The Roles of Ubiquitin in Mediating Autophagy
Abstract
:1. Introduction
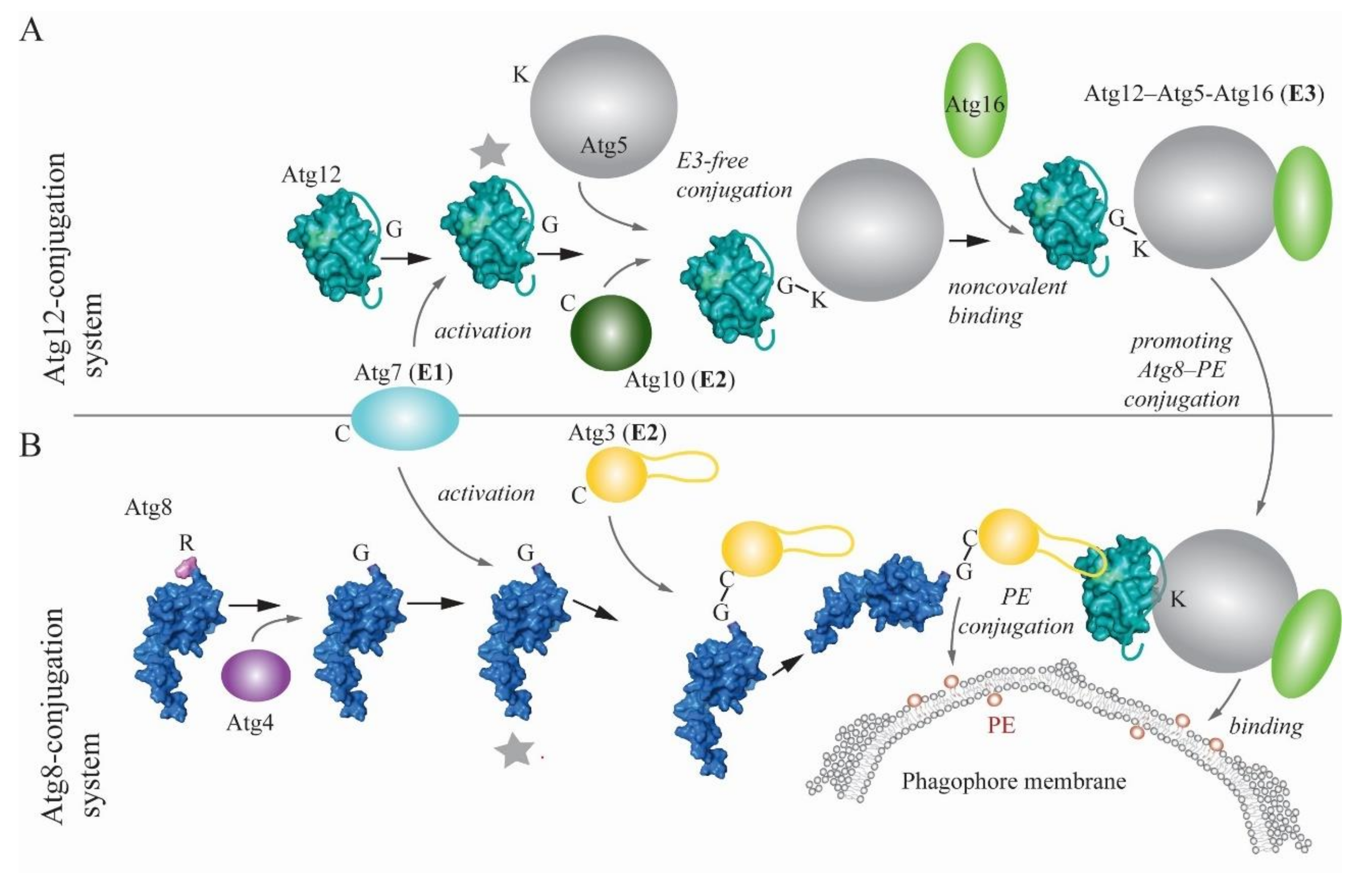
2. Ubiquitin-like Conjugation Systems in Autophagy
3. Unexplored Ubiquitin-Like Folds in Atg5
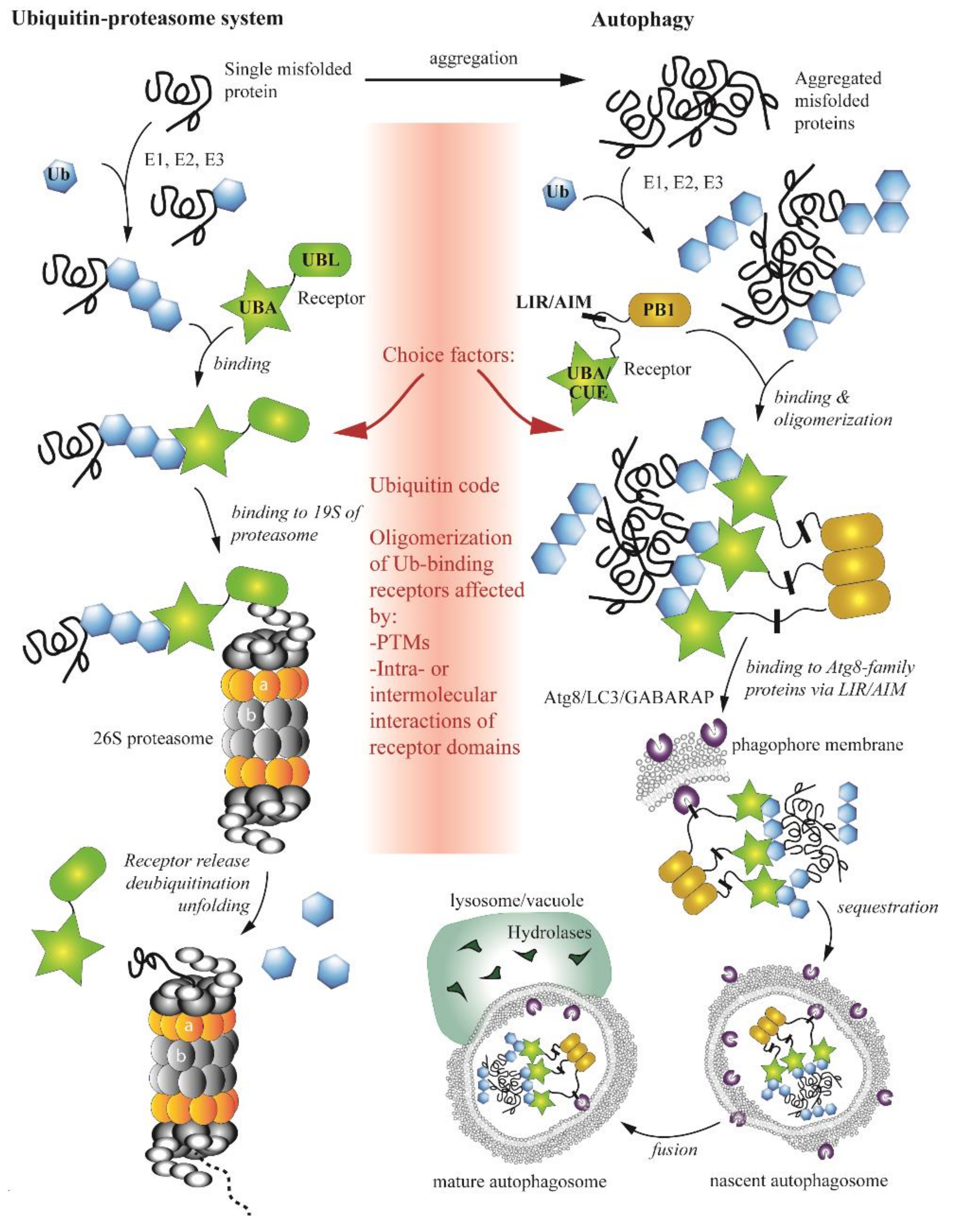
4. Complex Interplay between the Ubiquitin-Proteasome System and Autophagy
5. Ubiquitin and Selective Autophagy in Yeast
5.1. Mitophagy
5.2. Cvt Pathway
5.3. Pexophagy
5.4. Aggrephagy
5.5. Ribophagy
5.6. Proteaphagy
6. Ubiquitination in the Regulation of Autophagy
6.1. The Ubiquitination System and Autophagy Initiation
6.1.1. Ubiquitination, Tor and MTOR Complex 1
6.1.2. Ubiquitination of ULK1
6.2. The Ubiquitination System and Autophagy Nucleation
6.2.1. Ubiquitination of PIK3C3/VPS34
6.2.2. Ubiquitination of BECN1
6.3. The Ubiquitination System and Phagophore Expansion
6.4. The Ubiquitination System and Autophagosome Maturation
6.5. The Ubiquitination System and Autophagy Termination
6.6. Ubiquitination of Autophagy Regulators
7. Conclusions and Perspectives
Funding
Conflicts of Interest
Abbreviations
| AIM | Atg8-interacting motif |
| Atg | autophagy-related |
| CPs | core particles |
| CUE | coupling of ubiquitin conjugation to ER degradation |
| Cvt | cytoplasm-to-vacuole targeting |
| LIR | LC3-interacting region |
| MTORC1 | mechanistic target of rapamycin kinase complex 1 |
| PAS | phagophore assembly site |
| prApe1 | precursor aminopeptidase I |
| PtdIns3K | phosphatidylinositol 3-kinase |
| PtdIns3P | phosphatidylinositol-3-phosphate |
| PTMs | post-translational modifications |
| RPs | regulatory particles |
| UBA | ubiquitin-associated domain |
| UBD | ubiquitin-binding domain |
| UBL | ubiquitin-like |
| UPS | ubiquitin-proteasome system |
References
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klionsky, D.J.; Cregg, J.M.; A Dunn, W.; Emr, S.D.; Sakai, Y.; Sandoval, I.V.; Sibirny, A.; Subramani, S.; Thumm, M.; Veenhuis, M.; et al. A Unified Nomenclature for Yeast Autophagy-Related Genes. Dev. Cell 2003, 5, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Rogov, V.; Dötsch, V.; Johansen, T.; Kirkin, V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callis, J. The Ubiquitination Machinery of the Ubiquitin System. Arab. Book 2014, 12, e0174. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Herr, R.A.; Chua, W.-J.; Lybarger, L.; Wiertz, E.J.; Hansen, T.H. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 2007, 177, 613–624. [Google Scholar] [CrossRef]
- Cadwell, K.; Coscoy, L. Ubiquitination on Nonlysine Residues by a Viral E3 Ubiquitin Ligase. Science 2005, 309, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Husnjak, K.; Dikic, I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Grabbe, C.; Dikic, I. Functional Roles of Ubiquitin-Like Domain (ULD) and Ubiquitin-Binding Domain (UBD) Containing Proteins. Chem. Rev. 2009, 109, 1481–1494. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Schulman, B.A. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 2014, 21, 336–345. [Google Scholar] [CrossRef]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [Green Version]
- Sakoh-Nakatogawa, M.; Matoba, K.; Asai, E.; Kirisako, H.; Ishii, J.; Noda, N.N.; Inagaki, F.; Nakatogawa, H.; Ohsumi, Y. Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 2013, 20, 433–439. [Google Scholar] [CrossRef]
- Yamada, Y.; Suzuki, N.N.; Hanada, T.; Ichimura, Y.; Kumeta, H.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. The Crystal Structure of Atg3, an Autophagy-related Ubiquitin Carrier Protein (E2) Enzyme that Mediates Atg8 Lipidation. J. Biol. Chem. 2007, 282, 8036–8043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taherbhoy, A.M.; Tait, S.W.; Kaiser, S.E.; Williams, A.H.; Deng, A.; Nourse, A.; Hammel, M.; Kurinov, I.; Rock, C.O.; Green, D.R.; et al. Atg8 Transfer from Atg7 to Atg3: A Distinctive E1-E2 Architecture and Mechanism in the Autophagy Pathway. Mol. Cell 2011, 44, 451–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, S.E.; Mao, K.; Taherbhoy, A.M.; Yu, S.; Olszewski, J.L.; Duda, D.M.; Kurinov, I.; Deng, A.; Fenn, T.D.; Klionsky, D.J.; et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7–Atg3 and Atg7–Atg10 structures. Nat. Struct. Mol. Biol. 2012, 19, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, N.N.; Fujioka, Y.; Hanada, T.; Ohsumi, Y.; Inagaki, F. Structure of the Atg12–Atg5 conjugate reveals a platform for stimulating Atg8–PE conjugation. EMBO Rep. 2012, 14, 206–211. [Google Scholar] [CrossRef] [Green Version]
- Metlagel, Z.; Otomo, C.; Takaesu, G.; Otomo, T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc. Natl. Acad. Sci. USA 2013, 110, 18844–18849. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Qiu, Y.; Grace, C.R.R.; Liu, X.; Klionsky, D.J.; Schulman, B.A. A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade. Nat. Commun. 2019, 10, 3600. [Google Scholar] [CrossRef]
- Hervás, J.H.; Landajuela, A.; Antón, Z.; Shnyrova, A.V.; Goñi, F.M.; Alonso, A. Human ATG3 binding to lipid bilayers: Role of lipid geometry, and electric charge. Sci. Rep. 2017, 7, 15614. [Google Scholar] [CrossRef] [Green Version]
- Lystad, A.H.; Carlsson, S.R.; De La Ballina, L.R.; Kauffman, K.J.; Nag, S.; Yoshimori, T.; Melia, T.J.; Simonsen, A. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell. Biol. 2019, 21, 372–383. [Google Scholar] [CrossRef]
- Fracchiolla, D.; Sawa-Makarska, J.; Zens, B.; De Ruiter, A.; Zaffagnini, G.; Brezovich, A.; Romanov, J.; Runggatscher, K.; Kraft, C.; Zagrovic, B.; et al. Mechanism of cargo-directed Atg8 conjugation during selective autophagy. eLife 2016, 5, e18544. [Google Scholar] [CrossRef]
- Harada, K.; Kotani, T.; Kirisako, H.; Sakoh-Nakatogawa, M.; Oikawa, Y.; Kimura, Y.; Hirano, H.; Yamamoto, H.; Ohsumi, Y.; Nakatogawa, H. Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. eLife 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Popelka, H.; Klionsky, D.J. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy 2015, 11, 2153–2159. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Suzuki, N.N.; Obara, K.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of Atg5.Atg16, a Complex Essential for Autophagy. J. Biol. Chem. 2007, 282, 6763–6772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef]
- Park, C.; Cuervo, A.M. Selective autophagy: Talking with the UPS. Cell Biophys. 2013, 67, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin–proteasome system. Biochim. Biophys. Acta 2014, 1843, 163–181. [Google Scholar] [CrossRef] [Green Version]
- Ji, C.H.; Kwon, Y.T. Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Nam, T.; Han, J.H.; Devkota, S.; Lee, H.-W. Emerging Paradigm of Crosstalk between Autophagy and the Ubiquitin-Proteasome System. Mol. Cells 2017, 40, 897–905. [Google Scholar]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Subramani, S. The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy. Cells 2019, 8, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, W.H.; Park, J.H.; Chung, K.C. The central regulator p62 between ubiquitin proteasome system and autophagy and its role in the mitophagy and Parkinson’s disease. BMB Rep. 2020, 53, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.K.; Cho, C.H.; Lee, C.W.; Wu, Y.C.; Yu, L.; Li, Z.J.; Wong, C.C.M.; Li, H.T.; Zhang, L.; Ren, S.X.; et al. Macroautophagy and ERK phosphorylation counteract the antiproliferative effect of proteasome inhibitor in gastric cancer cells. Autophagy 2010, 6, 228–238. [Google Scholar] [CrossRef] [Green Version]
- Demishtein, A.; Fraiberg, M.; Berko, D.; Tirosh, B.; Elazar, Z.; Navon, A. SQSTM1/p62-mediated autophagy compensates for loss of proteasome polyubiquitin recruiting capacity. Autophagy 2017, 13, 1697–1708. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Chevet, E.; Oakes, S.A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 2015, 17, 829–838. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.J.; Yu, J.; Wong, S.; Cheng, A.S.; Chan, F.K.; Ng, S.S.-M.; Cho, C.H.; Sung, J.J.; Wu, W.K.K. A novel crosstalk between two major protein degradation systems: Regulation of proteasomal activity by autophagy. Autophagy 2013, 9, 1500–1508. [Google Scholar] [CrossRef]
- Athané, A.; Buisson, A.; Challier, M.; Beaumatin, F.; Manon, S.; Bhatia-Kiššová, I.; Camougrand, N. Insights into the relationship between the proteasome and autophagy in human and yeast cells. Int. J. Biochem. Cell Biol. 2015, 64, 167–173. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Mansilla, A.; Menzies, F.M.; Rubinsztein, D.C. Autophagy Inhibition Compromises Degradation of Ubiquitin-Proteasome Pathway Substrates. Mol. Cell 2009, 33, 517–527. [Google Scholar] [CrossRef]
- Xiong, Q.; Fischer, S.; Karow, M.; Müller, R.; Meßling, S.; Eichinger, L. ATG16 mediates the autophagic degradation of the 19S proteasomal subunits PSMD1 and PSMD2. Eur. J. Cell Biol. 2018, 97, 523–532. [Google Scholar] [CrossRef]
- Njomen, E.; Tepe, J. Regulation of Autophagic Flux by the 20S Proteasome. Cell Chem. Biol. 2019, 26, 1283–1294.e5. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Park, S.; Lee, J.H.; Mun, J.Y.; Choi, W.H.; Yun, Y.; Lee, J.; Kim, J.H.; Kang, M.-J.; Lee, M.J. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, Y.; Lee, S.; Kim, M.-J.; Chun, E.; Lee, K.-Y. Ubiquitin-Specific Protease 14 Negatively Regulates Toll-Like Receptor 4-Mediated Signaling and Autophagy Induction by Inhibiting Ubiquitination of TAK1-Binding Protein 2 and Beclin 1. Front. Immunol. 2017, 8, 1827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef] [Green Version]
- Minoia, M.; Boncoraglio, A.; Vinet, J.; Morelli, F.F.; Brunsting, J.F.; Poletti, A.; Krom, S.; Reits, E.; Kampinga, H.H.; Carra, S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 2014, 10, 1603–1621. [Google Scholar] [CrossRef] [Green Version]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The Ubiquitin–Proteasome System of Saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef] [Green Version]
- Dargemont, C.; Ossareh-Nazari, B. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim. Biophys. Acta 2012, 1823, 138–144. [Google Scholar] [CrossRef]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef] [Green Version]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.M.; Wong, E.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.-P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Lamark, T.; Johansen, T.; Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009, 5, 732–733. [Google Scholar] [CrossRef] [Green Version]
- Kravtsova-Ivantsiv, Y.; Sommer, T.; Ciechanover, A. The Lysine48-Based Polyubiquitin Chain Proteasomal Signal: Not a Single Child Anymore. Angew. Chem. Int. Ed. Engl. 2013, 44, 192–198. [Google Scholar] [CrossRef]
- Walinda, E.; Morimoto, D.; Sugase, K.; Konuma, T.; Tochio, H.; Shirakawa, M. Solution Structure of the Ubiquitin-associated (UBA) Domain of Human Autophagy Receptor NBR1 and Its Interaction with Ubiquitin and Polyubiquitin. J. Biol. Chem. 2014, 289, 13890–13902. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Brave, F.D.; Jentsch, S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 2017, 19, 732–739. [Google Scholar] [CrossRef]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife 2015, 4, 08941. [Google Scholar] [CrossRef]
- Sasaki, T.; Funakoshi, M.; Endicott, J.A.; Kobayashi, H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem. Biophys. Res. Commun. 2005, 336, 530–535. [Google Scholar] [CrossRef]
- Lowe, E.D.; Hasan, N.; Trempe, J.-F.; Fonso, L.; Noble, M.E.; Endicott, J.A.; Johnson, L.N.; Brown, N.R. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 177–188. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Klionsky, D.J. Autophagy and disease: Unanswered questions. Cell Death Differ. 2020, 27, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhang, J.; Zhu, N.; Ding, Q.; Zhang, X.; Yu, J.; Qiang, W.; Zhang, Z.; Ma, Y.; Huang, D.; et al. Ubiquitin ligase SYVN1/HRD1 facilitates degradation of the SERPINA1 Z variant/α-1-antitrypsin Z variant via SQSTM1/p62-dependent selective autophagy. Autophagy 2017, 13, 686–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wauer, T.; Simicek, M.; Schubert, A.; Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 2015, 524, 370–374. [Google Scholar] [CrossRef] [Green Version]
- Matscheko, N.; Mayrhofer, P.; Rao, Y.; Beier, V.; Wollert, T. Atg11 tethers Atg9 vesicles to initiate selective autophagy. PLoS Biol. 2019, 17, e3000377. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Subramani, S. Mechanistic Insights into the Role of Atg11 in Selective Autophagy. J. Mol. Biol. 2020, 432, 104–122. [Google Scholar] [CrossRef]
- Kanki, T.; Wang, K.; Cao, Y.; Baba, M.; Klionsky, D.J. Atg32 Is a Mitochondrial Protein that Confers Selectivity during Mitophagy. Dev. Cell 2009, 17, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Gladkova, C.; Maslen, S.L.; Skehel, J.M.; Komander, D. Mechanism of parkin activation by PINK1. Nature 2018, 559, 410–414. [Google Scholar] [CrossRef]
- Vigie, P.; Gonzalez, C.; Manon, S.; Bhatia-Kissova, I.; Camougrand, N. Ubiquitination of the yeast receptor Atg32 modulates mitophagy. bioRxiv 2019, 652933. [Google Scholar] [CrossRef]
- Muller, M.; Kötter, P.; Behrendt, C.; Walter, E.; Scheckhuber, C.Q.; Entian, K.-D.; Reichert, A.S. Synthetic Quantitative Array Technology Identifies the Ubp3-Bre5 Deubiquitinase Complex as a Negative Regulator of Mitophagy. Cell Rep. 2015, 10, 1215–1225. [Google Scholar] [CrossRef] [Green Version]
- Scott, S.V.; Guan, J.; Hutchins, M.U.; Kim, J.; Klionsky, D.J. Cvt19 Is a Receptor for the Cytoplasm-to-Vacuole Targeting Pathway. Mol. Cell 2001, 7, 1131–1141. [Google Scholar] [CrossRef] [Green Version]
- Yuga, M.; Gomi, K.; Klionsky, D.J.; Shintani, T. Aspartyl Aminopeptidase Is Imported from the Cytoplasm to the Vacuole by Selective Autophagy inSaccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 13704–13713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Noda, N.N.; Kumeta, H.; Suzuki, K.; Ohsumi, Y.; Inagaki, F. Selective Transport of α-Mannosidase by Autophagic Pathways STRUCTURAL BASIS FOR CARGO RECOGNITION BY Atg19 AND Atg34. J. Biol. Chem. 2010, 285, 30026–30033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, B.K.; Abeliovich, H.; Zhang, X.; Stirling, A.G.; Burlingame, A.L.; Goldfarb, D.S. Atg19p Ubiquitination and the Cytoplasm to Vacuole Trafficking Pathway in Yeast. J. Biol. Chem. 2005, 280, 39067–39076. [Google Scholar] [CrossRef] [Green Version]
- Motley, A.M.; Nuttall, A.J.M.; Hettema, E.H. Pex3-anchored Atg36 tags peroxisomes for degradation inSaccharomyces cerevisiae. EMBO J. 2012, 31, 2852–2868. [Google Scholar] [CrossRef] [Green Version]
- Burnett, S.F.; Farré, J.-C.; Nazarko, T.Y.; Subramani, S. Peroxisomal Pex3 Activates Selective Autophagy of Peroxisomes via Interaction with the Pexophagy Receptor Atg30. J. Biol. Chem. 2015, 290, 8623–8631. [Google Scholar] [CrossRef] [Green Version]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2013, 126, 939–952. [Google Scholar] [CrossRef] [Green Version]
- Nuttall, J.M.; Motley, A.M.; Hettema, E.H. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy 2014, 10, 835–845. [Google Scholar] [CrossRef] [Green Version]
- Germain, K.; Kim, P.K. Pexophagy: A Model for Selective Autophagy. Int. J. Mol. Sci. 2020, 21, 578. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef] [Green Version]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008, 10, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Ossareh-Nazari, B.; Bonizec, M.; Cohen, M.M.; Dokudovskaya, S.; Delalande, F.; Schaeffer, C.; Van Dorsselaer, A.; Dargemont, C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010, 11, 548–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatehashi, Y.; Watanabe, D.; Takagi, H. gamma-Glutamyl kinase is involved in selective autophagy of ribosomes in Saccharomyces cerevisiae. FEBS Lett. 2016, 590, 2906–2914. [Google Scholar] [CrossRef] [PubMed]
- Wyant, G.A.; Abu-Remaileh, M.; Frenkel, E.M.; Laqtom, N.N.; Dharamdasani, V.; Lewis, C.A.; Chan, S.H.; Heinze, I.; Ori, A.; Sabatini, D.M. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 2018, 360, 751–758. [Google Scholar] [CrossRef] [Green Version]
- Nemec, A.A.; Howell, L.A.; Peterson, A.K.; Murray, M.A.; Tomko, R.J. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J. Biol. Chem. 2017, 292, 21466–21480. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781.e24. [Google Scholar] [CrossRef] [Green Version]
- Waite, K.A.; De La Mota-Peynado, A.; Vontz, G.; Roelofs, J. Starvation Induces Proteasome Autophagy with Different Pathways for Core and Regulatory Particles. J. Biol. Chem. 2016, 291, 3239–3253. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.S.; Vierstra, R.D. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife 2018, 7, e34532. [Google Scholar] [CrossRef]
- Cohen-Kaplan, V.; Livneh, I.; Avni, N.; Fabre, B.; Ziv, T.; Kwon, Y.T.; Ciechanover, A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, E7490–E7499. [Google Scholar] [CrossRef] [Green Version]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [Green Version]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.-I.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a Binding Partner of Target of Rapamycin (TOR), Mediates TOR Action. Cell 2002, 110, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.P.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.-I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 Association with the ULK1–Atg13–FIP200 Complex Required for Autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Mercer, C.A.; Kaliappan, A.; Dennis, P.B. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009, 5, 649–662. [Google Scholar] [CrossRef] [Green Version]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wilson, W.A.; Fujino, M.A.; Roach, P.J. Antagonistic Controls of Autophagy and Glycogen Accumulation by Snf1p, the Yeast Homolog of AMP-Activated Protein Kinase, and the Cyclin-Dependent Kinase Pho85p. Mol. Cell. Biol. 2001, 21, 5742–5752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, C.; Tong, J.; Lu, P.; Wang, Y.; Zhang, J.; Sun, C.; Yuan, K.; Xue, R.; Zou, B.; Li, N.; et al. Formation of a Snf1-Mec1-Atg1 Module on Mitochondria Governs Energy Deprivation-Induced Autophagy by Regulating Mitochondrial Respiration. Dev. Cell 2017, 41, 59–71.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef] [Green Version]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Méry, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.; Guo, S.; Yan, G.; Yuan, W.; Zheng, Y.; Jiang, Y. Ubiquitin regulates TORC1 in yeast Saccharomyces cerevisiae. Mol. Microbiol. 2016, 100, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y. Regulation of TORC1 by ubiquitin through non-covalent binding. Curr. Genet. 2016, 62, 553–555. [Google Scholar] [CrossRef]
- Linares, J.F.; Duran, A.; Yajima, T.; Pasparakis, M.; Moscat, J.; Diaz-Meco, M.T. K63 Polyubiquitination and Activation of mTOR by the p62-TRAF6 Complex in Nutrient-Activated Cells. Mol. Cell 2013, 51, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ou, Y.; Yang, Y.; Li, W.; Xu, Y.; Xie, Y.; Liu, Y. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 2018, 557, 585–589. [Google Scholar] [CrossRef]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR Is an mTOR Inhibitor Frequently Overexpressed in Multiple Myeloma Cells and Required for Their Survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Xiong, X.; Sun, Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 2011, 44, 304–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Inuzuka, H.; Tan, M.-K.M.; Fukushima, H.; Locasale, J.W.; Liu, P.; Wan, L.; Zhai, B.; Chin, Y.R.; Shaik, S.; et al. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 2011, 44, 290–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, S.; Skaar, J.R.; Kuchay, S.; Toschi, A.; Kanarek, N.; Ben-Neriah, Y.; Pagano, M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell 2011, 44, 317–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonioli, M.; Albiero, F.; Nazio, F.; Vescovo, T.; Perdomo, A.B.; Corazzari, M.; Marsella, C.; Piselli, P.; Gretzmeier, C.; Dengjel, J.; et al. AMBRA1 Interplay with Cullin E3 Ubiquitin Ligases Regulates Autophagy Dynamics. Dev. Cell 2014, 31, 734–746. [Google Scholar] [CrossRef]
- McEwan, D.G.; Dikic, I. Cullins keep autophagy under control. Dev. Cell 2014, 31, 675–676. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Wang, X.; Yu, Y.; Deng, L.; Chen, L.; Peng, X.; Jiao, C.; Gao, G.; Tan, X.; Pan, W.; et al. OTUB1 protein suppresses mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1 inhibitor DEPTOR. J. Biol. Chem. 2018, 293, 4883–4892. [Google Scholar] [CrossRef] [Green Version]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Antonioli, M.; Fusco, C.; Liu, Y.; Mari, M.; Orhon, I.; Refolo, G.; Germani, F.; Corazzari, M.; Romagnoli, A.; et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci. Adv. 2019, 5, eaau8857. [Google Scholar] [CrossRef] [Green Version]
- Jiao, H.; Su, G.-Q.; Dong, W.; Zhang, L.; Xie, W.; Yao, L.-M.; Chen, P.; Wang, Z.-X.; Liou, Y.-C.; You, H. Chaperone-like protein p32 regulates ULK1 stability and autophagy. Cell Death Differ. 2015, 22, 1812–1823. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-E.; Yoo, J.E.; Kim, J.; Kim, S.; Kim, S.; Lee, H.; Cheong, H. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Crawley, O.; Opperman, K.J.; Desbois, M.; Adrados, I.; Borgen, M.A.; Giles, A.C.; Duckett, D.R.; Grill, B. Autophagy is inhibited by ubiquitin ligase activity in the nervous system. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Seo, D.; Kim, S.; Choi, D.W.; Park, J.S.; Ha, J.; Choi, J.; Lee, J.; Jung, S.M.; Seo, K.; et al. The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep. 2018, 19, e44378. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.; Cesselli, D.; Di Loreto, C.; La Marra, F.; Schneider, C.; Demarchi, F. USP1 (ubiquitin specific peptidase 1) targets ULK1 and regulates its cellular compartmentalization and autophagy. Autophagy 2018, 15, 613–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.A.; Koutelou, E.; Hirsch, C.; Akdemir, K.; Schibler, A.; Barton, M.; Dent, S.R. Ubp8 and SAGA Regulate Snf1 AMP Kinase Activity. Mol. Cell. Biol. 2011, 31, 3126–3135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pineda, C.T.; Ramanathan, S.; Tacer, K.F.; Weon, J.L.; Potts, M.B.; Ou, Y.-H.; White, M.A.; Potts, P.R. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.; Kang, Y.; Yan, C.; Yang, C.; Zhang, T.; Huo, H.; Liu, Y. LUBAC and OTULIN regulate autophagy initiation and maturation by mediating the linear ubiquitination and the stabilization of ATG13. Autophagy 2020, 1–16. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two Distinct Vps34 Phosphatidylinositol 3–Kinase Complexes Function in Autophagy and Carboxypeptidase Y Sorting inSaccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [Green Version]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1–VPS34 complex—At the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Furuya, T.; Kim, M.; Lipinski, M.M.; Li, J.; Kim, D.; Lu, T.; Shen, Y.; Rameh, L.; Yankner, B.; Tsai, L.-H.; et al. Negative Regulation of Vps34 by Cdk Mediated Phosphorylation. Mol. Cell 2010, 38, 500–511. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg-Lerner, A.; Kimchi, A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012, 19, 788–797. [Google Scholar] [CrossRef]
- Yang, Y.; Fiskus, W.; Yong, B.; Atadja, P.; Takahashi, Y.; Pandita, T.K.; Wang, H.-G.; Bhalla, K.N. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 6841–6846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.C.; Wei, Y.; An, Z.; Zou, Z.; Xiao, G.; Bhagat, G.; White, M.; Reichelt, J.; Levine, B. Akt-Mediated Regulation of Autophagy and Tumorigenesis Through Beclin 1 Phosphorylation. Science 2012, 338, 956–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Zou, Z.; Becker, N.; Anderson, M.; Sumpter, R.; Xiao, G.; Kinch, L.; Koduru, P.; Christudass, C.S.; Veltri, R.W.; et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013, 154, 1269–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Zhang, T.; Xu, D.; Wang, H.; Cai, Y.; Jin, T.; Liu, M.; Jin, M.; Wu, K.; Yuan, J. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015, 29, 184–196. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, M.; Li, L.; Chen, S.; Wang, X. Ubiquitination of the PI3-kinase VPS-34 promotes VPS-34 stability and phagosome maturation. J. Cell Biol. 2018, 217, 347–360. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Jin, S.; Wu, Y.; Xian, H.; Tian, S.; Liu, D.-A.; Guo, Z.; Cui, J. Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. Cell Rep. 2020, 30, 2807–2819.e4. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.-S.; Kehrl, J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010, 3, ra42. [Google Scholar] [CrossRef]
- Han, T.; Guo, M.; Gan, M.; Yu, B.; Tian, X.; Wang, J.-B. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy 2018, 14, 2035–2048. [Google Scholar] [CrossRef] [Green Version]
- Fusco, C.; Mandriani, B.; Di Rienzo, M.; Micale, L.; Malerba, N.; Cocciadiferro, D.; Sjøttem, E.; Augello, B.; Squeo, G.M.; Pellico, M.T.; et al. TRIM50 regulates Beclin 1 proautophagic activity. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, P.; Wang, S.; Du, Y.; Zhao, Z.; Shi, L.; Sun, L.; Huang, G.; Ye, B.; Li, C.; Dai, Z.; et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013, 32, 2685–2696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Feng, K.; Zhao, X.; Huang, S.; Cheng, Y.; Qian, L.; Wang, Y.; Sun, H.; Jin, M.; Chuang, T.-H.; et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy 2014, 10, 2239–2250. [Google Scholar] [CrossRef] [Green Version]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, G.; Buijze, H.; Liu, H.; Moura-Alves, P.; Goosmann, C.; Brinkmann, V.; Kawabe, H.; Dorhoi, A.; Kaufmann, S.H. The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy 2017, 13, 2041–2055. [Google Scholar] [CrossRef]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 Controls the Levels of p53 by Regulating the Deubiquitination Activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R.-F. USP 19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Shan, B.; Sun, H.; Xiao, J.; Zhu, K.; Xie, X.; Li, X.; Liang, W.; Lu, X.; Qian, L.; et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016, 30, 1718–1730. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Bento, C.F.; Ricketts, T.; Vicinanza, M.; Siddiqi, F.; Pavel, M.; Squitieri, F.; Hardenberg, M.C.; Imarisio, S.; Menzies, F.M.; et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 2017, 545, 108–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Dong, K.; Liang, W.; Xu, D.; Xia, H.; Geng, J.; Najafov, A.; Liu, M.; Li, Y.; Han, X.; et al. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. eLife 2015, 4, 06734. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Huang, G.; Du, Y.; Zhu, P.; Li, M.; Fan, Z. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014, 24, 943–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Gao, F.; Li, B.; Wang, H.; Xu, Y.; Zhu, C.; Wang, G. Parkin Mono-ubiquitinates Bcl-2 and Regulates Autophagy. J. Biol. Chem. 2010, 285, 38214–38223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodemann, B.O.; Orvedahl, A.; Cheng, T.; Ram, R.R.; Ou, Y.-H.; Formstecher, E.; Maiti, M.; Hazelett, C.C.; Wauson, E.M.; Balakireva, M.; et al. RalB and the Exocyst Mediate the Cellular Starvation Response by Direct Activation of Autophagosome Assembly. Cell 2011, 144, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simicek, M.; Lievens, S.; Laga, M.; Guzenko, D.; Aushev, V.N.; Kalev, P.; Baietti, M.F.; Strelkov, S.V.; Gevaert, K.; Tavernier, J.; et al. The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Nat. Cell Biol. 2013, 15, 1220–1230. [Google Scholar] [CrossRef]
- Wan, W.; You, Z.; Zhou, L.; Xu, Y.; Peng, C.; Zhou, T.; Yi, C.; Shi, Y.; Liu, W. mTORC1-Regulated and HUWE1-Mediated WIPI2 Degradation Controls Autophagy Flux. Mol. Cell 2018, 72, 303–315.e6. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.; Yi, J.; Gubas, A.; Wang, Y.-T.; Wu, Y.; Ren, Y.; Wu, M.; Shi, Y.; Ouyang, C.; Tan, H.W.S.; et al. Suppression of autophagy during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2 polyubiquitination and proteasomal degradation. Autophagy 2019, 15, 1917–1934. [Google Scholar] [CrossRef] [Green Version]
- Kuang, E.; Okumura, C.Y.M.; Sheffy-Levin, S.; Varsano, T.; Shu, V.C.-W.; Qi, J.; Niesman, I.R.; Yang, H.-J.; López-Otín, C.; Yang, W.Y.; et al. Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection. PLoS Genet. 2012, 8, e1003007. [Google Scholar] [CrossRef] [Green Version]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [Green Version]
- Scrivo, A.; Codogno, P.; Bomont, P. Gigaxonin E3 ligase governs ATG16L1 turnover to control autophagosome production. Nat. Commun. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Bonifacino, J.S. Negative regulation of autophagy by UBA6-BIRC6-mediated ubiquitination of LC3. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Joachim, J.; Razi, M.; Judith, D.; Wirth, M.; Calamita, E.; Encheva, V.; Dynlacht, B.D.; Snijders, A.P.; O’Reilly, N.; Jefferies, H.B.; et al. Centriolar Satellites Control GABARAP Ubiquitination and GABARAP-Mediated Autophagy. Curr. Biol. 2017, 27, 2123–2136.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Lee, J.-S.; Inn, K.-S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Jung, C.H.; Seo, M.; Kim, E.K.; Park, J.-M.; Bae, S.S.; Kim, D.-H. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 2015, 57, 207–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Jia, Y.; Zhang, Y.; Ma, F.; Zhu, Y.; Hong, X.; Zhou, Q.; He, R.; Zhang, H.; Jin, J.; et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 2019, 15, 1130–1149. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, G.; Xue, X.; Guo, X.; Yuan, C.; Wang, Z.; Zhang, G.; Chen, Y.; Feng, D.; Hu, J.; et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell 2016, 63, 781–795. [Google Scholar] [CrossRef] [Green Version]
- Gu, H.; Shi, X.; Liu, C.; Wang, C.; Sui, N.; Zhao, Y.; Gong, J.; Wang, F.; Zhang, H.; Li, W.; et al. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Nat. Commun. 2019, 10, 1465. [Google Scholar] [CrossRef]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-C.; Lin, Y.-C.; Chen, Y.-H.; Chen, C.-M.; Pang, L.-Y.; Chen, H.-A.; Wu, P.-R.; Lin, M.-Y.; Jiang, S.-T.; Tsai, T.-F.; et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Carinci, M.; Valacca, C.; Bielli, P.; Strappazzon, F.; Antonioli, M.; Ciccosanti, F.; Rodolfo, C.; Campello, S.; Fimia, G.M.; et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 2016, 215, 841–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartholomew, C.R.; Suzuki, T.; Du, Z.; Backues, S.K.; Jin, M.; Lynch-Day, M.A.; Umekawa, M.; Kamath, A.; Zhao, M.; Xie, Z.; et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, 11206–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, J.S.; Yeh, Y.-Y.; Ramachandran, V.V.; Deminoff, S.J.; Herman, P.K. The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 2010, 6, 294–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, M.J.; Cooper, K.F.; Strich, R. Meiosis-Specific Destruction of the Ume6p Repressor by the Cdc20-Directed APC/C. Mol. Cell 2007, 27, 951–961. [Google Scholar] [CrossRef] [Green Version]
- White, E. Autophagy and p53. Cold Spring Harb. Perspect. Med. 2016, 6, a026120. [Google Scholar] [CrossRef]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef]
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri-Mergny, M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687. [Google Scholar] [CrossRef] [Green Version]
- Di Malta, C.; Cinque, L.; Settembre, C. Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front. Cell Dev. Biol. 2019, 7, 114. [Google Scholar] [CrossRef]
- Huang, H.; Regan, K.M.; Wang, F.; Wang, D.; I Smith, D.; Van Deursen, J.M.A.; Tindall, D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 2005, 102, 1649–1654. [Google Scholar] [CrossRef] [Green Version]
- Kato, S.; Ding, J.; Pisck, E.; Jhala, U.S.; Du, K. COP1 Functions as a FoxO1 Ubiquitin E3 Ligase to Regulate FoxO1-mediated Gene Expression. J. Biol. Chem. 2008, 283, 35464–35473. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Ma, Q.; Chen, L.; Li, P.; Zhang, M.; Ramamoorthy, S.; Nawaz, Z.; Shimojima, T.; Wang, H.; Yang, Y.; et al. MDM2 Acts Downstream of p53 as an E3 Ligase to Promote FOXO Ubiquitination and Degradation. J. Biol. Chem. 2009, 284, 13987–14000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polager, S.; Ofir, M.; Ginsberg, D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008, 27, 4860–4864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Tindall, D.J. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 2011, 1813, 1961–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Ma, A.; Zhang, L.; Jin, W.-L.; Qian, Y.; Xu, G.; Qiu, B.; Yang, Z.; Liu, Y.; Xia, Q.; et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 2015, 6, 8704. [Google Scholar] [CrossRef]
- Mahanic, C.S.; Budhavarapu, V.; Graves, J.D.; Li, G.; Lin, W.-C. Regulation of E2 Promoter Binding Factor 1 (E2F1) Transcriptional Activity through a Deubiquitinating Enzyme, UCH37. J. Biol. Chem. 2015, 290, 26508–26522. [Google Scholar] [CrossRef] [Green Version]
- Glorian, V.; Allègre, J.; Berthelet, J.; Dumetier, B.; Boutanquoi, P.-M.; Droin, N.; Kayaci, C.; Cartier, J.; Gemble, S.; Marcion, G.; et al. DNA damage and S phase-dependent E2F1 stabilization requires the cIAP1 E3-ubiquitin ligase and is associated with K63-poly-ubiquitination on lysine 161/164 residues. Cell Death Dis. 2017, 8, e2816. [Google Scholar] [CrossRef] [Green Version]
- Herhaus, L.; Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015, 16, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Akopian, D.; Rape, M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef]
- Rioux, J.D.; Xavier, R.J.; Taylor, K.D.; Silverberg, M.S.; Goyette, P.; Huett, A.; Green, T.; Kuballa, P.; Barmada, M.M.; Datta, L.W.; et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007, 39, 596–604. [Google Scholar] [CrossRef]
- Musone, S.L.; Taylor, K.E.; Nititham, J.; Chu, C.; Poon, A.; Liao, W.; Lam, E.T.; Ma, A.; Kwok, P.-Y.; A Criswell, L. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes Immun. 2011, 12, 176–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slowicka, K.; Serramito-Gómez, I.; Boada-Romero, E.; Martens, A.; Sze, M.; Petta, I.; Vikkula, H.K.; De Rycke, R.; Parthoens, E.; Lippens, S.; et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat. Commun. 2019, 10, 1834. [Google Scholar] [CrossRef] [PubMed]


| Pathway | Cargo | Receptors | E3 Ligase |
|---|---|---|---|
| Cytoplasm-to-vacuole targeting (Cvt) pathway | prApe1, Ams1 and Ape4 | Atg19 and Atg34 | - |
| Mitophagy | Mitochondria | Atg32 | - |
| Pexophagy | Peroxisomes (S. cerevisiae) | Atg36 | - |
| Peroxisomes (K. phaffii/P. pastoris) | Atg30 | - | |
| Aggrephagy | Protein aggregates | Cue5 | Rsp5 |
| Ribophagy | Ribosome | - | - |
| Proteaphagy | Proteasome | Cue5 | - |
| Pathway | Cargo | Receptors | E3 Ligase |
|---|---|---|---|
| Mitophagy | Mitochondria | SQSTM1/p62, BNIP3L/Nix, OPTN | PRKN/PARK2/ parkin |
| FUNDC1, PHB2, CALCOCO2/NDP52 | |||
| Pexophagy | Peroxisomes | SQSTM1/p62, NBR1 | PEX2 |
| Aggrephagy | Protein aggregates | SQSTM1/p62, NBR1, TOLLIP | - |
| Ribophagy | Ribosome | NUFIP1 | - |
| Proteaphagy | Proteasome | SQSTM1/p62 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Z.; Popelka, H.; Lei, Y.; Yang, Y.; Klionsky, D.J. The Roles of Ubiquitin in Mediating Autophagy. Cells 2020, 9, 2025. https://doi.org/10.3390/cells9092025
Yin Z, Popelka H, Lei Y, Yang Y, Klionsky DJ. The Roles of Ubiquitin in Mediating Autophagy. Cells. 2020; 9(9):2025. https://doi.org/10.3390/cells9092025
Chicago/Turabian StyleYin, Zhangyuan, Hana Popelka, Yuchen Lei, Ying Yang, and Daniel J. Klionsky. 2020. "The Roles of Ubiquitin in Mediating Autophagy" Cells 9, no. 9: 2025. https://doi.org/10.3390/cells9092025
APA StyleYin, Z., Popelka, H., Lei, Y., Yang, Y., & Klionsky, D. J. (2020). The Roles of Ubiquitin in Mediating Autophagy. Cells, 9(9), 2025. https://doi.org/10.3390/cells9092025






