Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction
Abstract
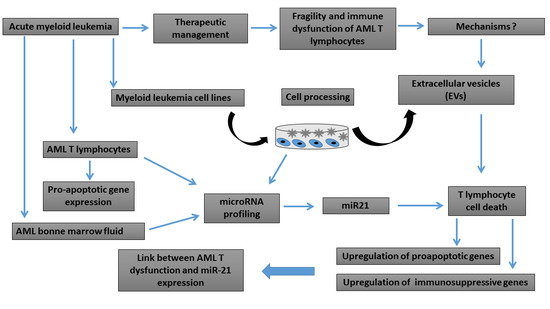
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Bone Marrow and Peripheral Blood T Lymphocyte Isolation
2.3. Cell Culture and Cell Death Assay
2.4. Cell Transduction
2.5. Extracellular Vesicle Purification and Analysis
2.6. Bone Marrow Bodily Fluid Sampling and miRNA Extraction
2.7. MicroRNA Expression Profile
2.8. TaqMan miRNA Assay for Individual miRNAs
2.9. Transcriptomic Profile Array Analysis
2.10. Reverse Transcription-Quantitative Polymerase Chain Reaction
2.11. Gene Expression Array Data Analysis
2.12. Statistical Analysis
3. Results
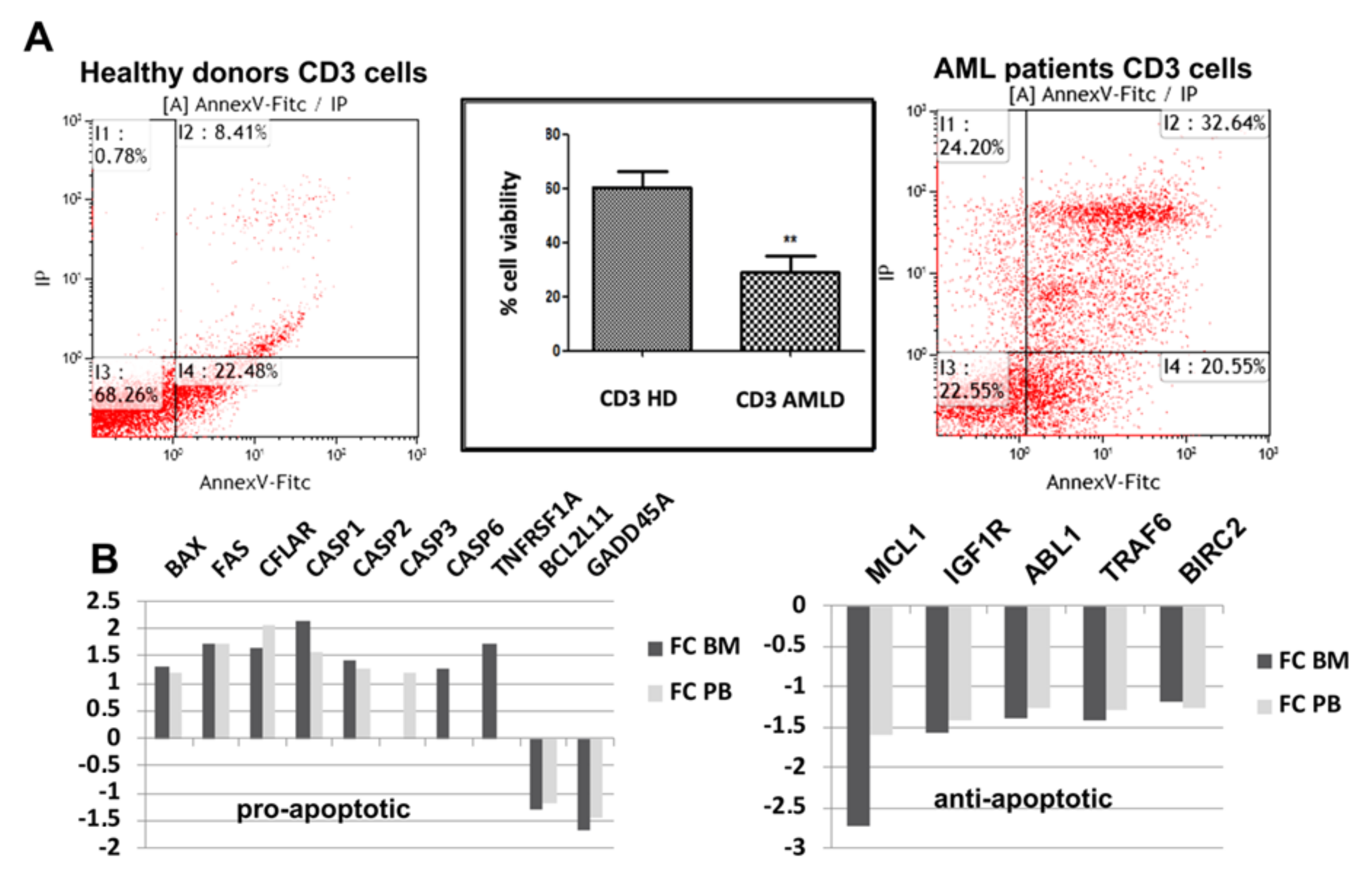
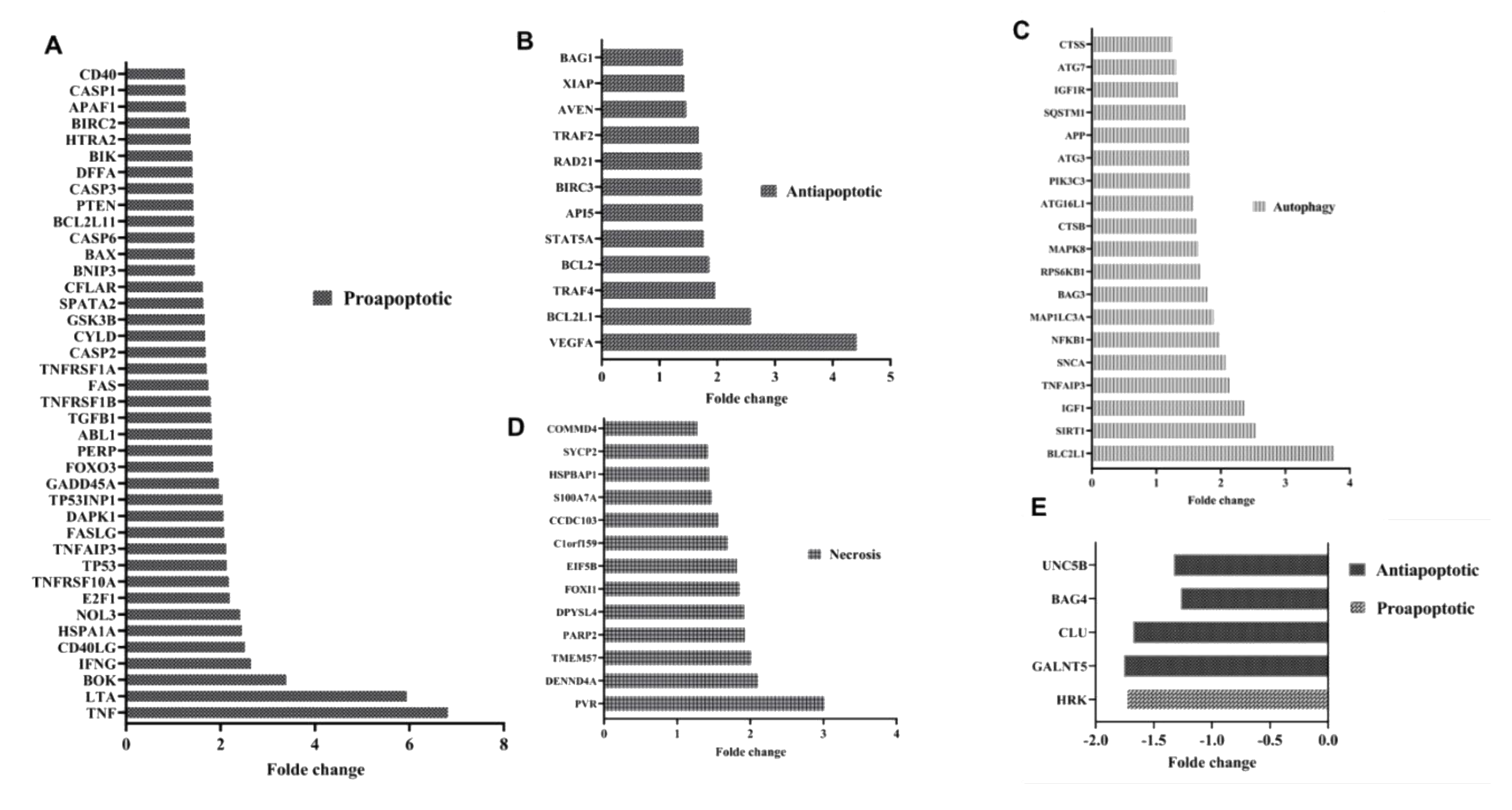
3.1. Apoptotic Pathways Induced Fragility of Aml T Lymphocytes
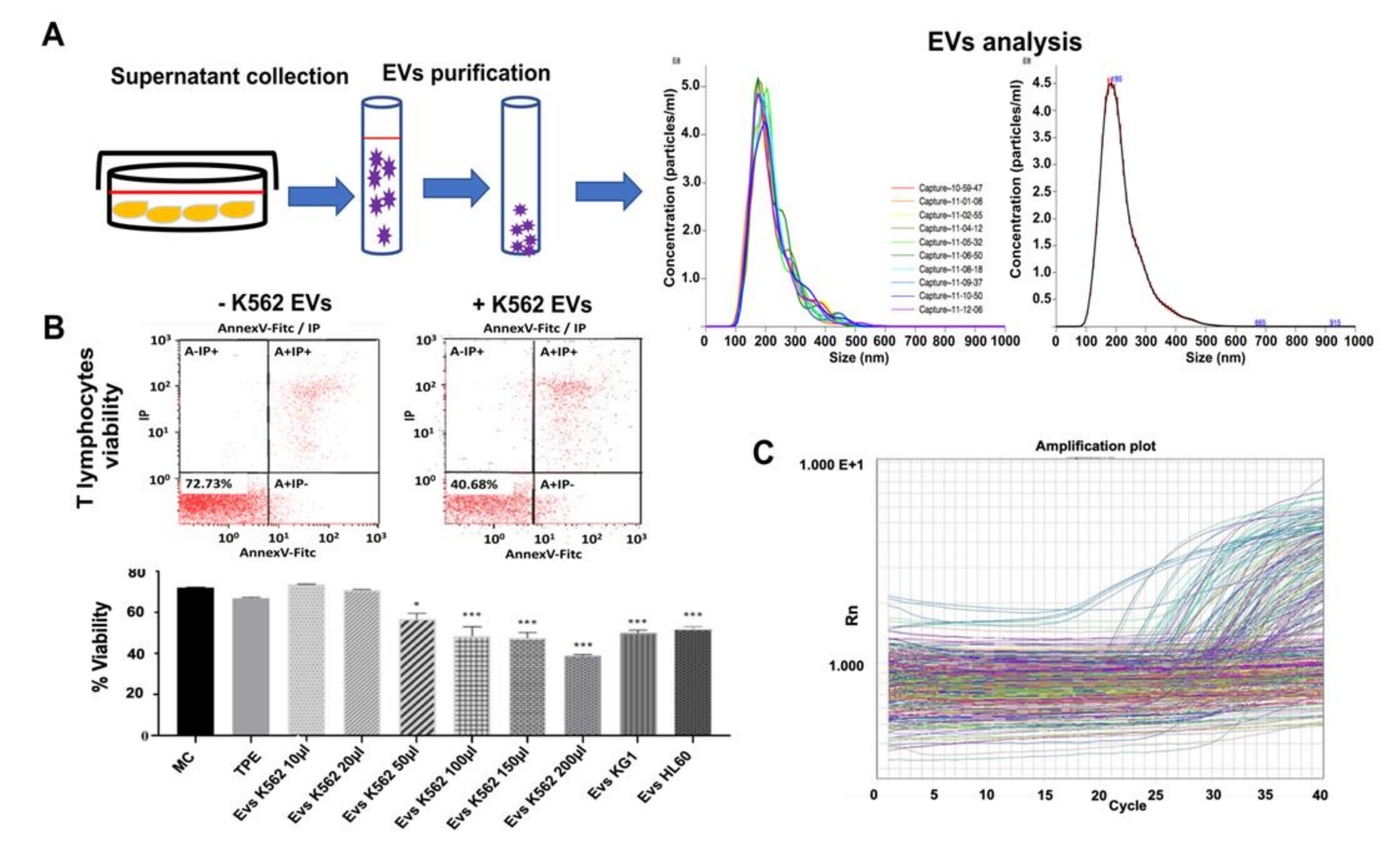
3.2. Myeloid Leukemia Cell Line-Derived Extracellular Vesicles Induce T Lymphocyte Cell Death
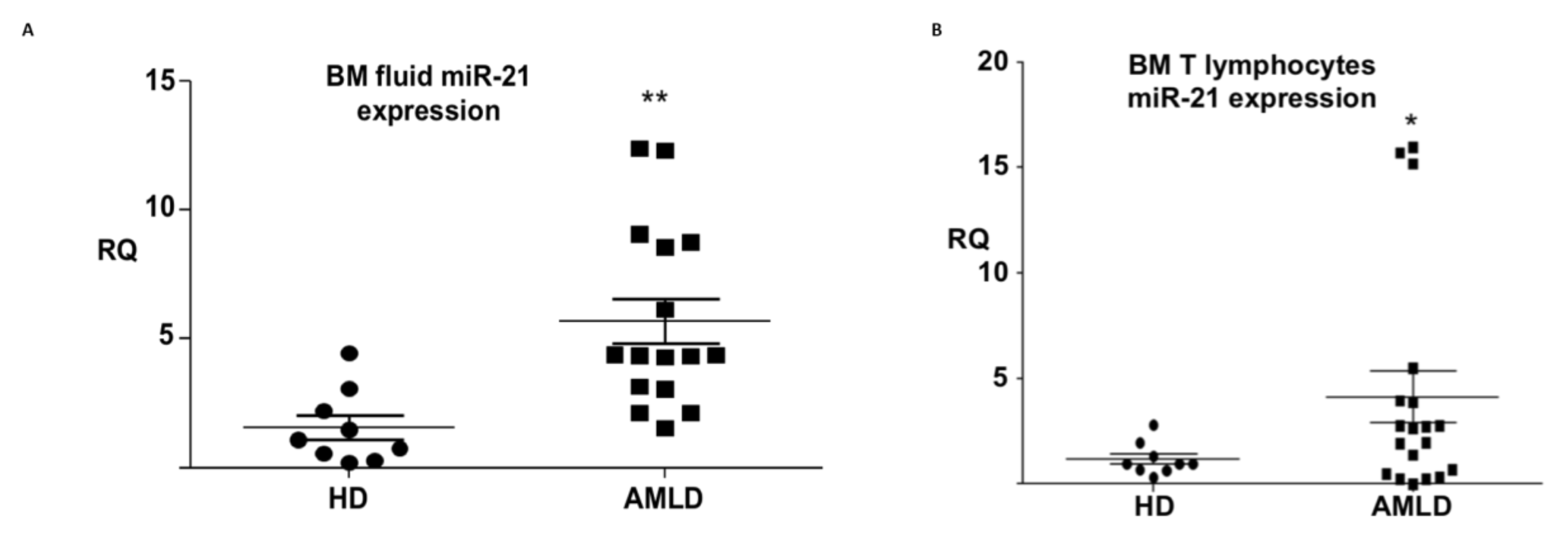
3.3. mir-21 Expression in the Aml Bone Marrow Tumor Microenvironment
3.4. mir-21 Induces T Lymphocyte Death Through Apoptotic Pathways
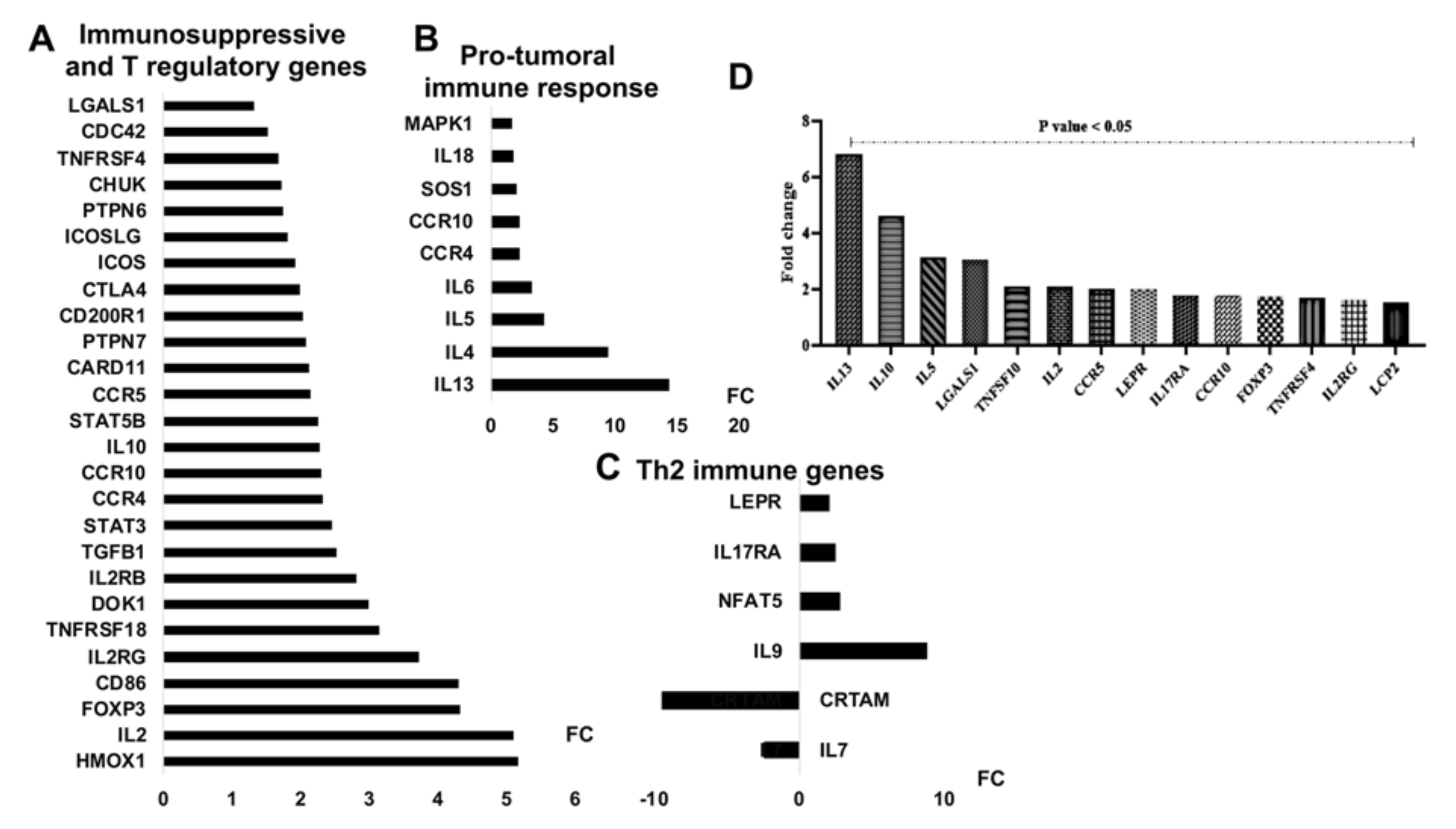
3.5. mir-21 Expression Increases T Cell Immunosuppressive Phenotypic Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson-Moncada, J.; Viboch, E.; Church, S.E.; Warren, S.E.; Rutella, S. Dissecting the Immune Landscape of Acute Myeloid Leukemia. Biomedicines 2018, 6, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molodtsov, A.; Turk, M.J. Tissue Resident CD8 Memory T Cell Responses in Cancer and Autoimmunity. Front. Immunol. 2018, 29, 2810. [Google Scholar] [CrossRef] [PubMed]
- Baxevanis, C.N.; Fortis, S.P.; Perez, S.A. The balance between breast cancer and the immune system: Challenges for prognosis and clinical benefit from immunotherapies. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Terme, M.; Tanchot, C. Immune system and tumors. Ann. Pathol. 2017, 37, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Toffalori, C.; Zito, L.; Gambacorta, V.; Riba, M.; Oliveira, G.; Bucci, G.; Barcella, M.; Spinelli, O.; Greco, R.; Crucitti, L.; et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat. Med. 2019, 25, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Vago, L.; Gojo, I. Immune escape and immunotherapy of acute myeloid leukemia. J. Clin. Investig. 2020, 130, 1552–1564. [Google Scholar] [CrossRef]
- Shang, Y.; Zhou, F. Current Advances in Immunotherapy for Acute Leukemia: An Overview of Antibody, Chimeric Antigen Receptor, Immune Checkpoint, and Natural Killer. Front. Oncol. 2019, 19, 917. [Google Scholar] [CrossRef]
- Mardiana, S.; Gill, S. CAR T Cells for Acute Myeloid Leukemia: State of the Art and Future Directions. Front. Oncol. 2020, 6, 697. [Google Scholar] [CrossRef]
- Ingenito, F.; Roscigno, G.; Affinito, A.; Nuzzo, S.; Scognamiglio, I.; Quintavalle, C.; Condorelli, G. The Role of Exo-miRNAs in Cancer: A Focus on Therapeutic and Diagnostic Applications. Int. J. Mol. Sci. 2019, 20, 4687. [Google Scholar] [CrossRef] [Green Version]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Emamgolizadeh Gurt Tapeh, B.; Mosayyebi, B.; Samei, M.; Beyrampour Basmenj, H.; Mohammadi, A.; Alivand, M.R.; Hassanpour, P.; Solali, S. microRNAs involved in T-cell development, selection, activation, and hemostasis. J. Cell Physiol. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.A.; O’Connell, R.M. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood 2017, 130, 1290–1301. [Google Scholar] [CrossRef] [Green Version]
- Gabra, M.M.; Salmena, L. microRNAs and Acute Myeloid Leukemia Chemoresistance: A Mechanistic Overview. Front. Oncol. 2017, 30, 255. [Google Scholar] [CrossRef]
- Panagal, M.; Senthil Kumar, S.R.; Sivakurunathan, P.; Biruntha, M.; Karthigeyan, M.; Gopinathe, V.; Sivakumare, P.; Sekar, D. MicroRNA21 and the various types of myeloid leukemia. Cancer Gene Therapy 2018, 25, 161–166. [Google Scholar] [CrossRef]
- Li, C.; Yan, H.; Yin, J.; Ma, J.; Liao, A.; Yang, S.; Wang, L.; Huang, Y.; Lin, C.; Dong, Z.; et al. MicroRNA-21 promotes proliferation in acute myeloid leukemia by targeting Kruppel-like factor 5. Oncol. Lett. 2019, 18, 3367–3372. [Google Scholar]
- Rouas, R.; Fayyad-Kazan, H.; El Zein, N.; Lewalle, P.; Rothe, F.; Simion, A.; Akl, H.; Mourtada, M.; El Rifai, M.; Burny, A.; et al. Human natural Treg microRNA signature: Role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur. J. Immunol. 2009, 39, 1608–1618. [Google Scholar] [CrossRef]
- Fayyad-Kazan, H.; Rouas, R.; Merimi, M.; El Zein, N.; Lewalle, P.; Jebbawi, F.; Mourtada, M.; Badran, H.; Ezzeddine, M.; Salaun, B.; et al. Valproate treatment of human cord blood CD4-positive effector T cells confers on them the molecular profile (microRNA signature and FOXP3 expression) of natural regulatory CD4-positive cells through inhibition of histone deacetylase. J. Biol. Chem. 2010, 285, 20481–20491. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Wang, C.; Li, Y.; Zhao, J.; Chen, C.; Zhou, Y.; Tao, Y.; Guo, M.; Qin, N.; Ren, T.; et al. MiR-21 controls in situ expansion of CCR6( + ) regulatory T cells through PTEN/AKT pathway in breast cancer. Immunol. Cell Biol. 2015, 93, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Caescu, C.I.; Guo, X.; Tesfa, L.; Bhagat, T.D.; Verma, A.; Zheng, D.; Stanley, E.R. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood 2015, 125, e1-13. [Google Scholar] [CrossRef] [PubMed]
- Samiei, H.; Sadighi-Moghaddam, B.; Mohammadi, S.; Gharavi, A.; Abdolmaleki, S.; Khosravi, A.; Kokhaei, P.; Bazzazi, H.; Memarian, A. Dysregulation of helper T lymphocytes in esophageal squamous cell carcinoma (ESCC) patients is highly associated with aberrant production of miR-21. Immunol. Res. 2019, 67, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Rouas, R.; Merimi, M.; Najar, M.; El Zein, N.; Fayyad-Kazan, M.; Berehab, M.; Agha, D.; Bron, D.; Burny, A.; Rachidi, W.; et al. Human CD8(+) CD25 (+) CD127 (low) regulatory T cells: microRNA signature and impact on TGF-beta and IL-10 expression. J. Cell Physiol. 2019, 234, 17459–17472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeets, A.; Daemen, A.; Vanden Bempt, I.; Gevaert, O.; Claes, B.; Wildiers, H.; Drijkoningen, R.; Van Hummelen, P.; Lambrechts, D.; De Moor, B.; et al. Prediction of lymph node involvement in breast cancer from primary tumor tissue using gene expression profiling and miRNAs. Breast Cancer Res. Treat. 2011, 129, 767–776. [Google Scholar] [CrossRef]
- Ortis, F.; Naamane, N.; Flamez, D.; Ladriere, L.; Moore, F.; Cunha, D.A.; Colli, M.L.; Thykjaer, T.; Thorsen, K.; Orntoft, T.F.; et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes 2010, 59, 358–374. [Google Scholar] [CrossRef] [Green Version]
- Reading, J.L.; Galvez-Cancino, F.; Swanton, C.; Lladser, A.; Peggs, K.S.; Quezada, S.A. The function and dysfunction of memory CD8 + T cells in tumor immunity. Immunol. Rev. 2018, 283, 194–212. [Google Scholar] [CrossRef]
- Zhu, J.; Petit, P.F.; Van den Eynde, B.J. Apoptosis of tumor-infiltrating T lymphocytes: A new immune checkpoint mechanism. Cancer Immunol. Immunother. 2019, 68, 835–847. [Google Scholar] [CrossRef]
- Salazar, Y.; Zheng, X.; Brunn, D.; Raifer, H.; Picard, F.; Zhang, Y.; Winter, H.; Guenther, S.; Weigert, A.; Weigmann, B.; et al. Microenvironmental Th9 and Th17 lymphocytes induce metastatic spreading in lung cancer. J. Clin. Investig. 2020, 130, 3560–3575. [Google Scholar] [CrossRef] [Green Version]
- Lamble, A.J.; Lind, E.F. Targeting the Immune Microenvironment in Acute Myeloid Leukemia: A Focus on T Cell Immunity. Front. Oncol. 2018, 13, 213. [Google Scholar] [CrossRef]
- Brenner, A.K.; Nepstad, I.; Bruserud, O. Mesenchymal Stem Cells Support Survival and Proliferation of Primary Human Acute Myeloid Leukemia Cells through Heterogeneous Molecular Mechanisms. Front. Immunol. 2017, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- Daud, A.I.; Loo, K.; Pauli, M.L.; Sanchez-Rodriguez, R.; Sandoval, P.M.; Taravati, K.; Tsai, K.; Nosrati, A.; Nardo, L.; Alvarado, M.D.; et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Investig. 2016, 126, 3447–3452. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.B.; Horton, B.L.; Zheng, Y.; Duan, Y.; Powell, J.D.; Gajewski, T.F. The EGR2 targets LAG-3 and 4-1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J. Exp. Med. 2017, 214, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Knaus, H.A.; Berglund, S.; Hackl, H.; Blackford, A.L.; Zeidner, J.F.; Montiel-Esparza, R.; Mukhopadhyay, R.; Vanura, K.; Blazar, B.R.; Karp, J.E.; et al. Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight. 2018, 3, e120974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, S.; Schubert, M.L.; Wang, L.; He, B.; Neuber, B.; Dreger, P.; Muller-Tidow, C.; Schmitt, M. Chimeric Antigen Receptor (CAR) T Cell Therapy in Acute Myeloid Leukemia (AML). J. Clin. Med. 2019, 8, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aurelius, J.; Thoren, F.B.; Akhiani, A.A.; Brune, M.; Palmqvist, L.; Hansson, M.; Hellstrand, K.; Martner, A. Monocytic AML cells inactivate antileukemic lymphocytes: Role of NADPH oxidase/gp91(phox) expression and the PARP-1/PAR pathway of apoptosis. Blood 2012, 119, 5832–5837. [Google Scholar] [CrossRef] [Green Version]
- Clayton, A.; Mitchell, J.P.; Court, J.; Mason, M.D.; Tabi, Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007, 67, 7458–7466. [Google Scholar] [CrossRef] [Green Version]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef] [Green Version]
- Nehrbas, J.; Butler, J.T.; Chen, D.W.; Kurre, P. Extracellular Vesicles and Chemotherapy Resistance in the AML Microenvironment. Front. Oncol. 2020, 10, 90. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Chen, S.N.; Macleod, R.A.F.; Drexler, H.G.; Nagel, S.; Wu, D.P.; Sun, A.N.; Dai, H.P. MiR-130a is aberrantly overexpressed in adult acute myeloid leukemia with t(8;21) and its suppression induces AML cell death. Upsala J. Med. Sci 2018, 123, 19–27. [Google Scholar] [CrossRef]
- Wang, J.; Yu, M.; Guo, Q.; Ma, Q.; Hu, C.; Ma, Z.; Yin, X.; Li, X.; Wang, Y.; Pan, H.; et al. Prognostic significance of huntingtin interacting protein 1 expression on patients with acute myeloid leukemia. Sci. Rep. 2017, 28, 45960. [Google Scholar] [CrossRef] [PubMed]
- Riccioni, R.; Lulli, V.; Castelli, G.; Biffoni, M.; Tiberio, R.; Pelosi, E.; Lo-Coco, F.; Testa, U. miR-21 is overexpressed in NPM1-mutant acute myeloid leukemias. Leuk. Res. 2015, 39, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Ling, Q.; Lv, Y.; Ye, W.; Huang, J.; Li, X.; Guo, Q.; Wang, J.; Li, Z.; Jin, J. Plasma exosome-derived microRNA-532 as a novel predictor for acute myeloid leukemia. Cancer Biomark. 2020, 28, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Rommer, A.; Steinleitner, K.; Hackl, H.; Schneckenleithner, C.; Engelmann, M.; Scheideler, M.; Vlatkovic, I.; Kralovics, R.; Cerny-Reiterer, S.; Valent, P.; et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer 2013, 29, 364. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhu, L.; Wang, Y.; Zhou, D.; Zhu, J.; Xie, W.; Ye, X. Profiling of microRNAs in AML cells following overexpression or silencing of the VEGF gene. Oncol. Lett. 2017, 13, 105–110. [Google Scholar] [CrossRef]
- Li, G.; Song, Y.; Li, G.; Ren, J.; Xie, J.; Zhang, Y.; Gao, F.; Mu, J.; Dai, J. Downregulation of microRNA21 expression inhibits proliferation, and induces G1 arrest and apoptosis via the PTEN/AKT pathway in SKM1 cells. Mol. Med. Rep. 2018, 18, 2771–2779. [Google Scholar]
- Hsieh, C.H.; Tai, S.K.; Yang, M.H. Snail-overexpressing Cancer Cells Promote M2-Like Polarization of Tumor-Associated Macrophages by Delivering MiR-21-Abundant Exosomes. Neoplasia 2018, 20, 775–788. [Google Scholar] [CrossRef]
- He, S.; Chu, J.; Wu, L.C.; Mao, H.; Peng, Y.; Alvarez-Breckenridge, C.A.; Hughes, T.; Wei, M.; Zhang, J.; Yuan, S.; et al. MicroRNAs activate natural killer cells through Toll-like receptor signaling. Blood 2013, 121, 4663–4671. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, K.; Nakajima, S.; Hosojima, S.; Thi Nguyen, D.; Hattori, T.; Manh Le, T.; Hori, O.; Mahib, M.R.; Yamaguchi, Y.; Miura, M.; et al. Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 2019, 10, 2091. [Google Scholar] [CrossRef] [Green Version]
- Mydlarz, W.; Uemura, M.; Ahn, S.; Hennessey, P.; Chang, S.; Demokan, S.; Sun, W.; Shao, C.; Bishop, J.; Krosting, J.; et al. Clusterin is a gene-specific target of microRNA-21 in head and neck squamous cell carcinoma. Clin. Cancer Res. 2014, 20, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Koltai, T. Clusterin: A key player in cancer chemoresistance and its inhibition. Onco. Targets 2014, 20, 447–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carissimi, C.; Carucci, N.; Colombo, T.; Piconese, S.; Azzalin, G.; Cipolletta, E.; Citarella, F.; Barnaba, V.; Macino, G.; Fulci, V. miR-21 is a negative modulator of T-cell activation. Biochimie 2014, 107, 319–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dessarthe, B.; Thedrez, A.; Latouche, J.B.; Cabillic, F.; Drouet, A.; Daniel, P.; de La Pintiere, C.T.; Catros, V.; Toutirais, O. CRTAM receptor engagement by Necl-2 on tumor cells triggers cell death of activated Vgamma9Vdelta2 T cells. J. Immunol. 2013, 190, 4868–4876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, A.; Badr Mel, S.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138. [Google Scholar] [CrossRef]
- Cui, G. TH9, TH17, and TH22 Cell Subsets and Their Main Cytokine Products in the Pathogenesis of Colorectal Cancer. Front. Oncol. 2019, 9, 1002. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.Y.; Meng, S.J.; Shi, S.F.; Liu, L.J.; Lv, J.C.; Zhu, L.; Zhang, H. MicroRNA-21-5p participates in IgA nephropathy by driving T helper cell polarization. J. Nephrol. 2020, 33, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Bautista-Sanchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velazquez, I.A.; Gonzalez-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manriquez, R.; Castro-Hernandez, C.; Fragoso-Ontiveros, V.; Alvarez-Gomez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Jongen-Lavrencic, M.; Sun, S.M.; Dijkstra, M.K.; Valk, P.J.; Lowenberg, B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008, 111, 5078–5085. [Google Scholar] [CrossRef] [Green Version]
- He, X.P.; Chen, P.; Yang, K.; Liu, B.; Zhang, Y.; Wang, F.; Guo, Z.; Liu, X.D.; Lou, J.X.; Chen, H.R. Overexpression of miR21 is involved in acute monocytic leukemia associated angiogenesis by targeting IL12. Mol. Med. Rep. 2018, 18, 4122–4128. [Google Scholar] [PubMed] [Green Version]
- Leone, E.; Morelli, E.; Di Martino, M.T.; Amodio, N.; Foresta, U.; Gulla, A.; Rossi, M.; Neri, A.; Giordano, A.; Munshi, N.C.; et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013, 19, 2096–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Sahraei, M.; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; Garcia-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef]

| Clinical details of AML patients (27) | Number |
|---|---|
| Sex and age | |
| Males | 13 |
| Females | 14 |
| Age < 55y | 9 |
| Age ≥ 55y | 18 |
| Classification of AML patients | |
| Cytogenetic abnormalities | |
| Normal Karyotype | 3 |
| Chromosome 9 deletion | 2 |
| Inversion (16) MYH11-CBFB | 2 |
| Translocation (t8;21) | 2 |
| Translocation (t15;17) | 3 |
| Translocation (X:21) (p11;q22) | 1 |
| trisomy 8 | 1 |
| complex Karyotype | 1 |
| WHO 2016 system | |
| AML with recurrent genetic abnormalities (t8;21) | 2 |
| AML with recurrent genetic abnormalities (inv 16) | 2 |
| AML with recurrent genetic abnormalities (t15;17) | 3 |
| Acute monoblastic/monocytic leukemia | 1 |
| Pure erythroid leukemia | 1 |
| AML not otherwise specified (NOS) | 15 |
| unknown | 3 |
| ELN 2017 genetic risk stratification | |
| Favorable | 10 |
| Intermediate | 3 |
| Adverse | 10 |
| unknown | 4 |
| EV-Derived miRNAs | K562 Mean Ct (SD) | KG1 Mean Ct (SD) | HL60 Mean Ct (SD) |
|---|---|---|---|
| hsa-miR-145 | 28.547 (1.034) | 29.522 (1.064) | 29.522 (1.064) |
| hsa-miR-484 | 27.792 (0.812) | 28.481 (1.114) | 28.481 (1.114) |
| hsa-miR-29a | 28.463 (1.235) | 27.747 (0.848) | 27.747 (0.848) |
| hsa-miR-25 | 27.750 (1.350) | 27.739 (0.707) | 27.739 (0.707) |
| hsa-miR-425-5p | 29.410 (1.167) | 27.454 (1.213) | 27.454 (1.213) |
| hsa-miR-186 | 27.534 (1.156) | 26.930 (1.056) | 26.930 (1.056) |
| hsa-miR-532 | 29.952 (1.000) | 26.685 (0.619) | 26.685 (0.619) |
| hsa-miR-210 | 26.659 (1.228) | 26.656 (0.,904) | 26.656 (0.904) |
| hsa-miR-27a | 29.696 (1.184) | 26.572 (0.798) | 26.572 (0.798) |
| hsa-miR-320 | 25.836 (0.844) | 26.540 (1.093) | 26.540 (1.093) |
| hsa-miR-15b | 28.530 (1.134) | 25.739 (0.715) | 25.739 (0.715) |
| hsa-miR-494 | 26.762 (1.276) | 25.598 (1.387) | 25.598 (1.387) |
| hsa-let-7e | 29.536 (1.014) | 25.354 (0.876) | 25.354 (0.876) |
| hsa-miR-19a | 29.703 (1.226) | 24.629 (0.756) | 24.629 (0.756) |
| hsa-miR-483-5p | 26.502 (1.077) | 24.592 (0.913) | 24.592 (0.913) |
| hsa-miR-20b | 28.791 (0.746) | 23.807 (0.999) | 23.807 (0.999) |
| hsa-miR-30c | 24.754 (0.762) | 23.778 (0.975) | 23.778 (0.975) |
| hsa-miR-21 | 27.538 (1.215) | 23.750 (1.082) | 23.750 (1.082) |
| hsa-miR-16 | 26.564 (1.331) | 23.693 (0.867) | 23.693 (0.867) |
| hsa-miR-30b | 26.688 (0.844) | 23.656 (0.811) | 23.656 (0.811) |
| hsa-miR-92a | 24.688 (1.311) | 23.539 (1.175) | 23.539 (1.175) |
| hsa-miR-221 | 29.812 (1.008) | 23.398 (1.106) | 23.398 (1.106) |
| hsa-miR-146b | 21.782 (1.058) | 22.850 (1.152) | 22.850 (1.152) |
| hsa-miR-142-3p | 28.549 (1.094) | 22.804 (0.799) | 22.804 (0.799) |
| hsa-miR-191 | 22.488 (0.917) | 22.626 (1.328) | 22.626 (1.328) |
| hsa-miR-342-3p | 24.720 (0.773) | 22.442 (1.177) | 22.442 (1.177) |
| hsa-miR-222 | 27.684 (0.892) | 21.850 (1.156) | 21.850 (1.156) |
| hsa-miR-24 | 26.898 (1.107) | 21.815 (1.257) | 21.815 (1.257) |
| hsa-miR-20a | 24.698 (0.668) | 21.463 (1.113) | 21.463 (1.113) |
| hsa-miR-17 | 23.745 (1.036) | 19.910 (1.037) | 19.910 (1.037) |
| hsa-miR-106a | 23.721 (0.986) | 19.773 (0.746) | 19.773 (0.746) |
| hsa-miR-223 | 20.705 (0.886) | 18.706 (1.167) | 18.706 (1.167) |
| hsa-miR-19b | 23.768 (1.284) | 18.478 (0.980) | 18.478 (0.980) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moussa Agha, D.; Rouas, R.; Najar, M.; Bouhtit, F.; Fayyad-Kazan, H.; Lagneaux, L.; Bron, D.; Meuleman, N.; Lewalle, P.; Merimi, M. Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction. Cells 2020, 9, 2053. https://doi.org/10.3390/cells9092053
Moussa Agha D, Rouas R, Najar M, Bouhtit F, Fayyad-Kazan H, Lagneaux L, Bron D, Meuleman N, Lewalle P, Merimi M. Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction. Cells. 2020; 9(9):2053. https://doi.org/10.3390/cells9092053
Chicago/Turabian StyleMoussa Agha, Douâa, Redouane Rouas, Mehdi Najar, Fatima Bouhtit, Hussein Fayyad-Kazan, Laurence Lagneaux, Dominique Bron, Nathalie Meuleman, Philippe Lewalle, and Makram Merimi. 2020. "Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction" Cells 9, no. 9: 2053. https://doi.org/10.3390/cells9092053
APA StyleMoussa Agha, D., Rouas, R., Najar, M., Bouhtit, F., Fayyad-Kazan, H., Lagneaux, L., Bron, D., Meuleman, N., Lewalle, P., & Merimi, M. (2020). Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction. Cells, 9(9), 2053. https://doi.org/10.3390/cells9092053