Association of the Hepatitis B Virus Large Surface Protein with Viral Infectivity and Endoplasmic Reticulum Stress-mediated Liver Carcinogenesis
Abstract
1. Introduction
2. HBV Surface Proteins
3. The Pre-S1 Domain of the HBV Large Surface Protein in Viral Entry
4. Glycosylations of the LHBs Pre-S Regions that Regulate Protein Topology
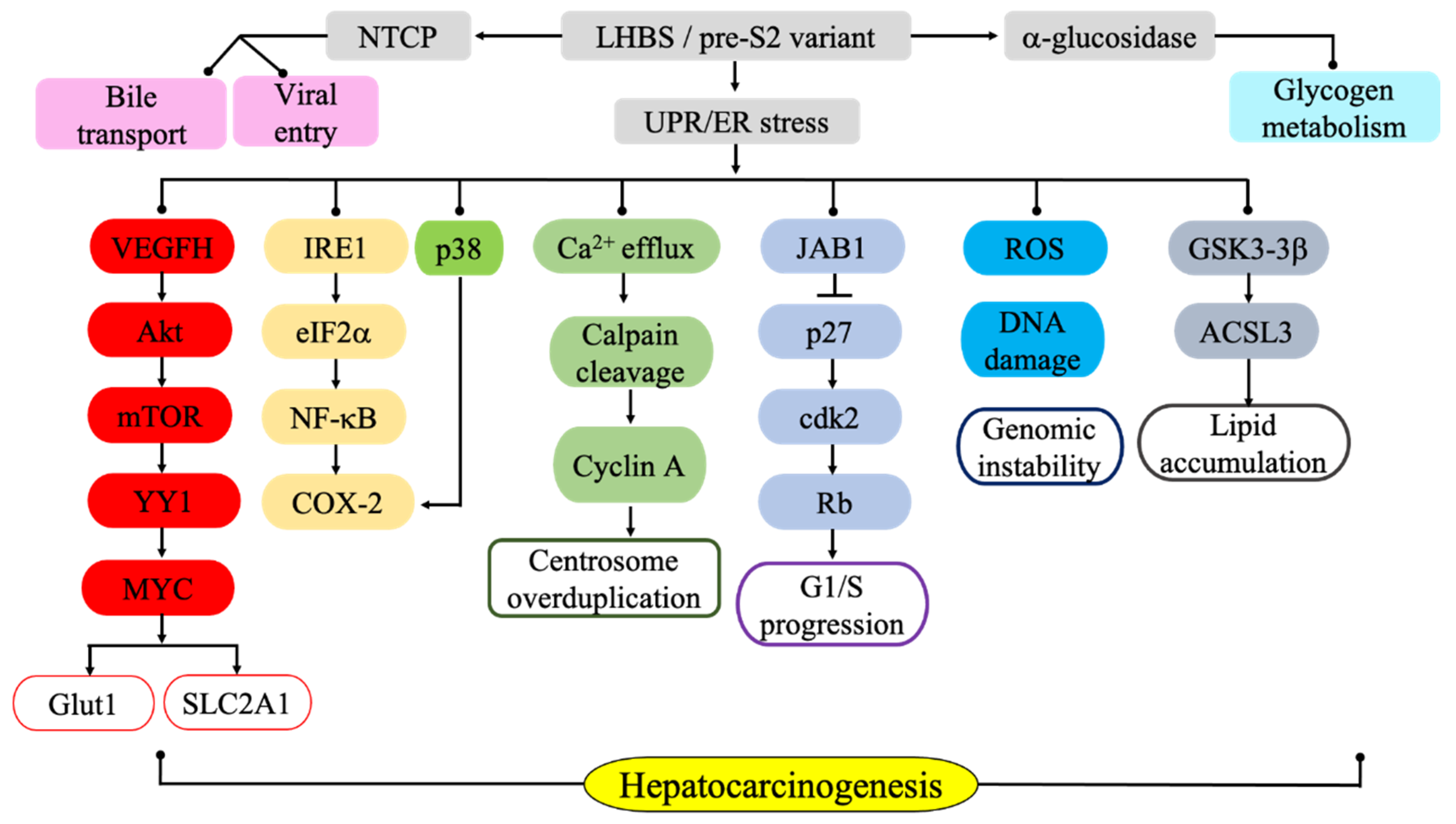
5. LHBs-induced Activation of Endoplasmic Reticulum Stress and Associated Carcinogenic Signaling Pathways
6. ER Stress-mediated Metabolic Pathways Associated with LHBs
7. LHBs Pre-S Genetic Variations in Ground Glass Hepatocytes Contribute to the ER Stress-mediated Carcinogenic Processes
7.1. Pre-S1 Deletion Variants
7.2. Pre-S2 Deletion Variants
8. LHBs in Infection Phases and HCC: Detection and Applications
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACSL | acyl-CoA synthetase |
| ACLY | ATP citrate lyase |
| AJCC | American Joint Committee on Cancer |
| C/EBPα/β | CCAAT-enhancer-binding protein α/β |
| CHOP | C/EBP homologous protein |
| CHB | Chronic hepatitis B |
| COX-2 | Cyclooxygenase-2 |
| CREB | cAMP-responsive element-binding |
| EOR | ER overload response |
| ER | Endoplasmic reticulum |
| ERAD | ER-associated protein degradation |
| ERSE | ER stress element |
| FADS2 | Fatty acid desaturase 2 |
| GGH | ground glass hepatocyte |
| GLUT | Glucose transporter |
| GSK | Glycogen synthase kinase |
| HBV | Hepatitis B virus |
| HIF-1 | Hypoxia-inducible factor 1 |
| HCC | hepatocellular carcinoma |
| JAB1 | c-jun activation domain binding protein 1 |
| LHBS | Large hepatitis B virus surface |
| MHBS | medium hepatitis B virus surface |
| NA | Nucleotide analog |
| NTCP | Sodium taurocholate cotransporting protein |
| OGG1 | Oxoguanine glycosylase 1 |
| PERK | Protein kinase R-like ER kinase |
| pg | Pregenomic |
| PTM | posttranslational modification |
| ROI | Reactive oxygen ion |
| ROS | Reactive oxygen species |
| SHBS | small hepatitis B virus surface |
| SNP | single nucleotide polymorphism |
| SLC2A1 | Solute carrier family 2 member 1 |
| SVP | Subviral particle |
| TM | transmembrane |
| UPR | unfolded protein response |
| VEGF | Vascular endothelial growth factor |
| XBP1 | X-box-binding protein 1 |
| XRCC1 | X-ray cross-complementation 1 |
| YY1 | Yin Yang 1 |
References
- World Health Organization (WHO). Global Hepatitis Report. 2017. Available online: https://apps.who.int/iris/bitstream/handle/10665/255016/9789241565455-eng.pdf (accessed on 1 August 2018).
- Arbuthnot, P.; Kew, M. Hepatitis B virus and hepatocellular carcinoma. Int. J. Exp. Pathol. 2001, 82, 77–100. [Google Scholar] [CrossRef]
- Inoue, T.; Tanaka, Y. Novel biomarkers for the management of chronic hepatitis B. Clin. Mol. Hepatol. 2020, 26, 261–279. [Google Scholar] [CrossRef]
- Chang, M.H.; Chen, C.J.; Lai, M.S.; Hsu, H.M.; Wu, T.C.; Kong, M.S.; Liang, D.C.; Shau, W.Y.; Chen, D.S. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N. Engl. J. Med. 1997, 336, 1855–1859. [Google Scholar] [CrossRef]
- Chang, M.H.; You, S.L.; Chen, C.J.; Liu, C.J.; Lee, C.M.; Lin, S.M.; Chu, H.C.; Wu, T.C.; Yang, S.S.; Kuo, H.S.; et al. Taiwan Hepatoma Study Group. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: A 20-year follow-up study. J. Natl. Cancer Inst. 2009, 101, 1348–1355. [Google Scholar] [CrossRef]
- Beasley, J.; Hwang, L.Y.; Lin, C.C.; Chien, C.S. Hepatocellular carcinoma and HBV. A prospective study of 22,707 men in Taiwan. Lancet 1981, 2, 1129–1133. [Google Scholar] [CrossRef]
- Blumberg, B.S.; Alter, H.J.; Visnich, S. A ‘‘new’’ antigen in leukemic sera. JAMA 1965, 191, 541–546. [Google Scholar] [CrossRef]
- Patient, R.; Hourioux, C.; Roingeard, P. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol. 2009, 11, 1561–1570. [Google Scholar] [CrossRef]
- Zuckerman, A.J. Hepatitis viruses. In Baron’s Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Seeger, C.; Zoulin, F.; Mason, W.S. Hepadnaviruses. In Field’s Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Churin, Y.; Roderfeld, M.; Roeb, E. Hepatitis B virus large surface protein: Function and fame. Hepatobiliary Surg. Nutr. 2015, 4, 1–10. [Google Scholar]
- Siegler, V.D.; Bruss, V. Role of transmembrane domains of hepatitis B virus small surface proteins in subviral-particle biogenesis. J. Virol. 2013, 87, 1491–1496. [Google Scholar] [CrossRef]
- Le Seyec, J.; Chouteau, P.; Cannie, I.; Guguen-Guillouzo, C.; Gripon, P. Role of the Pre-S2 Domain of the Large Envelope Protein in Hepatitis B Virus Assembly and Infectivity. J. Virol. 1998, 72, 5573–5578. [Google Scholar] [CrossRef]
- Bruss, V. Hepatitis B virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar] [CrossRef]
- Dobrica, M.-O.; Lazar, C.; Branza-Nichita, N. N-glycosylation and N-glycan processing in HBV biology and pathogenesis. Cells 2020, 9, 1404. [Google Scholar] [CrossRef]
- Hadziyannis, E.; Laras, A. Viral biomarkers in chronic HBeAg negative HBV infection. Genes 2018, 9, 469. [Google Scholar] [CrossRef]
- Hu, J.; Liu, K. Complete and incomplete hepatitis B virus particles: Formation, function, and application. Viruses 2017, 9, 56. [Google Scholar] [CrossRef]
- Lazarevic, I.; Banko, A.; Miljanovic, D.; Cupic, M. Immune-escape hepatitis B virus mutations associated with viral reactivation upon immunosuppression. Viruses 2019, 11, 778. [Google Scholar] [CrossRef]
- Heim, K.; Neumann-Haefelin, C.; Thimme, R.; Hofmann, M. Heterogeneity of HBV-specific CD8+ T-cell failure: Implications for immunotherapy. Front. Immunol. 2019, 10, 2240. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Wakita, T. Hepatitis B virus and hepatitis D virus entry, species specificity, and tissue tropism. Cold Spring Harb. Perspect. Med. 2015, 5, a021378. [Google Scholar] [CrossRef]
- Sumiya, M.; Liu, Q.; Yoshimoto, N.; Lijima, M.; Tatematsu, K.; Nakai, T.; Okajima, T.; Kuroki, K.; Ueda, K.; Kuroda, S. Cellular uptake of hepatitis B virus envelope L particles is independent of sodium taurocholate cotransporting polypeptide, but dependent on heparan sulfate proteoglycan. Virology 2016, 497, 23–32. [Google Scholar] [CrossRef]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, e00049. [Google Scholar] [CrossRef]
- Sun, Y.; Qi, Y.; Peng, B.; Li, W. NTCP-reconstituted in vitro HBV infection system. Methods Mol. Biol. 2017, 1540, 1–14. [Google Scholar]
- Falco, S.D.; Ruvo, M.; Verdoliva, A.; Scarallo, A.; Raimondo, D.; Raucci, A.; Fassina, G. N-terminal myristylation of HBV preS1 domain enhances receptor recognition. J. Pept. Res. 2001, 57, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Glebe, D.; Urban, S. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 2007, 13, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Falth, M.; Stindt, J.; Koniger, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Peng, B.; Liu, Y.; Xu, G.; He, W.; Ren, B.; Jling, Z.; Sui, J.; Li, W. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J. Virol. 2014, 88, 3273–3284. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wu, L.; Xu, W.; Liu, Y.; Zhen, L.; Ning, G.; Song, J.; Jiao, Q.; Zheng, Y.; Chen, T. Diverse effects of the NTCP p.Ser267Phe variant on disease progression during chronic HBV infection and on HBV preS1 variability. Front. Cell. Infect. Microbiol. 2019, 9, 18. [Google Scholar] [CrossRef]
- Lee, H.W.; Park, H.J.; Jin, B.; Dezhbord, M.; Kim, D.Y.; Han, K.-H.; Ryu, W.-S.; Kim, S.; Ahn, S.H. Effect of S267F variant of NTCP on the patients with chronic hepatitis B. Sci. Rep. 2017, 7, 17634. [Google Scholar] [CrossRef]
- König, A.; Döring, B.; Mohr, C.; Geipel, A.; Geyer, J.; Glebe, D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J. Hepatol. 2014, 61, 867–875. [Google Scholar] [CrossRef]
- Yang, F. Post-translational modification control of HBV biological processes. Front. Microbiol. 2018, 9, 2661. [Google Scholar] [CrossRef]
- Lambert, C.; Prange, R. Posttranslational N-glycosylation of the hepatitis B virus large envelope protein. Virol. J. 2007, 4, 45. [Google Scholar] [CrossRef]
- Hyakumura, M.; Walsh, R.; Thaysen-Andersen, M.; Kingston, N.J.; La, M.; Lu, L.; Lovrecz, G.; Packer, N.H.; Locarnini, S.; Netter, H.J. Modification of asparagine-linked glycan density for the design of hepatitis B virus virus-like particles with enhanced immunogenicity. J. Virol. 2015, 89, 11312–11322. [Google Scholar] [CrossRef]
- Suffner, S.; Gerstenberg, N.; Patra, M.; Ruibal, P.; Orabi, A.; Schindler, M.; Bruss, V. Domains of the hepatitis B Virus Small Surface Protein S Mediating Oligomerization. J. Virol. 2018, 92, e02232-17. [Google Scholar] [CrossRef] [PubMed]
- Le, S.I.; Thedja, M.D.; Roni, M.; Muljono, D.H. Prediction of conformational changes by single mutation in the hepatitis B virus surface antigen (HBsAg) identified in HBsAg-negative blood donors. Virol. J. 2010, 7, 326. [Google Scholar]
- Purdy, M.A. Hepatitis B virus S gene escape mutants. Asian J. Transfus. Sci. 2007, 1, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Sureau, C.; Salisse, J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology 2013, 57, 985–994. [Google Scholar] [CrossRef]
- Julithe, R.; Abou-Jaoudé, G.; Sureau, C. Modification of the hepatitis B virus envelope protein glycosylation pattern interferes with secretion of viral particles, infectivity, and susceptibility to neutralizing antibodies. J. Virol. 2014, 88, 9049–9059. [Google Scholar] [CrossRef]
- Cai, X.; Zheng, W.; Pan, S.; Zhang, S.; Xie, Y.; Guo, H.; Wang, G.; Li, Z.; Luo, M. A virus-like particle of the hepatitis B virus preS antigen elicits robust neutralizing antibodies and T cell responses in mice. Antivir. Res. 2018, 149, 48–57. [Google Scholar] [CrossRef]
- Pastor, F.; Herrscher, C.; Patient, R.; Eymieux, S.; Moreau, A.; Burlaud-Gaillard, J.; Seigneuret, F.; de Rocquigny, H. Direct interaction between the hepatitis B virus core and envelope proteins analyzed in a cellular context. Sci. Rep. 2019, 9, 16178. [Google Scholar] [CrossRef]
- Choi, Y.-M.; Lee, S.-Y.; Kim, B.-J. Naturally occurring hepatitis B virus mutations leading to endoplasmic reticulum stress and their contribution to the progression of hepatocellular carcinoma. Int. J. Mol. Sci. 2019, 20, 597. [Google Scholar] [CrossRef]
- Liu, W.; Cao, Y.; Wang, T.; Xiang, G.; Lu, J.; Zhang, J.; Hou, P. The N-glycosylation modification of LHBs (large surface proteins of HBV) effects on endoplasmic reticulum stress, cell proliferation and its secretion. Hepat. Mon. 2013, 13, e12280. [Google Scholar] [CrossRef]
- Lazar, C.; Uta, M.; Branza-Nichita, N. Modulation of the unfolded protein response by the human hepatitis B virus. Front. Microbiol. 2014, 5, 433. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kyaw, Y.Y.; Cheong, J. Functional interaction of endoplasmic reticulum stress and hepatitis B virus in the pathogenesis of liver diseases. World J. Gastroenterol. 2017, 23, 7657–7665. [Google Scholar] [CrossRef]
- Wang, H.C.; Huang, W.; Lai, M.D.; Su, I.J. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006, 97, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Wu, H.C.; Chen, C.F.; Fausto, N.; Lei, H.Y.; Su, I.J. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am. J. Pathol. 2003, 163, 2441–2449. [Google Scholar] [CrossRef]
- Lazar, C.; Macovei, A.; Petrescu, S.; Branza-Nichita, N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS ONE 2012, 7, e34169. [Google Scholar] [CrossRef]
- Tang, H.; Da, L.; Mao, Y.; Li, Y.; Li, D.; Xu, Z.; Li, F.; Wang, Y.; Tiollais, P.; Li, T.; et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology 2009, 49, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.F.; Lu, C.C.; Chen, W.C.; Yao, W.J.; Wang, H.C.; Chang, T.T.; Lei, H.Y.; Shiau, A.L.; Su, I.J. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology 2001, 33, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Su, I.-J.; Wang, H.-C.; Wu, H.-C.; Huang, W. Ground glass hepatocytes contain pre-S mutants and represent preneoplastic lesions in chronic hepatitis B virus infection. J. Gastroenterol. Hepatol. 2008, 23, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Su, I.J.; Wang, H.C.; Chang, W.W.; Lei, H.Y.; Lai, M.D.; Chang, W.T.; Huang, W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis 2004, 25, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Siddiquey, M.N.A.; Zhang, H.; Nguyen, C.C.; Domma, A.J.; Kamil, J.P. The human cytomegalovirus endoplasmic reticulum-resident glycoprotein UL148 activates the unfolded protein response. J. Virol. 2018, 92, e00896-18. [Google Scholar] [CrossRef]
- Yao, W.; Cai, H.; Li, X.; Li, T.; Hu, L.; Peng, T. Endoplasmic reticulum stress links hepatitis C virus RNA replication to wild-type PGC-1α/liver-specific PGC-1α upregulation. J. Virol. 2014, 88, 8361–8374. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Endoplasmic reticulum and the unfolded protein response: Dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 2013, 301, 215–290. [Google Scholar] [PubMed]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef]
- Ruggiano, A.; Foresti, O.; Carvalho, P. ER-associated degradation: Protein quality control and beyond. J. Cell Biol. 2014, 204, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, H.; Nishitoh, H. Signaling pathways from the endoplasmic reticulum and their roles in disease. Genes 2013, 4, 306–333. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.H.; Su, I.J.; Lei, H.Y.; Wang, H.C.; Lin, W.C.; Chang, W.T.; Huang, W.; Chang, W.C.; Chang, Y.S.; Chen, C.C.; et al. Endoplasmic reticulum stress stimulates the expression of cyclooxygenase-2 through activation of NF-kappaB and pp38 mitogen-activated protein kinase. J. Biol. Chem. 2004, 279, 46384–46392. [Google Scholar] [CrossRef]
- Liu, H.; Xu, J.; Zhou, L.; Yun, X.; Chen, L.; Wang, S.; Sun, L.; Wen, Y.; Gu, J. Hepatitis B virus large surface antigen promotes liver carcinogenesis by activating the Src/PI3K/Akt pathway. Cancer Res. 2011, 71, 7547–7557. [Google Scholar] [CrossRef] [PubMed]
- Tai, P.-C.; Suk, F.M.; Gerlich, W.; Neurath, R.; Shih, C. Hypermodification and immune escape of an internally deleted middle envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology 2002, 292, 44–58. [Google Scholar] [CrossRef]
- Chua, P.K.; Wang, Y.L.; Lin, M.H.; Masuda, T.; Suk, F.M.; Shih, C. Reduced secretion of virions and hepatitis B virus (HBV) surface antigen of a naturally occurring HBV variant correlates with the accumulation of the small S envelope protein in the endoplasmic reticulum and Golgi apparatus. J. Virol. 2005, 79, 13483–13496. [Google Scholar] [CrossRef]
- Schoeman, J.C.; Hou, J.; Harms, A.C.; Vreeken, R.J.; Berger, R.; Hankemeier, T.; Boonstra, A. Metabolic characterization of the natural progression of chronic hepatitis B. Genome Med. 2016, 8, 64. [Google Scholar] [CrossRef]
- Ghemrawi, R.; Battaglia-Hsu, S.-F.; Arnold, C. Endoplasmic reticulum stress in metabolic disorders. Cells 2018, 7, 63. [Google Scholar] [CrossRef]
- Hung, J.H.; Yan, C.W.; Su, I.J.; Wang, H.C.; Lei, H.Y.; Lin, W.C.; Chang, W.T.; Huang, W.; Lu, T.J.; Lai, M.D. Hepatitis B virus surface antigen interacts with acid alpha-glucosidase and alters glycogen metabolism. Hepatol. Res. 2010, 40, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Uemura, A.; Oku, M.; Mori, K.; Yoshida, H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 2009, 122, 2877–2886. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, B.; Ceña, V.; Posadas, I. The endoplasmic reticulum stress and the HIF-1 signalling pathways are involved in the neuronal damage caused by chemical hypoxia. Br. J. Pharmacol. 2015, 172, 2838–2851. [Google Scholar] [CrossRef] [PubMed]
- Gonen, N.; Meller, A.; Sabath, N.; Shalgi, R. Amino acid biosynthesis regulation during endoplasmic reticulum sgtress is coupled to protein expression demands. iScience 2019, 19, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Kasai, S.; Yamazaki, H.; Tanji, K.; Engler, M.J.; Matsumiya, T.; Itoh, K. Role of the ISR-ATF4 pathway and its cross talk with Nrf2 in mitochondrial quality control. J. Clin. Biochem. Nutr. 2019, 64, 1–12. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Hinoi, E.; Jung, D.Y.; Kajimura, D.; Ferron, M.; Seo, J.; Graff, J.M.; Kim, J.K.; Karsenty, G. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. J. Clin. Investig. 2009, 119, 2807–2817. [Google Scholar] [CrossRef]
- Teng, C.-F.; Hsieh, W.-C.; Wu, H.-C.; Lin, Y.-J.; Tsai, H.-W.; Huang, W.; Su, I.-J. Hepatitis B virus pre-S2 mutant induces aerobic glycolysis through mammalian target of rapamycin signal cascade. PLoS ONE 2015, 10, e0122373. [Google Scholar] [CrossRef]
- Teng, C.-F.; Wu, H.-C.; Tsai, H.-W.; Shiah, H.-S.; Huang, W.; Su, I.-H. Novel feedback inhibition of surface antigen synthesis by mammalian target of rapamycin (mTOR) signal and its implication for hepatitis B virus tumorigenesis and therapy. Hepatology 2011, 54, 1199–1207. [Google Scholar] [CrossRef]
- van der Harg, J.M.; van Heest, J.C.; Bangel, F.N.; Patiwael, S.; van Weering, J.R.; Scheper, W. The UPR reduces glucose metabolism via IRE1 signaling. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 655–665. [Google Scholar] [CrossRef]
- Amen, O.M.; Sarker, S.D.; Ghildyal, R.; Arya, A. Endoplasmic reticulum stress activates unfolded protein response signaling and mediates inflammation, obesity, and cardiac dysfunction: Therapeutic and molecular approach. Front. Pharmacol. 2019, 10, 977. [Google Scholar] [CrossRef]
- Oyadomari, S.; Harding, H.P.; Zhang, Y.; Oyadomari, M.; Ron, D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008, 7, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Tsai, C.T.; Huangfu, C.A.; Huang, W.; Lei, H.Y.; Lin, C.F.; Su, I.J.; Chang, W.T.; Wu, P.H.; Chen, Y.T.; et al. ACSL3 and GSK-3β are essential for lipid upregulation induced by endoplasmic reticulum stress in liver cells. J. Cell Biochem. 2011, 112, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.F.; Wu, H.C.; Hsieh, W.C.; Tsai, H.W.; Su, I.J. Activation of ATP citrate lyase by mTOR signal induces disturbed lipid metabolism in hepatitis B virus pre-S2 mutant tumorigenesis. J. Virol. 2015, 89, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Stieger, B. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb. Exp. Pharmacol. 2011, 201, 205–259. [Google Scholar]
- Chiang, J.Y.L. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar]
- Slijepcevic, D.; Kaufman, C.; Wichers, C.G.K.; Gilglioni, E.H.; Lempp, F.A.; Duijst, S.; de Waart, D.R.; Elferink, R.P.J.O.; Mier, W.; Stieger, B.; et al. Impaired uptake of conjugated bile acids and hepatitis b virus pres1-binding in na(+) -taurocholate cotransporting polypeptide knockout mice. Hepatology 2015, 62, 207–219. [Google Scholar] [CrossRef]
- Blank, A.; Eidam, A.; Haag, M.; Hohmann, N.; Burhenne, J.; Schwab, M.; de Graaf, S.V.; Meyer, M.R.; Maurer, H.H.; Meier, K.; et al. The NTCP-inhibitor Myrcludex B: Effects on bile acid disposition and tenofovir pharmacokinetics. Clin. Pharmacol. Ther. 2018, 103, 341–348. [Google Scholar] [CrossRef]
- Osiowy, C.; Giles, E.; Tanaka, Y.; Mizokami, M.; Minuk, G.Y. Molecular evolution of hepatitis B virus over 25 years. J. Virol. 2006, 80, 10307–10314. [Google Scholar] [CrossRef]
- Clark, D.N.; Hu, J. Unveiling the roles of HBV polymerase for new antiviral strategies. Future Virol. 2015, 10, 283–295. [Google Scholar] [CrossRef]
- Coffin, C.S.; Mulrooney-Cousins, P.M.; Peters, M.G.; van Marle, G.; Roberts, J.P.; Michalak, T.I.; Terrault, N.A. Molecular characterization of intrahepatic and extrahepatic hepatitis B virus (HBV) reservoirs in patients on suppressive antiviral therapy. J. Viral Hepat. 2011, 18, 415–423. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Wu, C.-C.; Chen, X.-W.; Li, X.; Li, J.; Lu, M.-J. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J. Gastroenterol. 2016, 22, 126–144. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.-A.; Do, S.Y.; Kim, B.-J. Precore/core region mutations of hepatitis B virus related to clinical severity. World J. Gastroenterol. 2016, 22, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Su, I.J.; Yen, C.J.; Liu, Y.R.; Liu, R.J.; Hsieh, W.C.; Tsai, H.W.; Wang, L.H.C.; Hsu, C.C.; Huang, W. Hepatitis B virus pre-S2 mutant surface protein inhibits DNA double strand break repair and leads to genome instability in hepatitis B virus hepatocarcinogenesis. J. Pathol. 2015, 236, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Su, I.J.; Wang, H.C.; Tsai, J.H.; Huang, Y.J.; Chang, W.W.; Lai, M.D.; Lei, H.Y.; Huang, W. Hepatitis B virus pre-S2 mutant surface antigen induces degradation of cyclin-dependent kinase inhibitor p27Kip1 through c-Jun activation domain-binding protein 1. Mol. Cancer Res. 2007, 5, 1063–1072. [Google Scholar] [CrossRef]
- Chen, B.-F.; Liu, C.-J.; Jow, G.-M.; Cheng, P.-J.; Kao, J.-H.; Chen, D.-S. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology 2006, 130, 1153–1168. [Google Scholar] [CrossRef]
- Hadziyannis, S.; Gerber, M.A.; Vissoulis, C.; Popper, H. Cytoplasmic hepatitis B antigen in “ground-glass” hepatocytes of carriers. Arch. Pathol. 1973, 96, 327–330. [Google Scholar]
- Callea, F.; De Vos, R.; Togni, R.; Tardanico, R.; Vanstapel, M.J.; Desmet, V.J. Fibrinogen inclusions in liver cells: A new type of ground-glass hepatocyte. Immune light and electron microscopic characterization. Histopathology 1986, 10, 65–73. [Google Scholar] [CrossRef]
- Fan, Y.F.; Lu, C.C.; Chang, Y.C.; Lin, P.W.; Su, I.J. Identification of a pre-S2 mutant in hepatocytes expressing a novel marginal pattern of surface antigen in advanced diseases of chronic hepatitis B virus infection. J. Gastroenterol. Hepatol. 2000, 15, 519–528. [Google Scholar] [CrossRef]
- Prange, R.; Streeck, R.E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995, 14, 247–256. [Google Scholar] [CrossRef]
- Shen, F.-C.; Su, I.-J.; Wu, H.-C.; Hsieh, Y.-H.; Yao, W.-J.; Young, K.-C.; Chang, T.-C.; Hsieh, H.-C.; Tsai, H.-N.; Huang, W. A Pre-S Gene Chip to detect the pre-S deletions in the hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in the chronic HBV carriers. J. Biomed. Sci. 2009, 16, 84–91. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chang, W.-T.; Chang, W.-W.; Wu, H.-C.; Huang, W.; Lei, H.-Y.; Lai, M.-D.; Fausto, N.; Su, I.-J. Hepatitis B virus pre-S2 mutant upregulates cyclin A expression and induces nodular proliferation of hepatocytes. Hepatology 2005, 41, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-C.; Teng, C.-F.; Wu, H.-C.; Tsai, H.-W.; Chuang, H.-C.; Tsai, T.-F.; Hsu, Y.-H.; Huang, W.; Wu, L.-W.; Su, I.-J. Enhanced expression of vascular endothelial growth factor-A in ground glass hepatocytes and its implication in hepatitis B virus hepatocarcinogenesis. Hepatology 2009, 49, 1962–1971. [Google Scholar] [CrossRef]
- Viganò, M.; Lampertico, P. Clinical implications of HBsAg quantification in patients with chronic hepatitis B. Saudi J. Gastroenterol. 2012, 18, 81–86. [Google Scholar] [PubMed]
- Pfefferkorn, M.; Böhm, S.; Schott, T.; Deichsel, D.; Bremer, C.M.; Schröder, K.; Gerlich, W.H.; Glebe, D.; Berg, T.; van Bömmel, F. Quantification of large and middle proteins of hepatitis B virus surface antigen (HBsAg) as a novel tool for the identification of inactive HBV carriers. Gut 2018, 67, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Patient, R.; Hourioux, C.; Sizaret, P.-Y.; Trassard, S.; Sureau, C.; Roingeard, P. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 2007, 81, 3842–3851. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, J.; Lu, Y.; Luo, S.; Zhang, J.; Zhu, P. Cryo-EM structure of native spherical subviral particles isolated from HBV carriers. Virus Res. 2019, 259, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V.; Gerhardt, E.; Vieluf, K.; Wunderlich, G. Functions of the large hepatitis B virus surface protein in viral particle morphogenesis. Intervirology 1996, 39, 23–31. [Google Scholar] [CrossRef]
- Raney, A.K.; Easton, A.J.; Milich, D.R.; McLachlan, A. Promoter-specific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. J. Virol. 1991, 65, 5774–5781. [Google Scholar] [CrossRef]
- Hildt, E.; Hofschneider, P.H. The PreS2 activators of the hepatitis B virus: Activators of tumour promoter pathways. Recent Res. Cancer Res. 1998, 154, 315–329. [Google Scholar]
- Chen, L.; Hou, J.; Wei, H.-S.; Liu, A.-X.; Li, P.-R.; Zhao, J.; Liu, J.; Li, B.-A. Characterization and clinical application of a monoclonal antibody to hepatitis B virus large surface proteins. Front. Lab. Med. 2017, 1, 31–35. [Google Scholar] [CrossRef]
- Zhu, X.; Gong, Q.; Yu, D.; Zhang, D.; Gu, L.; Han, Y.; Chen, J.; Zhang, Y.; Zhang, X. Early serum hepatitis B virus large surface protein level: A stronger predictor of virological response to peginterferon alfa-2a than that to entecavir in HBeAg-positive patients with chronic hepatitis B. J. Clin. Virol. 2013, 57, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.J.; Ai, Y.L.; Tsai, H.W.; Chan, S.H.; Yen, C.S.; Cheng, K.H.; Lee, Y.P.; Kao, C.W.; Wang, Y.C.; Chen, Y.L.; et al. Hepatitis B virus surface gene pre-S2 mutant as a high-risk serum marker for hepatoma recurrence after curative hepatic resection. Hepatology 2018, 68, 815–826. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, W.-L.; Hung, J.-H.; Huang, W. Association of the Hepatitis B Virus Large Surface Protein with Viral Infectivity and Endoplasmic Reticulum Stress-mediated Liver Carcinogenesis. Cells 2020, 9, 2052. https://doi.org/10.3390/cells9092052
Lin W-L, Hung J-H, Huang W. Association of the Hepatitis B Virus Large Surface Protein with Viral Infectivity and Endoplasmic Reticulum Stress-mediated Liver Carcinogenesis. Cells. 2020; 9(9):2052. https://doi.org/10.3390/cells9092052
Chicago/Turabian StyleLin, Wei-Ling, Jui-Hsiang Hung, and Wenya Huang. 2020. "Association of the Hepatitis B Virus Large Surface Protein with Viral Infectivity and Endoplasmic Reticulum Stress-mediated Liver Carcinogenesis" Cells 9, no. 9: 2052. https://doi.org/10.3390/cells9092052
APA StyleLin, W.-L., Hung, J.-H., & Huang, W. (2020). Association of the Hepatitis B Virus Large Surface Protein with Viral Infectivity and Endoplasmic Reticulum Stress-mediated Liver Carcinogenesis. Cells, 9(9), 2052. https://doi.org/10.3390/cells9092052





