbot, V.D. APOE2 is associated with spatial navigational strategies and increased gray matter in the hippocampus. Front. Hum. Neurosci. 2016, 10. [Google Scholar] [CrossRef]
- Cashdan, E. Biogeography of human infectious diseases: A global historical analysis. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Guernier, V.; Hochberg, M.E.; Guégan, J.-F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004, 2, e141. [Google Scholar] [CrossRef]
- Barger, S.W.; Harmon, A.D. Microglial activation by Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature 1997, 388, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Bonacina, F.; Coe, D.; Wang, G.; Longhi, M.P.; Baragetti, A.; Moregola, A.; Garlaschelli, K.; Uboldi, P.; Pellegatta, F.; Grigore, L.; et al. Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat. Commun. 2018, 9, 3083. [Google Scholar] [CrossRef]
- Kelly, M.E.; Clay, M.A.; Mistry, M.J.; Hsieh-Li, H.-M.; Harmony, J.A.K. Apolipoprotein E inhibition of proliferation of mitogen-activated T lymphocytes: Production of interleukin 2 with reduced biological activity. Cell. Immunol. 1994, 159, 124–139. [Google Scholar] [CrossRef]
- Picardi, A.; Gentilucci, U.V.; Bambacioni, F.; Galati, G.; Spataro, S.; Mazzarelli, C.; D’Avola, D.; Fiori, E.; Riva, E. Lower schooling, higher hepatitis C virus prevalence in Italy: An association dependent on age. J. Clin. Virol. 2007, 40, 168–170. [Google Scholar] [CrossRef]
- Corbo, R.M.; Scacchi, R.; Mureddu, L.; Mulas, G.; Alfano, G. Apolipoprotein E polymorphism in Italy investigated in native plasma by a simple polyacrylamide gel isoelectric focusing technique. Comparison with frequency data of other European populations. Ann. Hum. Genet. 1995, 59, 197–209. [Google Scholar] [CrossRef]
- Kuhlmann, I.; Minihane, A.M.; Huebbe, P.; Nebel, A.; Rimbach, G. Apolipoprotein E genotype and hepatitis C, HIV and herpes simplex disease risk: A literature review. Lipids Health Dis. 2010, 9, 8. [Google Scholar] [CrossRef]
- Soares, H.D.; Potter, W.Z.; Pickering, E.; Kuhn, M.; Immermann, F.W.; Shera, D.M.; Ferm, M.; Dean, R.A.; Simon, A.J.; Swenson, F.; et al. Biomarkers associated with the apolipoprotein E genotype and Alzheimer disease. Arch. Neurol. 2012, 69, 1310–1317. [Google Scholar] [CrossRef]
- Gale, S.C.; Gao, L.; Mikacenic, C.; Coyle, S.M.; Rafaels, N.; Murray, T.; Madenspacher, J.H.; Draper, D.W.; Ge, W.; Aloor, J.J.; et al. APOε 4 is associated with enhanced in vivo innate immune responses in humans. J. Allergy Clin. Immunol. 2014, 134, 127–134. [Google Scholar] [CrossRef]
- Oosten, M.V.; Rensen, P.C.N.; Amersfoort, E.S.V.; Eck, M.V.; Dam, A.-M.V.; Brevé, J.J.P.; Vogel, T.; Panet, A.; Berkel, T.J.C.V.; Kuiper, J. Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality a new therapeutic approach to treat gram-negative sepsis. J. Biol. Chem. 2001, 276, 8820–8824. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Wang, R.; An, Y.; Gao, W.; Bai, L.; Li, Y.; Zhao, S.; Fan, J.; Liu, E. Western diet feeding influences gut microbiota profiles in apoE knockout mice. Lipids Health Dis. 2018, 17, 159. [Google Scholar] [CrossRef]
- Kasahara, K.; Tanoue, T.; Yamashita, T.; Yodoi, K.; Matsumoto, T.; Emoto, T.; Mizoguchi, T.; Hayashi, T.; Kitano, N.; Sasaki, N.; et al. Commensal bacteria at the crossroad between cholesterol homeostasis and chronic inflammation in atherosclerosis. J. Lipid Res. 2017, 58, 519–528. [Google Scholar] [CrossRef]
- Saita, D.; Ferrarese, R.; Foglieni, C.; Esposito, A.; Canu, T.; Perani, L.; Ceresola, E.R.; Visconti, L.; Burioni, R.; Clementi, M.; et al. Adaptive immunity against gut microbiota enhances apoE-mediated immune regulation and reduces atherosclerosis and western-diet-related inflammation. Sci. Rep. 2016, 6, 29353. [Google Scholar] [CrossRef]
- Schneeberger, M.; Everard, A.; Gómez-Valadés, A.G.; Matamoros, S.; Ramírez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef]
- Li, J.; Lin, S.; Vanhoutte, P.M.; Woo, C.W.; Xu, A. Akkermansia Muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in ApoE−/− mice. Circulation 2016, 133, 2434–2446. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, W.; Wang, J.; Shi, J.; Sun, Y.; Wang, W.; Ning, G.; Liu, R.; Hong, J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J. Mol. Endocrinol. 2017, 58, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yashin, A.I.; Wu, D.; Arbeev, K.G.; Ukraintseva, S.V. Polygenic effects of common single-nucleotide polymorphisms on life span: When association meets causality. Rejuvenation Res. 2012, 15, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Gnecchi-Ruscone, G.A.; Abondio, P.; De Fanti, S.; Sarno, S.; Sherpa, M.G.; Sherpa, P.T.; Marinelli, G.; Natali, L.; Di Marcello, M.; Peluzzi, D.; et al. Evidence of polygenic adaptation to high altitude from Tibetan and Sherpa genomes. Genome Biol. Evol. 2018, 10, 2919–2930. [Google Scholar] [CrossRef]
- McClellan, J.; King, M.-C. Genetic heterogeneity in human disease. Cell 2010, 141, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Bao, S.; Zhang, Z.; Zhou, X.; Wang, J.; Fan, Y.; Zhang, Y.; Li, Y.; Chen, L.; Jia, Y.; et al. A rare variant in MLKL confers susceptibility to ApoE ɛ4-negative Alzheimer’s disease in Hong Kong Chinese population. Neurobiol. Aging 2018, 68, 160.e1–160.e7. [Google Scholar] [CrossRef] [PubMed]
- Evolutionary Thinking in Medicine: From Research to Policy and Practice; Alvergne, A.; Jenkinson, C.; Faurie, C. (Eds.) Springer International Publishing: Basel, Switzerland, 2016. [Google Scholar]
- Allentoft, M.E.; Sikora, M.; Sjögren, K.-G.; Rasmussen, S.; Rasmussen, M.; Stenderup, J.; Damgaard, P.B.; Schroeder, H.; Ahlström, T.; Vinner, L.; et al. Population genomics of Bronze Age Eurasia. Nature 2015, 522, 167–172. [Google Scholar] [CrossRef]
- Olalde, I.; Brace, S.; Allentoft, M.E.; Armit, I.; Kristiansen, K.; Booth, T.; Rohland, N.; Mallick, S.; Szécsényi-Nagy, A.; Mittnik, A.; et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 2018, 555, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Olalde, I.; Schroeder, H.; Sandoval-Velasco, M.; Vinner, L.; Lobón, I.; Ramirez, O.; Civit, S.; García Borja, P.; Salazar-García, D.C.; Talamo, S.; et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol. Biol. Evol. 2015, 32, 3132–3142. [Google Scholar] [CrossRef]
- Olalde, I.; Allentoft, M.E.; Sánchez-Quinto, F.; Santpere, G.; Chiang, C.W.K.; DeGiorgio, M.; Prado-Martinez, J.; Rodríguez, J.A.; Rasmussen, S.; Quilez, J.; et al. Derived immune and ancestral pigmentation alleles in a 7000-year-old Mesolithic European. Nature 2014, 507, 225–228. [Google Scholar] [CrossRef]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Olalde, I.; Broomandkhoshbacht, N.; Candilio, F.; Cheronet, O.; et al. The genomic history of southeastern Europe. Nature 2018, 555, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.R.; Gonzalez-Fortes, G.; Connell, S.; Siska, V.; Eriksson, A.; Martiniano, R.; McLaughlin, R.L.; Gallego Llorente, M.; Cassidy, L.M.; Gamba, C.; et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 2015, 6, 8912. [Google Scholar] [CrossRef]
- Haak, W.; Lazaridis, I.; Patterson, N.; Rohland, N.; Mallick, S.; Llamas, B.; Brandt, G.; Nordenfelt, S.; Harney, E.; Stewardson, K.; et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 2015, 522, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Lipson, M.; Szécsényi-Nagy, A.; Mallick, S.; Pósa, A.; Stégmár, B.; Keerl, V.; Rohland, N.; Stewardson, K.; Ferry, M.; Michel, M.; et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 2017, 551, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, I.; Patterson, N.; Mittnik, A.; Renaud, G.; Mallick, S.; Kirsanow, K.; Sudmant, P.H.; Schraiber, J.G.; Castellano, S.; Lipson, M.; et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 2014, 513, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, I.; Mittnik, A.; Patterson, N.; Mallick, S.; Rohland, N.; Pfrengle, S.; Furtwängler, A.; Peltzer, A.; Posth, C.; Vasilakis, A.; et al. Genetic origins of the Minoans and Mycenaeans. Nature 2017, 548, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Schiffels, S.; Haak, W.; Paajanen, P.; Llamas, B.; Popescu, E.; Loe, L.; Clarke, R.; Lyons, A.; Mortimer, R.; Sayer, D.; et al. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 2016, 7, 10408. [Google Scholar] [CrossRef]
- Martiniano, R.; Caffell, A.; Holst, M.; Hunter-Mann, K.; Montgomery, J.; Müldner, G.; McLaughlin, R.L.; Teasdale, M.D.; van Rheenen, W.; Veldink, J.H.; et al. Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun. 2016, 7, 10326. [Google Scholar] [CrossRef]
- Broushaki, F.; Thomas, M.G.; Link, V.; López, S.; van Dorp, L.; Kirsanow, K.; Hofmanová, Z.; Diekmann, Y.; Cassidy, L.M.; Díez-del-Molino, D.; et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 2016, 353, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, L.M.; Martiniano, R.; Murphy, E.M.; Teasdale, M.D.; Mallory, J.; Hartwell, B.; Bradley, D.G. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. USA 2016, 113, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Skoglund, P.; Graf, K.E.; Metspalu, M.; Albrechtsen, A.; Moltke, I.; Rasmussen, S.; Stafford, T.W., Jr.; Orlando, L.; Metspalu, E.; et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 2014, 505, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, H.; Moorjani, P.; Jay, F.; Slepchenko, S.M.; Bondarev, A.A.; Johnson, P.L.F.; Aximu-Petri, A.; Prüfer, K.; de Filippo, C.; et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 2014, 514, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Valdiosera, C.; Malmström, H.; Ureña, I.; Rodriguez-Varela, R.; Sverrisdóttir, Ó.O.; Daskalaki, E.A.; Skoglund, P.; Naidoo, T.; Svensson, E.M.; et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. USA 2015, 112, 11917–11922. [Google Scholar] [CrossRef] [PubMed]
- Hofmanová, Z.; Kreutzer, S.; Hellenthal, G.; Sell, C.; Diekmann, Y.; Díez-del-Molino, D.; van Dorp, L.; López, S.; Kousathanas, A.; Link, V.; et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. USA 2016, 113, 6886–6891. [Google Scholar] [CrossRef] [PubMed]
- Kılınç, G.M.; Omrak, A.; Özer, F.; Günther, T.; Büyükkarakaya, A.M.; Bıçakçı, E.; Baird, D.; Dönertaş, H.M.; Ghalichi, A.; Yaka, R.; et al. The Demographic development of the First Farmers in Anatolia. Curr. Biol. 2016, 26, 2659–2666. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Graefen, A.; Ball, M.; Matzas, M.; Boisguerin, V.; Maixner, F.; Leidinger, P.; Backes, C.; Khairat, R.; Forster, M.; et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 2012, 3, 698. [Google Scholar] [CrossRef] [PubMed]
- Nesse, R.M. Evolution: Medicine’s most basic science. Lancet 2008, 372, S21–S27. [Google Scholar] [CrossRef]
- Nesse, R.M. Ten questions for evolutionary studies of disease vulnerability. Evol. Appl. 2011, 4, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Nesse, R.M.; Bergstrom, C.T.; Ellison, P.T.; Flier, J.S.; Gluckman, P.; Govindaraju, D.R.; Niethammer, D.; Omenn, G.S.; Perlman, R.L.; Schwartz, M.D.; et al. Making evolutionary biology a basic science for medicine. Proc. Natl. Acad. Sci. USA 2010, 107, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.C.K.; Nesse, R.M.; Sear, R.; Johnstone, R.A.; Stearns, S.C. Evolutionary public health: Introducing the concept. Lancet 2017, 390, 500–509. [Google Scholar] [CrossRef]
- Randolph, M.; Nesse, M.D.; George, C.W. Why We Get Sick. Available online: https://www.penguinrandomhouse.com/books/120768/why-we-get-sick-by-randolph-m-nesse/9780679746744 (accessed on 1 March 2019).

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abondio, P.; Sazzini, M.; Garagnani, P.; Boattini, A.; Monti, D.; Franceschi, C.; Luiselli, D.; Giuliani, C. The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity. Genes 2019, 10, 222. https://doi.org/10.3390/genes10030222
Abondio P, Sazzini M, Garagnani P, Boattini A, Monti D, Franceschi C, Luiselli D, Giuliani C. The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity. Genes. 2019; 10(3):222. https://doi.org/10.3390/genes10030222
Chicago/Turabian StyleAbondio, Paolo, Marco Sazzini, Paolo Garagnani, Alessio Boattini, Daniela Monti, Claudio Franceschi, Donata Luiselli, and Cristina Giuliani. 2019. "The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity" Genes 10, no. 3: 222. https://doi.org/10.3390/genes10030222
APA StyleAbondio, P., Sazzini, M., Garagnani, P., Boattini, A., Monti, D., Franceschi, C., Luiselli, D., & Giuliani, C. (2019). The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity. Genes, 10(3), 222. https://doi.org/10.3390/genes10030222




