Abstract
Transfer RNA (tRNA) is subject to a multitude of posttranscriptional modifications which can profoundly impact its functionality as the essential adaptor molecule in messenger RNA (mRNA) translation. Therefore, dynamic regulation of tRNA modification in response to environmental changes can tune the efficiency of gene expression in concert with the emerging epitranscriptomic mRNA regulators. Several of the tRNA modifications are required to prevent human diseases and are particularly important for proper development and generation of neurons. In addition to the positive role of different tRNA modifications in prevention of neurodegeneration, certain cancer types upregulate tRNA modification genes to sustain cancer cell gene expression and metastasis. Multiple associations of defects in genes encoding subunits of the tRNA modifier complex Elongator with human disease highlight the importance of proper anticodon wobble uridine modifications (xm5U34) for health. Elongator functionality requires communication with accessory proteins and dynamic phosphorylation, providing regulatory control of its function. Here, we summarized recent insights into molecular functions of the complex and the role of Elongator dependent tRNA modification in human disease.
1. Epitranscriptomic Transfer RNA Regulation
Apart from the four standard nucleosides, cellular RNA contains a broad variety of posttranscriptional modifications which may have significant impact on its function [1]. The total RNA modification set of a cell is termed the epitranscriptome, and its dynamic changes are thought to represent a strategy to modulate RNA function involving “writers” (modifiers), “erasers” (demodifiers) and “readers”, proteins specifically recognizing RNA modifications [2,3]. Among the different types of RNA subject to epitranscriptomic changes, transfer RNA (tRNA) harbors by far the most abundant and chemically diverse modifications (Figure 1). While initially thought to be constitutively modified, numerous examples are known by now demonstrating dynamic changes in tRNA modification patterns in response to changing environments or conditions [4,5,6,7,8,9,10,11,12,13]. One of these involves a writer methylase and an eraser demethylase, as observed in epitranscriptomic regulation of messenger RNA (mRNA) translation [10]. In this case, mammalian ALKBH1 was identified as a demethylase removing the methyl-group in 1-methyladenosine (m1A), which is found in initiator and elongator tRNAs and introduced by TRMT6 (substrate binding subunit) and TRMT61 (catalytic subunit) [10]. It was demonstrated that epitranscriptomic modulation of m1A presence in both tRNA types (initiator and elongator) regulates tRNA function in translation initiation and elongation in a dynamic manner in response to glucose availability [10]. Importantly, the absence of the eraser protein causes embryonic lethality or neural defects in a mouse model system, indicating a direct or indirect role of epitranscriptomic changes in mammalian tRNA for prevention of disease [10].
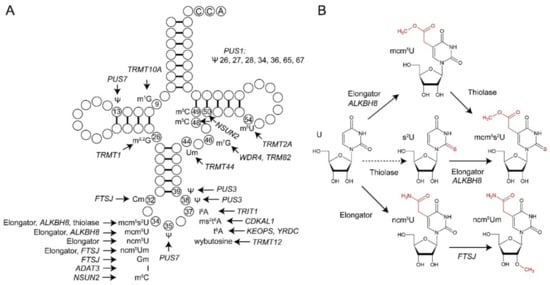
Figure 1.
Transfer RNA (tRNA) modifications associated with human disease. (A) Schematic representation of a cytoplasmic tRNA with disease linked modifications at indicated base positions. Modification genes linked to human diseases when mutated or upregulated are denoted (see Table 1 and references therein for details). (B) Overview of steps and genes involved in xm5U34 synthesis. A broken line between U34 and s2U34 indicates the fact that several lines of evidence support preferential action of the thiolase on mcm5U34 rather than unmodified U34. Elongator specifies different subunits of the Elongator complex, whereas thiolase represents the complex composed of subunits CTU1 and CTU2. Abbreviations for U34 modifications are according to the modomics database [14].
Despite the fact that m1A represents the only tRNA modification known so far to be actively removed by an eraser protein, a variety of additional modifications are altered in their relative abundance in response to changing environmental or physiological conditions. These triggers include elevated temperature, oxidative stress or availability of nutrients required for synthesis of modifications, e.g., sulfur, bicarbonate, queuine or taurine [4,5,6,9,11,13]. Due to the growing body of evidence for a dynamic, rather than static nature of tRNA modifications, additional examples for epitranscriptomic regulation of tRNA function should be envisioned.
2. Disease Related Transfer RNA Modifications
For tRNA to function as the adaptor molecule in translation, its folding into an l-shaped structure is essential [1]. The structure is characterized by tertiary base-pairing and the presence of stems formed by intramolecular base-pairing in addition to single-stranded loops. A number of posttranscriptional modifications in unpaired tRNA loop regions contribute to tRNA structure by preventing canonical base-pairing [15]. Not only cytoplasmic but also mitochondrial tRNA is subject to dynamic modification by enzymes encoded in the nucleus [9,11]. A variety of human diseases are linked to mutations in genes required for introduction of modifications [16,17,18,19,20] in both cytoplasmic and mitochondrial tRNAs (Figure 1 and Table 1). Diseases associated with defects in human tRNA modification genes include various neurological syndromes as well as metabolic and respiratory dysfunctions (Table 1). In addition, an upregulation of different tRNA modification genes is observed in various cancer types (Table 1), suggesting an increased demand for the activity of tRNA modifiers in different tumor cell types.

Table 1.
Transfer RNA (tRNA) modification genes linked to human disease.
Among disease-relevant modifications targeting cytoplasmic tRNA species, neurological disorders appear to be most common. These include intellectual disability, familial dysautonomia (FD), epilepsy and other syndromes. Interestingly, various mutations known to cause defects in different tRNA modifications such as 5-methylcytosine (m5C), 5-methoxycarbonylmethluridine (mcm 5U), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 5-carbamoylmethyluridine (ncm5U), pseudouridine (Ψ) at positions 38/39 and 13/35, 1-methylguanosine (m1G), inosine (I) and N2,N2-dimethylguanosine (m2,2G) are all associated with intellectual disability in humans (Table 1), suggesting normal brain function and development to be particularly dependent on the presence of these modifications.
Considering the position of critical modifications within the tRNA (Figure 1), a remarkable feature is that many of the disease-relevant tRNA modifications are naturally found within the anticodon loop and more specifically, at the wobble position (34) of the anticodon. Wobble base modification defects in mitochondrial tRNAs are also linked to various myopathies and may also induce neurological symptoms (Table 1) [19,20,37], highlighting the central role of wobble base modifications for normal functioning of tRNA in the different cellular compartments. Modifications at the wobble position are known to enable expanded base-pairing possibilities (inosine) [68] or to improve translational fidelity and elongation by optimizing codon translation rates (mcm5s2U) [69,70,71,72,73,74]. Wobble uridine modifications of the xm5U type (mcm5U, mcm5s2U and ncm5U; Figure 1B) belong to the most complex tRNA modifications in terms of involved genes (Table 2) and pathways as well as with respect to posttranslational modification of the tRNA modifier complexes themselves [66]. Multiple correlations of xm5U defects (Table 1) with human disease indicate this modification family to be of outstanding relevance for human health and development (Figure 2).

Table 2.
Yeast and human genes of the Elongator and ubiquitin related modifier 1 (Urm1) pathways. See text for references. Parts of the modification in which individual genes are involved are underlined.
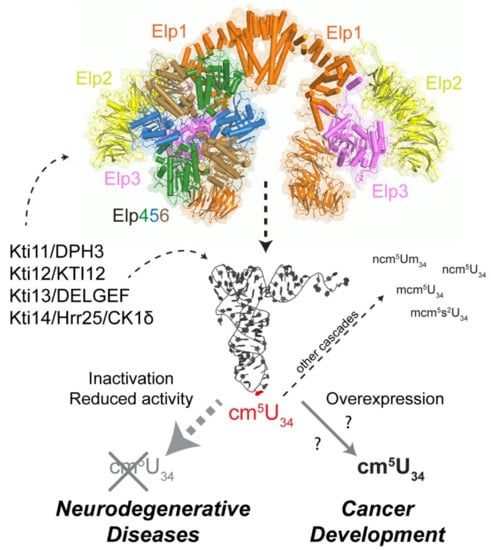
Figure 2.
U34 modifications in human diseases. Scheme showing the “Janus headed” nature of the Elongator complex that plays an important role in human health and disease. (Top) The pseudo-atomic model of the fully assembled Elongator complex is shown in cartoon and transparent surface representation (Elp1/orange, Elp2/yellow, Elp3/pink, Elp4/green, Elp5/blue, Elp6/brown). Additional regulatory factors (Kti11-Kti14, left) and subsequent modifications (right) are indicated and labeled (for further details, see text). (Bottom) The opposing roles of reduced or enhanced levels of cm5 modifications in neurodegenerative diseases and cancer are highlighted.
3. Elongator Dependent Transfer RNA Modifications
In anticodons of a subset of tRNAs, modifications of wobble uridines (U34) require the activity of the Elongator complex (for recent reviews, see [18,75,76]). Target tRNAs receiving Elongator dependent modifications are tRNALysUUU, tRNAGlnUUG, tRNAGluUUC, tRNAArgUCU, tRNAGlyUCC, tRNASerUGA, tRNAProUGG, tRNAThrUGU, tRNAAlaUGC and tRNALeuUAA [77,78]. Elongator is a multi-subunit (Elp1–Elp6) protein assembly that was originally isolated from yeast in association with elongating RNA polymerase II holo-enzyme [79,80] and hence implicated in mRNA transcription rather than translation [78,81,82].
However, Elongator is now known to assemble into an active holo-dodecameric complex [(Elp1–Elp6)2] that binds tRNAs with different subunits [83,84,85,86,87,88]. Accordingly, Elongator operates as a tRNA modifier in a conserved pathway that is found in all three domains of life and shown to be functionally exchangeable between eukaryotic model organisms [86,89,90,91,92,93]. In yeast, the complex attaches to U34 from eleven different tRNA species 5-carboxy-methyl (cm5) groups [77,78], which in concert with additional enzymatic U34 modifier cascades can be further converted to even more composite modifications including (but not limited to) ncm5, mcm5 or mcm5s2 (see Figure 1B, [77,78]). Formation of the cm5 side chain is directly catalyzed by Elp3, Elongator’s catalytic core subunit, which carries binding domains for tRNA and acetyl-CoA and a radical SAM (S-adenosyl-methionine) motif that coordinates an organometallic [4Fe–4S]SAM cluster shown to be crucial for U34 modification in vivo [78,81,89,94]. In support of this notion, archaeal Elp3 from Methanocaldococcus infernus has been shown to modify U34 in the anticodon loop of synthetic tRNAArgUCU substrates iin vitro in a radical SAM dependent reaction containing electron donor (Na2S2O4), SAM and acetyl-CoA [93]. Moreover, sophisticated insights into the basis and mechanism for Elongator’s tRNA modification capacity have been recently provided through elegant structure-functional work on prokaryotic Elp3 from Dehalococcoides mccartyi and holo-Elongator from the yeast Saccharomyces cerevisiae [86,87,88,95,96,97].
In addition to the biochemical isolation of Elongator (see above), genetic screens aimed at isolating yeast mutants that resist anticodon cleavage by zymocin, a U34 modification-dependent tRNase ribotoxin, were also instrumental in identifying Elongator subunits and accessory proteins with roles related to U34 modification and regulation [78,98,99,100,101,102,103,104]. Together with other strategies from many independent research labs, these approaches collectively led to the identification of the U34 wobble methylase complex (Trm9•Trm112) [101,105,106,107] and components of a pathway (Nfs1, Tum1, Uba4, Urm1, Ncs2, Ncs6) for Elongator related U34 thiolation [99,101,108,109,110,111,112,113,114,115,116]. The methyltransferase (MTase) activity of Trm9•Trm112 appears to depend on Elongator activity for the concerted formation of mcm5 and mcm5s2 in yeast and related modifications by homologs of wobble methylase (ALKBH8•TRMT112) in higher eukaryotic systems [117,118,119,120]. Intriguingly, Trm112 not only promotes the MTase activity of the catalytic Trm9 subunit but also acts as an activating hub or platform for three more SAM-dependent MTases modifying ribosomal RNA (Bud23), tRNA (Trm11) or eRF1 translation termination factor (Mtq2) [121,122]. Collectively, this illustrates the importance of methyltransferases for mRNA translation and de novo protein biosynthesis.
As for U34 thiolation and formation of the mcm5s2 modification, it has been shown that S-incorporation into U34 substrate tRNAs by yeast wobble thiolase (Ncs2•Ncs6 aka CTU1•CTU2, see Figure 1) requires a conserved S-relay system (Nfs1, Tum1, Uba4) for S-activation, S-mobilization and S-transfer onto Urm1, a ubiquitin related modifier protein [109,110,111,112,113,114]. Urm1 is unique in combining features typical of prokaryotic S-carriers with eukaryotic ubiquitin proteins [123,124]. In line with this notion, Urm1 can act as a non-canonical, lysine-directed protein modifier in a pathway known as protein urmylation and as a sulfur donor for U34 thiolation [112,113,114,125]. Both roles are strictly sulfur-dependent and exchangeable among eukaryotes [115,116,126] and based on Urm1-like proteins from archaea and bacteria [127,128]; they appear to be conserved and important in all domains of life. In line with this, their inactivation triggers stress-induced growth defects in microbes, organ underdevelopment in plants and, strikingly, lethality in flies [108,113,124,129,130,131]. In contrast to ubiquitin activation and conjugation by conventional E1-E2-E3 enzyme cascades, no E2/E3 activities for Urm1 are known to this end, and Urm1 activation by its E1-like enzyme Uba4 results in C-terminal thiocarboxylation (Urm1-COSH) that is crucial for both urmylation and tRNA thiolation [113,115,123,132,133].
The formation of Urm1-COSH is very similar if not identical to E1-like (MoeB or ThiF) activation of bacterial S-carrier proteins (MoaD or ThiS) that (rather than being involved in protein conjugation) solely donate sulfur for synthesis of molybdopterin or thiamine co-factors [134,135]. Thus, apart from its similarity to eukaryotic ubiquitin-like proteins, Urm1 indeed relates to prokaryotic S-carrier proteins, which is why the protein was coined a molecular fossil at the cross-road of protein and RNA modifications [123]. Its dual-functionality requires desulfurase Nfs1, which mobilizes sulfur from cysteine for direct S-transfer onto Uba4 [110,111] or indirectly via sulfur transferase Tum1 [109,112,113,114]. Uba4 is equipped with MoeB-like (MoeBD, see above) and rhodanese-type domains (RHD) that carry thiol-active cysteines [113,133]. S-transfer to the one in RHD results in a persulfide, which, following adenylation of Urm1 by the MoeBD, likely forms an acyl-disulfide with the Urm1 modifier [132,133,136]. Upon reductive cleavage of this bond, Urm1-COSH gets released [137] to be able to operate in urmylation or donate the activated sulfur species for S-insertion into tRNAs by thiolase Ncs2•Ncs6 [115,133]. Recently, in eukaryotic Ncs6 and TtuA (a related thiouridine synthetase from thermophilic bacteria and archaea) [3Fe-4S] and [4Fe-4S] clusters were identified, respectively, that appear to be involved (directly or indirectly) in the thio-modification reaction required for s2U formation.
Together with findings that tRNA thiolation is apparently not required for protein urmylation and vice versa, the two URM1 pathway branches—albeit mechanistically linked through sulfur activation—seem to be functionally separated from rather than dependent on each other [115]. Thus, a previously suggested concept, according to which the sulfur flow from Urm1-COSH to tRNA thiolation may be kept in-check by urmylation [138], seems less likely to date. However, in this context, it is noteworthy that human URM1 (hURM1) and Urm1-like proteins (SAMP, TtuB) have been shown to form urmylated conjugates with human and prokaryotic orthologs of yeast thiolase (CTU1, CTU2, NcsA, TtuA) [125,127,128]. Whether or not this implies that S-transfer (via Urm1-COSH) for tRNA thiolation may involve direct urmylation of thiolase subunits is unknown but attractive to support the option of interdependence among the two URM1 pathway branches, protein urmylation and U34 thiolation. Although the S-donor role of Urm1 for tRNA thiolation has been demonstrated iin vitro [137], we are not aware of sulfur transfer during lysine-directed urmylation of protein targets (including thiolase components, see above) in yeast or other model organisms. Nonetheless, it has been shown in human cell lines that IKAP (the homolog of yeast Elongator subunit Elp1) was urmylated under conditions of oxidative stress [116]. Additional urmylation target proteins, including human thiolase subunits CTU1 and CTU2 (see above) have been suggested as well [125]. Since the relevance of IKAP urmylation for Elongator’s U34 modification function in human cells has not been addressed and a functional link (if any) between urmylation of IKAP/Elp1 and thiolase (CTU1•CTU2) is not clear for the time being, these intriguing phenomena remain to be resolved in the future.
4. U34 Modifications and Neurodegeneration
Notably, neurons are known to be particularly sensitive to translational defects for some time [139]; however, it was only recently shown that tRNA modifications in combination with the specific usage of AA-ending over AG-ending codons fine tune the translation of specific neuronal transcripts [140]. In addition, a single point mutation in the Elongator subunit Elp6 was identified in a novel cerebellar ataxia mouse model that triggers the cell type specific degeneration of Purkinje neurons [141]. The neuronal decay is accompanied by proteotoxic stress and substantial microgliosis, which can be partially delayed by blocking the NLRP3 inflammasome. The Elp6 mutation leads to a destabilization of the Elp456 subcomplex and reduces the tRNA modification levels [141] in a similar extent as FD patients carrying Elp1 mutations (Table 1) [21].
These similarities indicate that a certain reduction of modification levels causes severe cellular malfunctions, whereas more dramatic alterations would not permit the survival of the patients. Additionally, patients of sporadic amyotrophic lateral sclerosis (ALS) show reduced levels of Elp3 protein and mcm5s2U in the motor cortex [27]. The authors of this study further demonstrated that the role of Elp3 in the pathogenesis of ALS is mediated through its tRNA modification activity, which provided yet another link between tRNA modification defects and neurodegeneration. However, the study surprisingly reported that the SAM domain of Elp3 seemed sufficient to rescue the effects. This observation is in stark contrast to biochemical [86,93] and functional [69,89] studies, showing that both the SAM and acetyl-CoA binding domains are necessary for Elp3’s tRNA modification activity (see above). In summary, the list of neurodegenerative diseases that are directly and indirectly connected to the disruption of the Elongator tRNA modifier complex is continuously growing.
5. A Role of Protein Aggregation in Neurodegeneration
Using ribosome profiling, the absence of mcm5s2U in yeast tRNAGlnUUG and tRNALysUUU was shown to result in a translational slow down at cognate glutamine and lysine codons [70]. This defect goes hand-in-hand with the accumulation of cellular protein aggregates [70]. While the direct mechanism of aggregate formation remains to be solved, a striking observation was that a large overlap exists between aggregates induced by tRNA hypomodification and those formed in the absence of a functional ribosome associated chaperone complex (ssb1/ssb2) [70,142]. This may indicate that ribosomal slow down perturbs folding of the nascent polypeptides. Genetic approaches in budding yeast further support that formation of protein aggregates in the absence of different important tRNA modifications represents a key trigger of pleiotropic cellular defects in cell polarity, morphogenesis and nuclear segregation during cell division [143].
Importantly, protein aggregation in the absence of U34 modification is not limited to the yeast model system but was also observed in nematodes, mice and human cells [27,32,33,70,144,145]. In addition, the absence of the mitochondria-specific taurine-derived wobble uridine modifications induce aggregation of mistargeted mitochondrial proteins [146]. Interestingly, protein aggregation is also a hallmark of various neurodegenerative diseases, and therefore, might represent a functional link between tRNA modification defects (affecting mcm5s2U and other modifications) and neurodegenerative or neurodevelopmental diseases. In support of this, it was already demonstrated that a conditional ELP3 knockout in mice induced the unfolded protein response (UPR) and caused reduced numbers of cortical projection neurons, leading to neurodevelopmental defects and microcephaly [144]. While UPR induction in tRNA modification mutants seems to be confined to higher eukaryotic cell systems, aggregation of endogenous cytoplasmic proteins is observed in both mammals and yeast cells [27,32,33,70,144,145,147].
Since mutations in ELP3 are also linked to the fatal degenerative motor neuron disorder ALS (see above and Table 1), protein aggregation in a mouse motor neuron-like cell line was also analyzed. Upon depletion of ELP3 from this cell line, protein aggregation including the ALS relevant mutant form of SOD1 was observed [27]. In addition, several lines of evidence indicated the neurodegenerative effects of the ELP6 mutation causing ataxia-like syndromes in a mouse model (see above) to be accompanied by protein aggregation [141]. Additionally, silencing of either ELP3 or CTU2 (s 2U) in human melanoma cells resulted in the induction of endogenous protein aggregates [32], suggesting the effect of Elongator and tRNA thiolation defects on protein solubility to be general and highly likely disease relevant. In the latter case, it was also observed that a key factor reprogramming cancer cell gene expression (see below) depends on wobble uridine modification for efficient translation and is present in the endogenous aggregates [32]. This observation is consistent with the proposal that reduced codon translation rates in the absence of mcm5s2U cause protein folding defects and aggregation of the nascent polypeptide. The finding that protein aggregation may also be induced in yeast by defects in other modifications, such as Ψ38/39 or t6A [143,148] and that the very same defects in human cells cause related neurodegenerative/neurodevelopmental syndromes [41,47,60] provide solid support for the assumption that protein aggregation may represent an important trigger of neurodegenerative disease correlated with mutations in tRNA modifiers.
6. U34 Modification in Cancer
In addition to their roles in prevention of neurodegeneration, enzymes introducing U34 tRNA modifications were also identified as key factors to sustain metastasis of breast and bladder cancer and survival of malignant melanoma cells [32,33,149]. It has been known for some time that genes in DNA damage repair pathways display a certain codon-bias [130,150] and that loss of Elongator or related pathways (e.g., Uba4,Urm1, see above) can induce DNA damage [151]. This knowledge has recently been extended by showing that the very same pathways are crucial determinants for the survival of therapy-resistant melanoma cells. In detail, BRAFV600E is the most prevalent mutation among human melanoma patients and responsible for resistance to targeted therapy. ELP1, ELP3, CTU1 and CTU2 are strongly upregulated in BRAFV600E cells and inactivation of ELP3 impaired the development of BRAFV600E melanoma in a zebrafish model [32]. Moreover, it was shown that the Elongator complex promotes glycolysis in melanoma cells through direct, codon-dependent, regulation of the translation of Hypoxia Induced factor 1 α (HIF1A) mRNA, and the maintenance of high levels of HIF1α protein providing strong resistance to anti-BRAF therapy [32]. In a previous study the migration and tumorigenicity of melanoma-derived cells was shown to be significantly decreased upon depletion of ELP1, ELP3, ELP5 or ELP6 in melanoma cells [31]. In another study, ELP3 was shown to be upregulated in human hepatocellular carcinoma (HCC) cells, which correlated well with the phosphorylation of protein kinase B (AKT) [152]. The Elongator mediated migration and invasion of HCC cells is further promoted by the induced expression of MMP-2 and MMP-9 through the PI3K (phosphoinositide 3-kinase)/AKT signaling pathway. ELP3 was also shown to drive Wnt-dependent tumor initiation and regeneration in the intestine by maintaining a subpopulation of cells expressing LGR5- and SOX9 cells [153]. Furthermore, genetic ablation of ELP3 strongly impaired invasion and metastasis formation in a model system of invasive breast cancer. In detail, ELP3 and CTU1•CTU2 are upregulated in human invasive breast cancer and support cellular invasion through the translation of the DEK oncoprotein, which subsequently promotes the IRES (internal ribosome entry site)-dependent translation of the pro-invasive transcription factor LEF1 [33]. In addition, ALKBH8, the methyltransferase implicated in the final step of mcm5s2U and mcm5U formation (Figure 1) is highly expressed in bladder cancer and ALHBH8 knockdown induces cancer cell death due to reduced expression of the anti-apoptotic protein survivin [29,149].
Hence, multiple lines of evidence indicate U34 modifications to promote cancer cell growth and metastasis by ensuring efficient translation reprograming upon transition from normal to cancer cell growth mode. In addition to upregulation of Elongator, ALKBH8 and thiolase genes CTU1 and CTU2 genes, other tRNA modification genes, including NSUN2 (m5C) and METTL1 (m 7G) also become overexpressed in human cancers [52,54,154,155] (Table 1). Moreover, the latter two genes are implicated in resistance against anti-cancer therapy [156] since NSUN2 upregulation is correlated with poor prognosis in patients with in Head and Neck Squamous Carcinoma [157]. Changes in epitranscriptomic tRNA modification in cancer cells may have a general broad significance for prognosis of disease progression and therapy. Hence, a greater understanding of the underlying pathways involved in the modification of U34 is necessary to further understand its two-faced character in human diseases. It remains to be shown if a specific inhibitor for Elongator can be identified and developed. First and foremost, a targeted therapy against Elongator must define a therapeutic window that permits the treatment of cancer cells and simultaneously avoids negative effects on neuronal tissues.
7. Phosphoregulation of Elongator Involving Kti12 and Kti14/Hrr25
Several studies in yeast and other model organisms have shown that tRNA modifications, including Elongator dependent ones can change in response to cell cycle progression and different environmental stresses. This indicates that tRNA modifications are subject to regulation rather than being constitutively formed [158,159,160]. In case of the Elongator complex from yeast, several accessory proteins have been described that influence its tRNA modification activity, namely Kti11-Kti14/Hrr25 and Sit4 (Figure 2). Consistent with this, a casein kinase 1 (CK1) isozyme (Kti14/Hrr25), type 2A protein phosphatases (Sit4·Sap185 and Sit4·Sap190) and an Elongator interactor (Kti12) were shown to affect the phosphorylation state of Elongator’s largest scaffold subunit Elp1 [98,161,162,163]. In principle, dynamic phosphorylation may have an impact on Elongator’s catalytic Elp3 subunit, its localization or its ability to interact with substrate tRNAs [164,165,166]. Although it is unclear what precise role Elp1 phosphorylation plays, it has been proposed as an ‘on/off’ switch for Elongator’s U34 modifier activity, for example, in response to growth conditions or cellular stress. Given that translation of some mRNAs are indeed dependent on proper U34 modification, and hence tunable by Elongator [130,167,168], and that tRNA modifications including Elongator-dependent ones do oscillate, this raises the option that Elongator is part of a translational control mechanism which functions through its role as a U34 modifier. Such a role, which reflects that Elp1 phosphorylation by casein kinase Kti14/Hrr25 is largely positive for Elongator’s performance, is consistent with loss-of-function phenotypes associated with kinase-dead hrr25/kti14 mutations, ablative Elp1 phosphosite substitutions and specific inhibition by ATP analogs of an analog-sensitive Hrr25 kinase variant (Hrr25-I82G) [164,165]. Although Hrr25 operates on many cellular functions [169], which complicates providing clear insights into Elp1 phosphorylation signals, its kinase activity is required for full functionality of ribosomes and U34 containing tRNAs demonstrating its importance in mRNA translation and protein synthesis [164,165,170,171,172]. Since dynamic IKAP/Elp1 phosphorylation was also observed in human melanoma cells [32] and change in response to insulin availability, pharmacological interference with this process may represent an attractive option to downregulate the Elongator function and possibly impair cancer cell proliferation.
A key component in the phosphoregulation of Elp1 seems to be the Elongator partner protein Kti12 (Figure 2). Although the precise role of Kti12 is ill-defined, the yeast protein and its plant ortholog (DRL1/ELO4) carry N-terminal P-loop motifs that are typical of nucleotide binding kinases and NTPases [173,174,175,176,177]. Consistent with a functional role for this domain, a P-loop truncation triggers defects typical of Elongator mutants including loss of U34 modification [98]. Importantly, Kti12 interacts with the Hrr25 kinase in an Elongator-dependent fashion and in doing so, apparently supports Elp1 phosphorylation [98,100,102,178]. This is based on data showing that KTI12 gene deletions abolish Elongator interaction with Hrr25, cause U34 modification defects typical of elp (and hrr25) mutants and trigger formation of hypo-phosphorylated Elp1 isoforms similar to kinase-dead hrr25 cells [101,161,165].
8. Elongator Regulation via Kti11/Dph3 and Kti13
Two additional factors involved in Elongator regulation are Kti11 (alias Dph3) and Kti13 (alias Ats1) [179] (Figure 2). Kti11 precipitates Elongator subunits (Elp1, Elp2, Elp3, Elp5) and associates with Kti13 [108,180,181] in a hetero-dimer shown not only to affect Elongator’s U34 modification activity but also to relate to a posttranslational protein modification pathway [108,180,181,182,183,184]. The Kti11/Dph3•Kti13 complex has been implicated in electron transfer to Elp3 for radical SAM dependent U34 modification by the Elongator complex [182,183]. Removal of Kti11 (i.e., no electron flow) was found to eliminate the U34 modifier activity of Elongator while loss of Kti13 significantly reduced the tRNA modification function to ~20% of wild-type levels [78,180,181]. Kti11/Dph3 partakes together with Dph1, Dph2, Dph4, Dph5, Dph6 and Dph7 in the synthesis of diphthamide, a posttranslationally modified histidine residue (His699 in yeast; His715 in humans) found on translation elongation factor 2 (EF2) [185,186]. EF2 is an essential translation factor that mediates the translocation of the ribosome on the mRNA during translation elongation. Strikingly, diphthamide modified EF2 is the target for diphtheria toxin (DT), which inactivates the translation factor via ADP (adenosine diphosphate)-ribosylation, and thereby, induces death of the intoxicated cell [187,188,189]. Thus, the absence of Dph3/Kti11 leads to full resistance to the lethal ADP-riboslyase activity of DT [180,182,185,186,190]. These data clearly indicate a pathological role for the diphthamide modification in cell growth and proliferation control. The physiological relevance of diphthamide-modified EF2 is less evident [191,192]. However, recent functional and structural analyses suggest diphthamide-modified EF2 supports reading frame maintenance and reduces ribosomal errors [190,193,194,195,196,197].
Based on structure-function analyses, Kti11/Dph3 carries a metal (iron, zinc) binding domain [182,183,198] that is essential for both the modification of U34 carrying tRNAs by Elongator and the synthesis of diphthamide on EF2 [182,183]. Similar to bacterial rubredoxins, Kti11/Dph3 is a redox-active protein and capable of electron transfer to the iron-sulfur clusters of Elongator’s core subunit Elp3 [86,89] and the radical SAM enzyme Dph1•Dph2, which is essential for the first step of diphthamide biosynthesis [108,180,186,199]. Dong et al. (2014) reconstituted a reaction iin vitro, in which Dph3/Kti11 was able to feed electrons into the [4Fe–4S]SAM cluster of the Dph1•Dph2 enzyme for reductive SAM cleavage and generation of a 3-amino-3-carboxypropyl radical (ACP) subsequently used for formation of ACP-EF2, the first intermediate of the diphthamide pathway [200,201,202,203]. Moreover, the Dph3/Kti11 reductase Cbr1 has been identified, which is required for recycling the electron carrier and transfer function of Dph3/Kti11 to both the U34 and the diphthamide modification pathways [204].
The formation of a stable Kti11/Dph3•Kti13 heterodimer suggested an involvement of not only Kti11, but also Kti13 in diphthamide formation [181,182,183]. Indeed, a yeast KTI13 gene deletion strain was shown to confer protection against growth inhibition by DT (see above) [182]. Apart from Kti11/Dph3 and Cbr1, Kti13 may, therefore, be yet another factor operating in both the radical SAM pathways for Elongator dependent tRNA modification and diphthamide synthesis on EF2 [182,183].
Table 1 lists up severe pathologies in humans that are linked with Elongator dependent and related tRNA modification defects. Similarly, defects in genes involved in the diphthamide synthesis pathway of higher eukaryotic model organisms can associate with a variety of diseases and syndromes (Table 3) such as tumorigenesis [205,206,207,208,209,210,211], intellectual disability, craniofacial abnormalities [212,213] and Miller-Dieker Syndrome (MDS) [214], as well as airway obstruction and external genital abnormalities [215] in humans. In mice, embryonic lethality, edema, polaydactyly, jaw shortening, cleft palate, necrosis, apoptosis and defects in placenta development as well as neuronal underdevelopment have been observed in conjunction with diphthamide defects [214,216,219,220]. Furthermore, sensitivity to oxidative stress was reported in chinese hamsters to correlate with failure to diphthamide-modify EF2 [218]. Moreover, failure of intestinal stem cell proliferation has been strictly associated with malfunctional Dph1 and Dph5 in Drosophila melanogaster [217]. Hence, defects in both the radical SAM pathways for tRNA and EF2 modifications, which have been shown to interfere with the fidelity and efficiency of mRNA translation elongation and trigger ribosomal errors, are potentially linked to a multitude of diseases (Table 1 and 3) from several eukaryotes. With the Kti11/Dph3•Kti13 heterodimer and the Cbr1 reductase qualifying as prime candidates to interconnect and possibly, cross-link U34 and diphthamide modifications, there is the emerging prospect for regulation of disease related radical SAM modifier enzymes by a common mechanism that involves control by electron flow to (and from) their iron-sulfur centers. Further studies will be required to address whether drug-based interference with Kti11/Dph3•Kti13 and/or Cbr1 functioning may provide new intervention schemes against disease syndromes linked with dynamic changes of epitranscriptomic tRNA modifications typical of defects in Elongator or the related diphthamide synthesis pathway.

Table 3.
Diphthamide synthesis genes and defects linked to disease syndromes in higher eukaryotes.
9. Conclusions
tRNA modifications are required for optimal translational efficiency and fidelity. A growing list of dynamic tRNA modifications suggests a role in adaptation of translational efficiency to changes in environmental conditions. Consequently, several of these modifications are necessary to prevent human disease. Among the diseases linked to modification/modifier defects, neurodevelopmental syndromes are common, pointing to a specific requirement of full tRNA modification sets in neuronal cells. Among the different modifications linked to human disease, wobble uridine modifications dependent on the Elongator complex are prevalent. Elongator dependent tRNA modifications are not only required for human brain cell function, but also for cancer cell function. Recent work strengthens the implication of Elongator dependent tRNA modifications in sustaining metastasis and uncovered potential regulatory inputs into the modifier complex. Further work will be required to determine whether drug-based interference with Elongator function or regulation opens new approaches to cancer therapy.
Funding
Support to S.G. by the First Team grant (FirstTEAM/2016-1/2) from the Foundation for Polish Science, to R.S. from the PhosMOrg (P/1082 232) research unit,the European Union Cost Action EPITRAN CA16120, and project funds from the DFG Priority Programs SPP1784 “Chemical Biology of Native Nucleic Acid Modifications” and SPP1927 “Iron-Sulfur for Life: Cooperative Function of Iron-Sulfur Centers in Assembly, Biosynthesis, Catalysis and Disease” to R.K. (KL2937/1) and R.S. (SCHA750/20 & SCHA750/21) are gratefully acknowledged.
Acknowledgments
We thank all researchers cited who contributed to the current fundus of knowledge and we apologize to colleagues in the field for not being able to cite many other important papers due to the space limitation of the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Laxman, S.; Sutter, B.M.; Wu, X.; Kumar, S.; Guo, X.; Trudgian, D.C.; Mirzaei, H.; Tu, B.P. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 2013, 154, 416–429. [Google Scholar] [CrossRef]
- Chan, C.T.; Pang, Y.L.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Deng, W.; Li, F.; DeMott, M.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem. Res. Toxicol. 2015, 28, 978–988. [Google Scholar] [CrossRef]
- Alings, F.; Sarin, L.P.; Fufezan, C.; Drexler, H.C.; Leidel, S.A. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 2015, 21, 202–212. [Google Scholar] [CrossRef]
- Han, L.; Kon, Y.; Phizicky, E.M. Functional importance of Psi38 and Psi39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 2015, 21, 188–201. [Google Scholar] [CrossRef]
- Lin, H.; Miyauchi, K.; Harada, T.; Okita, R.; Takeshita, E.; Komaki, H.; Fujioka, K.; Yagasaki, H.; Goto, Y.I.; Yanaka, K.; et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 2018, 9, 1875. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 2016, 167, 816.e16–828.e16. [Google Scholar] [CrossRef]
- Asano, K.; Suzuki, T.; Saito, A.; Wei, F.Y.; Ikeuchi, Y.; Numata, T.; Tanaka, R.; Yamane, Y.; Yamamoto, T.; Goto, T.; et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018, 46, 1565–1583. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Hartmann, M.; Schuster, I.; Bender, S.; Thuring, K.L.; Helm, M.; Katze, J.R.; Nellen, W.; Lyko, F.; Ehrenhofer-Murray, A.E. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015, 43, 10952–10962. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Legrand, C.; Cirzi, C.; Federico, G.; Liebers, R.; Muller, M.; Ehrenhofer-Murray, A.E.; Dittmar, G.; Grone, H.J.; Lyko, F. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Lunse, C.E.; Morl, M. tRNA modifications: Impact on structure and thermal adaptation. Biomolecules 2017, 7. [Google Scholar] [CrossRef]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Bednarova, A.; Hanna, M.; Durham, I.; VanCleave, T.; England, A.; Chaudhuri, A.; Krishnan, N. Lost in translation: Defects in transfer RNA modifications and neurological disorders. Front. Mol. Neurosci. 2017, 10, 135. [Google Scholar] [CrossRef]
- Sokolowski, M.; Klassen, R.; Bruch, A.; Schaffrath, R.; Glatt, S. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 409–418. [Google Scholar] [CrossRef]
- Kirino, Y.; Suzuki, T. Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol. 2005, 2, 41–44. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Sloan, K.E. The mitochondrial epitranscriptome: The roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. CMLS 2018, 75, 241–260. [Google Scholar] [CrossRef]
- Karlsborn, T.; Tukenmez, H.; Chen, C.; Bystrom, A.S. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem. Biophys. Res. Commun. 2014, 454, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Slaugenhaupt, S.A.; Blumenfeld, A.; Gill, S.P.; Leyne, M.; Mull, J.; Cuajungco, M.P.; Liebert, C.B.; Chadwick, B.; Idelson, M.; Reznik, L.; et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001, 68, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.L.; Coli, R.; Daly, I.W.; Kichula, E.A.; Rork, M.J.; Volpi, S.A.; Ekstein, J.; Rubin, B.Y. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001, 68, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P.; et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.S.; Srivastava, S.; Farwell, K.D.; Lu, H.M.; Zeng, W.; Lu, H.; Chao, E.C.; Fatemi, A. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med Genet. Part A 2015, 167, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.L.; Lemmens, R.; Miskiewicz, K.; Broom, W.J.; Hansen, V.K.; van Vught, P.W.; Landers, J.E.; Sapp, P.; Van Den Bosch, L.; Knight, J.; et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009, 18, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Bento-Abreu, A.; Jager, G.; Swinnen, B.; Rue, L.; Hendrickx, S.; Jones, A.; Staats, K.A.; Taes, I.; Eykens, C.; Nonneman, A.; et al. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum. Mol. Genet. 2018, 27, 1276–1289. [Google Scholar] [CrossRef]
- Begley, U.; Sosa, M.S.; Avivar-Valderas, A.; Patil, A.; Endres, L.; Estrada, Y.; Chan, C.T.; Su, D.; Dedon, P.C.; Aguirre-Ghiso, J.A.; et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO Mol. Med. 2013, 5, 366–383. [Google Scholar] [CrossRef]
- Shimada, K.; Nakamura, M.; Anai, S.; De Velasco, M.; Tanaka, M.; Tsujikawa, K.; Ouji, Y.; Konishi, N. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009, 69, 3157–3164. [Google Scholar] [CrossRef]
- Takeoka, S.; Unoki, M.; Onouchi, Y.; Doi, S.; Fujiwara, H.; Miyatake, A.; Fujita, K.; Inoue, I.; Nakamura, Y.; Tamari, M. Amino-acid substitutions in the IKAP gene product significantly increase risk for bronchial asthma in children. J. Hum. Genet. 2001, 46, 57–63. [Google Scholar] [CrossRef]
- Close, P.; Gillard, M.; Ladang, A.; Jiang, Z.; Papuga, J.; Hawkes, N.; Nguyen, L.; Chapelle, J.P.; Bouillenne, F.; Svejstrup, J.; et al. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J. Biol. Chem. 2012, 287, 32535–32545. [Google Scholar] [CrossRef] [PubMed]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, S.; Rapino, F.; Tharun, L.; Zhou, Z.; Heukamp, L.; Termathe, M.; Shostak, K.; Klevernic, I.; Florin, A.; Desmecht, H.; et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016, 213, 2503–2523. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Shaw, M.; Weiner, C.L.; Hobson, L.; Stark, Z.; Rose, K.; Kalscheuer, V.M.; Gecz, J.; Phizicky, E.M. Defects in tRNA anticodon loop 2′-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 2015, 36, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Goto, Y.; Campos, Y.; Arenas, J.; Suzuki, T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl. Acad. Sci. USA 2005, 102, 7127–7132. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Suzuki, T.; Ueda, T.; Ohta, S.; Watanabe, K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000, 275, 4251–4257. [Google Scholar] [CrossRef]
- Kopajtich, R.; Nicholls, T.J.; Rorbach, J.; Metodiev, M.D.; Freisinger, P.; Mandel, H.; Vanlander, A.; Ghezzi, D.; Carrozzo, R.; Taylor, R.W.; et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 2014, 95, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Suzuki, T.; Ishii, N.; Ueda, T.; Ohta, S.; Watanabe, K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA(Lys) with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000, 467, 175–178. [Google Scholar] [CrossRef]
- Guan, M.X.; Yan, Q.; Li, X.; Bykhovskaya, Y.; Gallo-Teran, J.; Hajek, P.; Umeda, N.; Zhao, H.; Garrido, G.; Mengesha, E.; et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 2006, 79, 291–302. [Google Scholar] [CrossRef]
- Zeharia, A.; Shaag, A.; Pappo, O.; Mager-Heckel, A.M.; Saada, A.; Beinat, M.; Karicheva, O.; Mandel, H.; Ofek, N.; Segel, R.; et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009, 85, 401–407. [Google Scholar] [CrossRef]
- Edvardson, S.; Prunetti, L.; Arraf, A.; Haas, D.; Bacusmo, J.M.; Hu, J.F.; Ta-Shma, A.; Dedon, P.C.; de Crecy-Lagard, V.; Elpeleg, O. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. EJHG 2017, 25, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Rao, J.; Mollet, G.; Schapiro, D.; Daugeron, M.C.; Tan, W.; Gribouval, O.; Boyer, O.; Revy, P.; Jobst-Schwan, T.; et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017, 49, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Potapov, V.A.; Smetanina, S.A.; Bel’chikova, L.N.; Suplotova, L.A.; Nosikov, V.V. The carriage of risk variants of CDKAL1 impairs beta-cell function in both diabetic and non-diabetic patients and reduces response to non-sulfonylurea and sulfonylurea agonists of the pancreatic KATP channel. Acta Diabetol. 2011, 48, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Voight, B.F.; Lyssenko, V.; Burtt, N.P.; de Bakker, P.I.; Chen, H.; Roix, J.J.; Kathiresan, S.; Hirschhorn, J.N.; Daly, M.J.; et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007, 316, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, V.; Chen, Y.; Elkahloun, A.; Dutra, A.; Pak, E.; Chandrasekharappa, S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosom. Cancer 2007, 46, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Han, L.; Faqeih, E.; Ewida, N.; Alobeid, E.; Phizicky, E.M.; Alkuraya, F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016, 135, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, H.A.; Al-Shamsi, A.M.; Ali, B.R.; Al-Gazali, L. A null variant in PUS3 confirms its involvement in intellectual disability and further delineates the associated neurodevelopmental disease. Clin. Genet. 2018, 94, 586–587. [Google Scholar] [CrossRef]
- De Brouwer, A.P.M.; Abou Jamra, R.; Kortel, N.; Soyris, C.; Polla, D.L.; Safra, M.; Zisso, A.; Powell, C.A.; Rebelo-Guiomar, P.; Dinges, N.; et al. Variants in PUS7 Cause Intellectual Disability with Speech Delay, Microcephaly, Short Stature, and Aggressive Behavior. Am. J. Hum. Genet. 2018, 103, 1045–1052. [Google Scholar] [CrossRef]
- Abbasi-Moheb, L.; Mertel, S.; Gonsior, M.; Nouri-Vahid, L.; Kahrizi, K.; Cirak, S.; Wieczorek, D.; Motazacker, M.M.; Esmaeeli-Nieh, S.; Cremer, K.; et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 847–855. [Google Scholar] [CrossRef]
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med Genet. 2012, 49, 380–385. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Almuriekhi, M.; Nawaz, Z.; Staffa, A.; Lepage, P.; Ali, R.; Hashim, L.; Schwartzentruber, J.; Abu Khadija, K.; Zaineddin, S.; et al. Whole exome sequencing unravels disease-causing genes in consanguineous families in Qatar. Clin. Genet. 2014, 86, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Watt, F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. CB 2006, 16, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Vachon, C.M.; Sellers, T.A.; Carlson, E.E.; Cunningham, J.M.; Hilker, C.A.; Smalley, R.L.; Schaid, D.J.; Kelemen, L.E.; Couch, F.J.; Pankratz, V.S. Strong evidence of a genetic determinant for mammographic density, a major risk factor for breast cancer. Cancer Res. 2007, 67, 8412–8418. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Dragoni, I.; Chin, S.F.; Spiteri, I.; Kurowski, A.; Provenzano, E.; Green, A.; Ellis, I.O.; Grimmer, D.; Teschendorff, A.; et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010, 289, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Hijazi, H.; Al-Dosari, M.S.; Shaheen, R.; Hashem, A.; Aldahmesh, M.A.; Mohamed, J.Y.; Kentab, A.; Salih, M.A.; Awaji, A.; et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J. Med. Genet. 2013, 50, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Yarham, J.W.; Lamichhane, T.N.; Pyle, A.; Mattijssen, S.; Baruffini, E.; Bruni, F.; Donnini, C.; Vassilev, A.; He, L.; Blakely, E.L.; et al. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 2014, 10, e1004424. [Google Scholar] [CrossRef] [PubMed]
- Spinola, M.; Falvella, F.S.; Galvan, A.; Pignatiello, C.; Leoni, V.P.; Pastorino, U.; Paroni, R.; Chen, S.; Skaug, V.; Haugen, A.; et al. Ethnic differences in frequencies of gene polymorphisms in the MYCL1 region and modulation of lung cancer patients’ survival. Lung Cancer 2007, 55, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Spinola, M.; Galvan, A.; Pignatiello, C.; Conti, B.; Pastorino, U.; Nicander, B.; Paroni, R.; Dragani, T.A. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 2005, 24, 5502–5509. [Google Scholar] [CrossRef]
- Yue, Z.; Li, H.T.; Yang, Y.; Hussain, S.; Zheng, C.H.; Xia, J.; Chen, Y. Identification of breast cancer candidate genes using gene co-expression and protein-protein interaction information. Oncotarget 2016, 7, 36092–36100. [Google Scholar] [CrossRef]
- Shaheen, R.; Abdel-Salam, G.M.; Guy, M.P.; Alomar, R.; Abdel-Hamid, M.S.; Afifi, H.H.; Ismail, S.I.; Emam, B.A.; Phizicky, E.M.; Alkuraya, F.S. Mutation in WDR4 impairs tRNA m7G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015, 16, 210. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Coffey, A.J.; Andrew, T.; Gregorio, S.P.; Dias-Neto, E.; Calafato, M.; Bentley, D.R.; Kinton, L.; Sander, J.W.; Johnson, M.R. Q8IYL2 is a candidate gene for the familial epilepsy syndrome of Partial Epilepsy with Pericentral Spikes (PEPS). Epilepsy Res. 2011, 96, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Igoillo-Esteve, M.; Genin, A.; Lambert, N.; Desir, J.; Pirson, I.; Abdulkarim, B.; Simonis, N.; Drielsma, A.; Marselli, L.; Marchetti, P.; et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013, 9, e1003888. [Google Scholar] [CrossRef] [PubMed]
- Gillis, D.; Krishnamohan, A.; Yaacov, B.; Shaag, A.; Jackman, J.E.; Elpeleg, O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J. Med. Genet. 2014, 51, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Zung, A.; Kori, M.; Burundukov, E.; Ben-Yosef, T.; Tatoor, Y.; Granot, E. Homozygous deletion of TRMT10A as part of a contiguous gene deletion in a syndrome of failure to thrive, delayed puberty, intellectual disability and diabetes mellitus. Am. J. Med. Genet. Part A 2015, 167, 3167–3173. [Google Scholar] [CrossRef] [PubMed]
- Yew, T.W.; McCreight, L.; Colclough, K.; Ellard, S.; Pearson, E.R. tRNA methyltransferase homologue gene TRMT10A mutation in young adult-onset diabetes with intellectual disability, microcephaly and epilepsy. Diabet. Med. 2016, 33, e21–e25. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.M.; Thomas, J.; Ross, D.T.; Seitz, R.S.; Ring, B.Z.; Beck, R.A.; Pedersen, H.C.; Munro, A.; Kunkler, I.H.; Campbell, F.M.; et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. BCR 2010, 12, R47. [Google Scholar] [CrossRef] [PubMed]
- Bykhovskaya, Y.; Casas, K.; Mengesha, E.; Inbal, A.; Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 2004, 74, 1303–1308. [Google Scholar] [CrossRef]
- Torres, A.G.; Pineyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef]
- Esberg, A.; Huang, B.; Johansson, M.J.; Bystrom, A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef]
- Nedialkova, D.D.; Leidel, S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef]
- Tukenmez, H.; Xu, H.; Esberg, A.; Bystrom, A.S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Bruch, A.; Schaffrath, R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2017, 14, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Bhatt, M.J.; Farabaugh, P.J. Codon-specific effects of tRNA anticodon loop modifications on translational misreading errors in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 10331–10339. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Schaffrath, R. Collaboration of tRNA modifications and elongation factor eEF1A in decoding and nonsense suppression. Sci. Rep. 2018, 8, 12749. [Google Scholar] [CrossRef] [PubMed]
- Schaffrath, R.; Leidel, S.A. Wobble uridine modifications—a reason to live, a reason to die?! RNA Biol. 2017, 14, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.O.; Xu, F.; Bystrom, A.S. Elongator—a tRNA modifying complex that promotes efficient translational decoding. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.; Esberg, A.; Huang, B.; Bjork, G.R.; Bystrom, A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef]
- Huang, B.; Johansson, M.J.; Bystrom, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef]
- Otero, G.; Fellows, J.; Li, Y.; de Bizemont, T.; Dirac, A.M.; Gustafsson, C.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 1999, 3, 109–118. [Google Scholar] [CrossRef]
- Winkler, G.S.; Petrakis, T.G.; Ethelberg, S.; Tokunaga, M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 2001, 276, 32743–32749. [Google Scholar] [CrossRef]
- Chen, C.; Huang, B.; Eliasson, M.; Ryden, P.; Bystrom, A.S. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011, 7, e1002258. [Google Scholar] [CrossRef] [PubMed]
- Karlsborn, T.; Tukenmez, H.; Mahmud, A.K.; Xu, F.; Xu, H.; Bystrom, A.S. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol. 2014, 11, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R.; Bandau, S.; Stark, M.J. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol. Microbiol. 2014, 92, 1227–1242. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Lin, Z.; Li, F.; Diao, W.; Dong, C.; Zhou, H.; Xie, X.; Wang, Z.; Shen, Y.; Long, J. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc. Natl. Acad. Sci. USA 2015, 112, 10697–10702. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Letoquart, J.; Faux, C.; Taylor, N.M.; Seraphin, B.; Muller, C.W. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Zabel, R.; Kolaj-Robin, O.; Onuma, O.F.; Baudin, F.; Graziadei, A.; Taverniti, V.; Lin, T.Y.; Baymann, F.; Seraphin, B.; et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016, 23, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Setiaputra, D.T.; Cheng, D.T.; Lu, S.; Hansen, J.M.; Dalwadi, U.; Lam, C.H.; To, J.L.; Dong, M.Q.; Yip, C.K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017, 18, 280–291. [Google Scholar] [CrossRef]
- Dauden, M.I.; Jaciuk, M.; Muller, C.W.; Glatt, S. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 2018, 592, 502–515. [Google Scholar] [CrossRef]
- Paraskevopoulou, C.; Fairhurst, S.A.; Lowe, D.J.; Brick, P.; Onesti, S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 2006, 59, 795–806. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, H.; Jablonowski, D.; Zhou, X.; Ren, X.; Hong, X.; Schaffrath, R.; Zhu, J.K.; Gong, Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006, 26, 6902–6912. [Google Scholar] [CrossRef]
- Lin, F.J.; Shen, L.; Jang, C.W.; Falnes, P.O.; Zhang, Y. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 2013, 9, e1003516. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Jablonowski, D.; Wrackmeyer, U.; Tschitschmann, S.; Sondermann, D.; Jager, G.; Gong, Z.; Bystrom, A.S.; Schaffrath, R.; Breunig, K.D. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010, 76, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Selvadurai, K.; Wang, P.; Seimetz, J.; Huang, R.H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014, 10, 810–812. [Google Scholar] [CrossRef] [PubMed]
- Wittschieben, B.O.; Otero, G.; de Bizemont, T.; Fellows, J.; Erdjument-Bromage, H.; Ohba, R.; Li, Y.; Allis, C.D.; Tempst, P.; Svejstrup, J.Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 1999, 4, 123–128. [Google Scholar] [CrossRef]
- Dalwadi, U.; Yip, C.K. Structural insights into the function of Elongator. Cell. Mol. Life Sci. CMLS 2018, 75, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Dauden, M.I.; Kosinski, J.; Kolaj-Robin, O.; Desfosses, A.; Ori, A.; Faux, C.; Hoffmann, N.A.; Onuma, O.F.; Breunig, K.D.; Beck, M.; et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017, 18, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Glatt, S. tRNA Modification by Elongator Protein 3 (Elp3). In Encyclopedia of Inorganic and Bioinorganic Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 1–9. [Google Scholar]
- Fichtner, L.; Frohloff, F.; Burkner, K.; Larsen, M.; Breunig, K.D.; Schaffrath, R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 2002, 43, 783–791. [Google Scholar] [CrossRef]
- Jablonowski, D.; Schaffrath, R. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 2007, 35, 1533–1537. [Google Scholar] [CrossRef]
- Fichtner, L.; Frohloff, F.; Jablonowski, D.; Stark, M.J.; Schaffrath, R. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol. Microbiol. 2002, 45, 817–826. [Google Scholar] [CrossRef][Green Version]
- Huang, B.; Lu, J.; Bystrom, A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008, 14, 2183–2194. [Google Scholar] [CrossRef]
- Frohloff, F.; Fichtner, L.; Jablonowski, D.; Breunig, K.D.; Schaffrath, R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001, 20, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Frohloff, F.; Jablonowski, D.; Fichtner, L.; Schaffrath, R. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (gamma-toxin target (TOT)) complex. J. Biol. Chem. 2003, 278, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Frohloff, F.; Fichtner, L.; Stark, M.J.; Schaffrath, R. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 2001, 42, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, H.R.; Clarke, S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003, 23, 9283–9292. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Zink, S.; Mehlgarten, C.; Daum, G.; Schaffrath, R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator’s key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006, 59, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Studte, P.; Zink, S.; Jablonowski, D.; Bar, C.; von der Haar, T.; Tuite, M.F.; Schaffrath, R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008, 69, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, L.; Jablonowski, D.; Schierhorn, A.; Kitamoto, H.K.; Stark, M.J.R.; Schaffrath, R. Elongator’s toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol. Microbiol. 2003, 49, 1297–1307. [Google Scholar] [CrossRef]
- Nakai, Y.; Nakai, M.; Hayashi, H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008, 283, 27469–27476. [Google Scholar] [CrossRef]
- Nakai, Y.; Nakai, M.; Lill, R.; Suzuki, T.; Hayashi, H. Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol. Cell. Biol. 2007, 27, 2841–2847. [Google Scholar] [CrossRef]
- Nakai, Y.; Umeda, N.; Suzuki, T.; Nakai, M.; Hayashi, H.; Watanabe, K.; Kagamiyama, H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 2004, 279, 12363–12368. [Google Scholar] [CrossRef]
- Schlieker, C.D.; Van der Veen, A.G.; Damon, J.R.; Spooner, E.; Ploegh, H.L. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 18255–18260. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Pedrioli, P.G.; Bucher, T.; Brost, R.; Costanzo, M.; Schmidt, A.; Aebersold, R.; Boone, C.; Hofmann, K.; Peter, M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009, 458, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef] [PubMed]
- Judes, A.; Bruch, A.; Klassen, R.; Helm, M.; Schaffrath, R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4*Urm1 link protein urmylation and tRNA thiolation in yeast. Microb. Cell 2016, 3, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Judes, A.; Ebert, F.; Bar, C.; Thuring, K.L.; Harrer, A.; Klassen, R.; Helm, M.; Stark, M.J.; Schaffrath, R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4.Urm1 systems are conserved from yeast to man. FEBS Lett. 2015, 589, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Leihne, V.; Kirpekar, F.; Vagbo, C.B.; van den Born, E.; Krokan, H.E.; Grini, P.E.; Meza, T.J.; Falnes, P.O. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 2011, 39, 7688–7701. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Phizicky, E.M. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014, 11, 1608–1618. [Google Scholar] [CrossRef]
- Letoquart, J.; van Tran, N.; Caroline, V.; Aleksandrov, A.; Lazar, N.; van Tilbeurgh, H.; Liger, D.; Graille, M. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015, 43, 10989–11002. [Google Scholar] [CrossRef]
- Lentini, J.M.; Ramos, J.; Fu, D. Monitoring the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) modification in eukaryotic tRNAs via the gamma-toxin endonuclease. RNA 2018, 24, 749–758. [Google Scholar] [CrossRef]
- Van Tran, N.; Muller, L.; Ross, R.L.; Lestini, R.; Letoquart, J.; Ulryck, N.; Limbach, P.A.; de Crecy-Lagard, V.; Cianferani, S.; Graille, M. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018, 46, 8483–8499. [Google Scholar] [CrossRef]
- Bourgeois, G.; Letoquart, J.; van Tran, N.; Graille, M. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 2017, 7. [Google Scholar] [CrossRef]
- Pedrioli, P.G.; Leidel, S.; Hofmann, K. Urm1 at the crossroad of modifications. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008, 9, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Mizushima, N.; Noda, T.; Ohsumi, Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000, 275, 7462–7465. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, A.G.; Schorpp, K.; Schlieker, C.; Buti, L.; Damon, J.R.; Spooner, E.; Ploegh, H.L.; Jentsch, S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. USA 2011, 108, 1763–1770. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Harada, A.; Hashiguchi, Y.; Nakai, M.; Hayashi, H. Arabidopsis molybdopterin biosynthesis protein Cnx5 collaborates with the ubiquitin-like protein Urm11 in the thio-modification of tRNA. J. Biol. Chem. 2012, 287, 30874–30884. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N. Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 2012, 287, 17568–17577. [Google Scholar] [CrossRef]
- Maupin-Furlow, J.A. Prokaryotic ubiquitin-like protein modification. Annu. Rev. Microbiol. 2014, 68, 155–175. [Google Scholar] [CrossRef]
- Scheidt, V.; Judes, A.; Bar, C.; Klassen, R.; Schaffrath, R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell 2014, 1, 416–424. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Gilbert, W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef]
- Khoshnood, B.; Dacklin, I.; Grabbe, C. Urm1: An essential regulator of JNK signaling and oxidative stress in Drosophila melanogaster. Cell. Mol. Life Sci. CMLS 2016, 73, 1939–1954. [Google Scholar] [CrossRef]
- Schmitz, J.; Chowdhury, M.M.; Hanzelmann, P.; Nimtz, M.; Lee, E.Y.; Schindelin, H.; Leimkuhler, S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 2008, 47, 6479–6489. [Google Scholar] [CrossRef] [PubMed]
- Termathe, M.; Leidel, S.A. The Uba4 domain interplay is mediated via a thioester that is critical for tRNA thiolation through Urm1 thiocarboxylation. Nucleic Acids Res. 2018, 46, 5171–5181. [Google Scholar] [CrossRef] [PubMed]
- Lake, M.W.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 2001, 414, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Iyer, L.M.; Aravind, L. Natural history of the E1-like superfamily: Implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins 2009, 75, 895–910. [Google Scholar] [CrossRef]
- Marelja, Z.; Stocklein, W.; Nimtz, M.; Leimkuhler, S. A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 2008, 283, 25178–25185. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.M.; Dosche, C.; Lohmannsroben, H.G.; Leimkuhler, S. Dual role of the molybdenum cofactor biosynthesis protein MOCS3 in tRNA thiolation and molybdenum cofactor biosynthesis in humans. J. Biol. Chem. 2012, 287, 17297–17307. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Salvesen, G.S.; Wolf, D.A. Urm1 couples sulfur transfer to ubiquitin-like protein function in oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1749–1750. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, R.; Moore, M.J.; Al-Chalabi, A.; Brown, R.H., Jr.; Robberecht, W. RNA metabolism and the pathogenesis of motor neuron diseases. Trends Neurosci. 2010, 33, 249–258. [Google Scholar] [CrossRef]
- Goffena, J.; Lefcort, F.; Zhang, Y.; Lehrmann, E.; Chaverra, M.; Felig, J.; Walters, J.; Buksch, R.; Becker, K.G.; George, L. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun. 2018, 9, 889. [Google Scholar] [CrossRef]
- Kojic, M.; Gaik, M.; Kiska, B.; Salerno-Kochan, A.; Hunt, S.; Tedoldi, A.; Mureev, S.; Jones, A.; Whittle, B.; Genovesi, L.A.; et al. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nat. Commun. 2018, 9, 3195. [Google Scholar] [CrossRef]
- Koplin, A.; Preissler, S.; Ilina, Y.; Koch, M.; Scior, A.; Erhardt, M.; Deuerling, E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010, 189, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Ciftci, A.; Funk, J.; Bruch, A.; Butter, F.; Schaffrath, R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016, 44, 10946–10959. [Google Scholar] [CrossRef] [PubMed]
- Laguesse, S.; Creppe, C.; Nedialkova, D.D.; Prevot, P.P.; Borgs, L.; Huysseune, S.; Franco, B.; Duysens, G.; Krusy, N.; Lee, G.; et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 2015, 35, 553–567. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Ruggero, D. A wobbly road to drug resistance in melanoma: tRNA-modifying enzymes in translation reprogramming. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Wei, F.Y.; Suzuki, T.; Asano, K.; Kaieda, T.; Omori, A.; Izumi, R.; Fujimura, A.; Kaitsuka, T.; Miyata, K.; et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 2018, 22, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Bruch, A.; Klassen, R.; Schaffrath, R. Unfolded protein response suppression in yeast by loss of tRNA modifications. Genes 2018, 9. [Google Scholar] [CrossRef]
- Thiaville, P.C.; Legendre, R.; Rojas-Benitez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crecy-Lagard, V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef]
- Ohshio, I.; Kawakami, R.; Tsukada, Y.; Nakajima, K.; Kitae, K.; Shimanoe, T.; Saigo, Y.; Hase, H.; Ueda, Y.; Jingushi, K.; et al. ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin. Biochem. Biophys. Res. Commun. 2016, 477, 413–418. [Google Scholar] [CrossRef]
- Fernandez-Vazquez, J.; Vargas-Perez, I.; Sanso, M.; Buhne, K.; Carmona, M.; Paulo, E.; Hermand, D.; Rodriguez-Gabriel, M.; Ayte, J.; Leidel, S.; et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013, 9, e1003647. [Google Scholar] [CrossRef]
- Dewez, M.; Bauer, F.; Dieu, M.; Raes, M.; Vandenhaute, J.; Hermand, D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. USA 2008, 105, 5459–5464. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, W.; Ji, Y.; Shen, J.; Zhu, X.; Yu, H.; Guo, J.; Pang, Z.; Wei, W. Elongator promotes the migration and invasion of hepatocellular carcinoma cell by the phosphorylation of AKT. Int. J. Biol. Sci. 2018, 14, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Ladang, A.; Rapino, F.; Heukamp, L.C.; Tharun, L.; Shostak, K.; Hermand, D.; Delaunay, S.; Klevernic, I.; Jiang, Z.; Jacques, N.; et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 2015, 212, 2057–2075. [Google Scholar] [CrossRef] [PubMed]
- Wikman, H.; Nymark, P.; Vayrynen, A.; Jarmalaite, S.; Kallioniemi, A.; Salmenkivi, K.; Vainio-Siukola, K.; Husgafvel-Pursiainen, K.; Knuutila, S.; Wolf, M.; et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes Chromosom. Cancer 2005, 42, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Hirata, S.; Sato, S.; Koga, S.; Fujii, M.; Qi, G.; Ogawa, I.; Takata, T.; Shimamoto, F.; Tatsuka, M. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012, 31, 660–671. [Google Scholar] [CrossRef]
- Okamoto, M.; Fujiwara, M.; Hori, M.; Okada, K.; Yazama, F.; Konishi, H.; Xiao, Y.; Qi, G.; Shimamoto, F.; Ota, T.; et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014, 10, e1004639. [Google Scholar] [CrossRef]
- Lu, L.; Zhu, G.; Zeng, H.; Xu, Q.; Holzmann, K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Investig. 2018, 36, 246–253. [Google Scholar] [CrossRef]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Patil, A.; Dyavaiah, M.; Joseph, F.; Rooney, J.P.; Chan, C.T.; Dedon, P.C.; Begley, T.J. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 2012, 11, 3656–3665. [Google Scholar] [CrossRef]
- Mehlgarten, C.; Schaffrath, R. Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactiszymocin in budding yeast. Mol. Genet. Genom. MGG 2003, 269, 188–196. [Google Scholar] [CrossRef]
- Jablonowski, D.; Butler, A.R.; Fichtner, L.; Gardiner, D.; Schaffrath, R.; Stark, M.J. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 2001, 159, 1479–1489. [Google Scholar] [PubMed]
- Jablonowski, D.; Taubert, J.E.; Bar, C.; Stark, M.J.; Schaffrath, R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryot. Cell 2009, 8, 1637–1647. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fattah, W.; Jablonowski, D.; Di Santo, R.; Thuring, K.L.; Scheidt, V.; Hammermeister, A.; Ten Have, S.; Helm, M.; Schaffrath, R.; Stark, M.J. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet. 2015, 11, e1004931. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Jablonowski, D.; Breunig, K.D.; Stark, M.J.; Schaffrath, R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009, 73, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Fichtner, L.; Stark, M.J.; Schaffrath, R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell 2004, 15, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Bauer, F.; Matsuyama, A.; Candiracci, J.; Dieu, M.; Scheliga, J.; Wolf, D.A.; Yoshida, M.; Hermand, D. Translational control of cell division by Elongator. Cell Rep. 2012, 1, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Dedon, P.C.; Begley, T.J. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014, 27, 330–337. [Google Scholar] [CrossRef]
- Cheong, J.K.; Virshup, D.M. Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell Biol. 2011, 43, 465–469. [Google Scholar] [CrossRef]
- Schafer, T.; Maco, B.; Petfalski, E.; Tollervey, D.; Bottcher, B.; Aebi, U.; Hurt, E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 2006, 441, 651–655. [Google Scholar] [CrossRef]
- Ray, P.; Basu, U.; Ray, A.; Majumdar, R.; Deng, H.; Maitra, U. The Saccharomyces cerevisiae 60S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J. Biol. Chem. 2008, 283, 9681–9691. [Google Scholar] [CrossRef]
- Ghalei, H.; Schaub, F.X.; Doherty, J.R.; Noguchi, Y.; Roush, W.R.; Cleveland, J.L.; Stroupe, M.E.; Karbstein, K. Hrr25/CK1delta-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 2015, 208, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.R.; White, J.H.; Folawiyo, Y.; Edlin, A.; Gardiner, D.; Stark, M.J. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 1994, 14, 6306–6316. [Google Scholar] [CrossRef] [PubMed]
- Leipe, D.D.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002, 317, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H.; Clarke, J.H.; De Block, M.; De Block, S.; Vanderhaeghen, R.; Zielinski, R.E.; Dyer, T.; Lust, S.; Inze, D.; Van Lijsebettens, M. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 2003, 15, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Prochaska, H.; Hammermeister, A.; Abdel-Fattah, W.; Wagner, M.; Krutyholowa, R.; Jun, S.E.; Kim, G.T.; Glatt, S.; Breunig, K.D.; et al. Use of a yeast tRNase killer toxin to diagnose Kti12 motifs required for tRNA modification by elongator. Toxins 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.E.; Cho, K.H.; Hwang, J.Y.; Abdel-Fattah, W.; Hammermeister, A.; Schaffrath, R.; Bowman, J.L.; Kim, G.T. Comparative analysis of the conserved functions of Arabidopsis DRL1 and yeast KTI12. Mol. Cells 2014. [Google Scholar] [CrossRef]
- Petrakis, T.G.; Sogaard, T.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Physical and functional interaction between Elongator and the chromatin-associated Kti12 protein. J. Biol. Chem. 2005, 280, 19454–19460. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, L.; Schaffrath, R. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 2002, 44, 865–875. [Google Scholar] [CrossRef]
- Bar, C.; Zabel, R.; Liu, S.; Stark, M.J.; Schaffrath, R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol. Microbiol. 2008, 69, 1221–1233. [Google Scholar] [CrossRef]
- Zabel, R.; Bar, C.; Mehlgarten, C.; Schaffrath, R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol. Microbiol. 2008, 69, 175–187. [Google Scholar] [CrossRef]
- Glatt, S.; Zabel, R.; Vonkova, I.; Kumar, A.; Netz, D.J.; Pierik, A.J.; Rybin, V.; Lill, R.; Gavin, A.C.; Balbach, J.; et al. Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure 2015, 23, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Kolaj-Robin, O.; McEwen, A.G.; Cavarelli, J.; Seraphin, B. Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification. FEBS J. 2015, 282, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Leppla, S.H. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol. Cell 2003, 12, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Bodley, J.W.; Livingston, D.M. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 1985, 5, 3357–3360. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Milne, G.T.; Kuremsky, J.G.; Fink, G.R.; Leppla, S.H. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 2004, 24, 9487–9497. [Google Scholar] [CrossRef] [PubMed]
- Pappenheimer, A.M., Jr. Diphtheria toxin. Annu. Rev. Biochem. 1977, 46, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Van Ness, B.G.; Barrowclough, B.; Bodley, J.W. Recognition of elongation factor 2 by diphtheria toxin is not solely defined by the presence of diphthamide. FEBS Lett. 1980, 120, 4–6. [Google Scholar] [CrossRef]
- Uthman, S.; Liu, S.; Giorgini, F.; Stark, M.J.; Costanzo, M.; Schaffrath, R. Diphtheria disease and genes involved in formation of diphthamide, key effector of the diphtheria toxin. In Insight and Control of Infectious Disease in Global Scenario; InTech: London, UK, 2012; pp. 333–356. [Google Scholar]
- Uthman, S.; Bar, C.; Scheidt, V.; Liu, S.; ten Have, S.; Giorgini, F.; Stark, M.J.; Schaffrath, R. The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet. 2013, 9, e1003334. [Google Scholar] [CrossRef]
- Schaffrath, R.; Abdel-Fattah, W.; Klassen, R.; Stark, M.J. The diphthamide modification pathway from Saccharomyces cerevisiae—revisited. Mol. Microbiol. 2014, 94, 1213–1226. [Google Scholar] [CrossRef]
- Tsuda-Sakurai, K.; Miura, M. The hidden nature of protein translational control by diphthamide—the secrets under the leather. J. Biochem. 2018. [Google Scholar] [CrossRef]
- Ortiz, P.A.; Ulloque, R.; Kihara, G.K.; Zheng, H.; Kinzy, T.G. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 2006, 281, 32639–32648. [Google Scholar] [CrossRef] [PubMed]
- Abeyrathne, P.D.; Koh, C.S.; Grant, T.; Grigorieff, N.; Korostelev, A.A. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Savva, C.G.; Shin, B.S.; Dever, T.E.; Ramakrishnan, V.; Fernandez, I.S. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, S.; Demeshkina, N.; Mancera-Martinez, E.; Melnikov, S.; Simonetti, A.; Myasnikov, A.; Yusupov, M.; Yusupova, G.; Hashem, Y. Structural insights into the role of diphthamide on elongation factor 2 in mRNA reading-frame maintenance. J. Mol. Biol. 2018, 430, 2677–2687. [Google Scholar] [CrossRef] [PubMed]
- Hawer, H.; Utkur, K.; Arend, M.; Mayer, K.; Adrian, L.; Brinkmann, U.; Schaffrath, R. Importance of diphthamide modified EF2 for translational accuracy and competitive cell growth in yeast. PLoS ONE 2018, 13, e0205870. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, J.; Wu, F.; Xu, C.; Li, S.; Zhao, W.; Wu, Z.; Wu, J.; Zhou, C.Z.; Shi, Y. Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module. Biochemistry 2005, 44, 8801–8809. [Google Scholar] [CrossRef]
- Abdel-Fattah, W.; Scheidt, V.; Uthman, S.; Stark, M.J.; Schaffrath, R. Insights into diphthamide, key diphtheria toxin effector. Toxins 2013, 5, 958–968. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, X.; Torelli, A.T.; Lee, M.; Dzikovski, B.; Koralewski, R.M.; Wang, E.; Freed, J.; Krebs, C.; Ealick, S.E.; et al. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 2010, 465, 891–896. [Google Scholar] [CrossRef]
- Zhu, X.; Dzikovski, B.; Su, X.; Torelli, A.T.; Zhang, Y.; Ealick, S.E.; Freed, J.H.; Lin, H. Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol. BioSyst. 2011, 7, 74–81. [Google Scholar] [CrossRef]
- Dong, M.; Horitani, M.; Dzikovski, B.; Freed, J.H.; Ealick, S.E.; Hoffman, B.M.; Lin, H. Substrate-dependent cleavage site selection by unconventional radical S-adenosylmethionine enzymes in diphthamide biosynthesis. J. Am. Chem. Soc. 2017, 139, 5680–5683. [Google Scholar] [CrossRef]
- Dong, M.; Kathiresan, V.; Fenwick, M.K.; Torelli, A.T.; Zhang, Y.; Caranto, J.D.; Dzikovski, B.; Sharma, A.; Lancaster, K.M.; Freed, J.H.; et al. Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis. Science 2018, 359, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Dong, M.; Zhang, Y.; Lee, E.A.; Lin, H. Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification. Nat. Chem. Biol. 2016, 12, 995–997. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Takahashi, T.; Kozaki, K.; Yatabe, Y.; Mitsudomi, T.; Fujii, Y.; Sugiura, T.; Matsuda, H.; Takahashi, T.; Takahashi, T. Detailed deletion mapping suggests the involvement of a tumor suppressor gene at 17p13.3, distal to p53, in the pathogenesis of lung cancers. Oncogene 1998, 17, 2095–2100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liscia, D.S.; Morizio, R.; Venesio, T.; Palenzona, C.; Donadio, M.; Callahan, R. Prognostic significance of loss of heterozygosity at loci on chromosome 17p13.3-ter in sporadic breast cancer is evidence for a putative tumour suppressor gene. Br. J. Cancer 1999, 80, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Hoff, C.; Mollenhauer, J.; Waldau, B.; Hamann, U.; Poustka, A. Allelic imbalance and fine mapping of the 17p13.3 subregion in sporadic breast carcinomas. Cancer Genet. Cytogenet. 2001, 129, 145–149. [Google Scholar] [CrossRef]
- Saxena, A.; Clark, W.C.; Robertson, J.T.; Ikejiri, B.; Oldfield, E.H.; Ali, I.U. Evidence for the involvement of a potential second tumor suppressor gene on chromosome 17 distinct from p53 in malignant astrocytomas. Cancer Res. 1992, 52, 6716–6721. [Google Scholar] [PubMed]
- Schultz, D.C.; Vanderveer, L.; Berman, D.B.; Hamilton, T.C.; Wong, A.J.; Godwin, A.K. Identification of two candidate tumor suppressor genes on chromosome 17p13.3. Cancer Res. 1996, 56, 1997–2002. [Google Scholar] [PubMed]
- Phillips, N.J.; Ziegler, M.R.; Radford, D.M.; Fair, K.L.; Steinbrueck, T.; Xynos, F.P.; Donis-Keller, H. Allelic deletion on chromosome 17p13.3 in early ovarian cancer. Cancer Res. 1996, 56, 606–611. [Google Scholar]
- Liu, M.; Yin, K.; Guo, X.; Feng, H.; Yuan, M.; Liu, Y.; Zhang, J.; Guo, B.; Wang, C.; Zhou, G.; et al. Diphthamide biosynthesis 1 is a novel oncogene in colorectal cancer cells and is regulated by MiR-218-5p. Cell. Physiol. Biochem. 2017, 44, 505–514. [Google Scholar] [CrossRef]
- Sekiguchi, F.; Nasiri, J.; Sedghi, M.; Salehi, M.; Hosseinzadeh, M.; Okamoto, N.; Mizuguchi, T.; Nakashima, M.; Miyatake, S.; Takata, A.; et al. A novel homozygous DPH1 mutation causes intellectual disability and unique craniofacial features. J. Hum. Genet. 2018, 63, 487–491. [Google Scholar] [CrossRef]
- Loucks, C.M.; Parboosingh, J.S.; Shaheen, R.; Bernier, F.P.; McLeod, D.R.; Seidahmed, M.Z.; Puffenberger, E.G.; Ober, C.; Hegele, R.A.; Boycott, K.M.; et al. Matching two independent cohorts validates DPH1 as a gene responsible for autosomal recessive intellectual disability with short stature, craniofacial, and ectodermal anomalies. Hum. Mutat. 2015, 36, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.R.; You, L.R.; Yan, Y.T.; Chen, C.M. Role of OVCA1/DPH1 in craniofacial abnormalities of Miller-Dieker syndrome. Hum. Mol. Genet. 2014, 23, 5579–5596. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, J.; Oana, S.; Sakaguchi, T.; Nakashima, M.; Numabe, H.; Kawashima, H.; Matsumoto, N.; Miyake, N. Novel compound heterozygous DPH1 mutations in a patient with the unique clinical features of airway obstruction and external genital abnormalities. J. Hum. Genet. 2018, 63, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Behringer, R.R. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004, 18, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Obata, F.; Tsuda-Sakurai, K.; Yamazaki, T.; Nishio, R.; Nishimura, K.; Kimura, M.; Funakoshi, M.; Miura, M. Nutritional control of stem cell division through S-adenosylmethionine in Drosophila intestine. Dev. Cell 2018, 44, 741.e3–751.e3. [Google Scholar] [CrossRef] [PubMed]
- Arguelles, S.; Camandola, S.; Cutler, R.G.; Ayala, A.; Mattson, M.P. Elongation factor 2 diphthamide is critical for translation of two IRES-dependent protein targets, XIAP and FGF2, under oxidative stress conditions. Free Radic. Biol. Med. 2014, 67, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bachran, C.; Gupta, P.; Miller-Randolph, S.; Wang, H.; Crown, D.; Zhang, Y.; Wein, A.N.; Singh, R.; Fattah, R.; et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc. Natl. Acad. Sci. USA 2012, 109, 13817–13822. [Google Scholar] [CrossRef]
- Webb, T.R.; Cross, S.H.; McKie, L.; Edgar, R.; Vizor, L.; Harrison, J.; Peters, J.; Jackson, I.J. Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J. Cell Sci. 2008, 121, 3140–3145. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).