Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase
Abstract
1. Introduction
1.1. Cyclin Dependent Kinase 1 (CDK1) Is a Subunit of the M Phase-Promoting Factor (MPF)
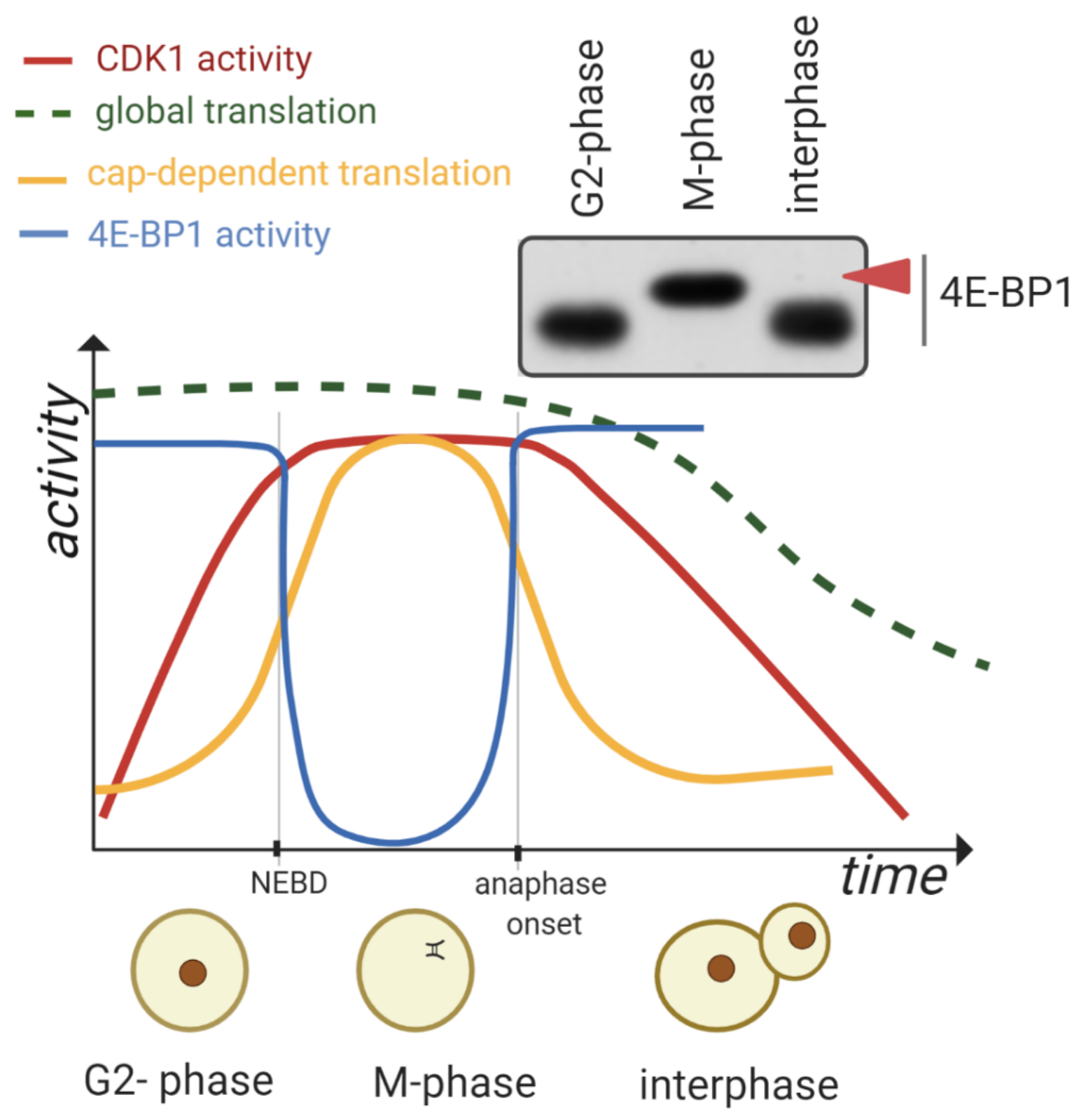
1.2. Global mRNA Translation during the M-Phase
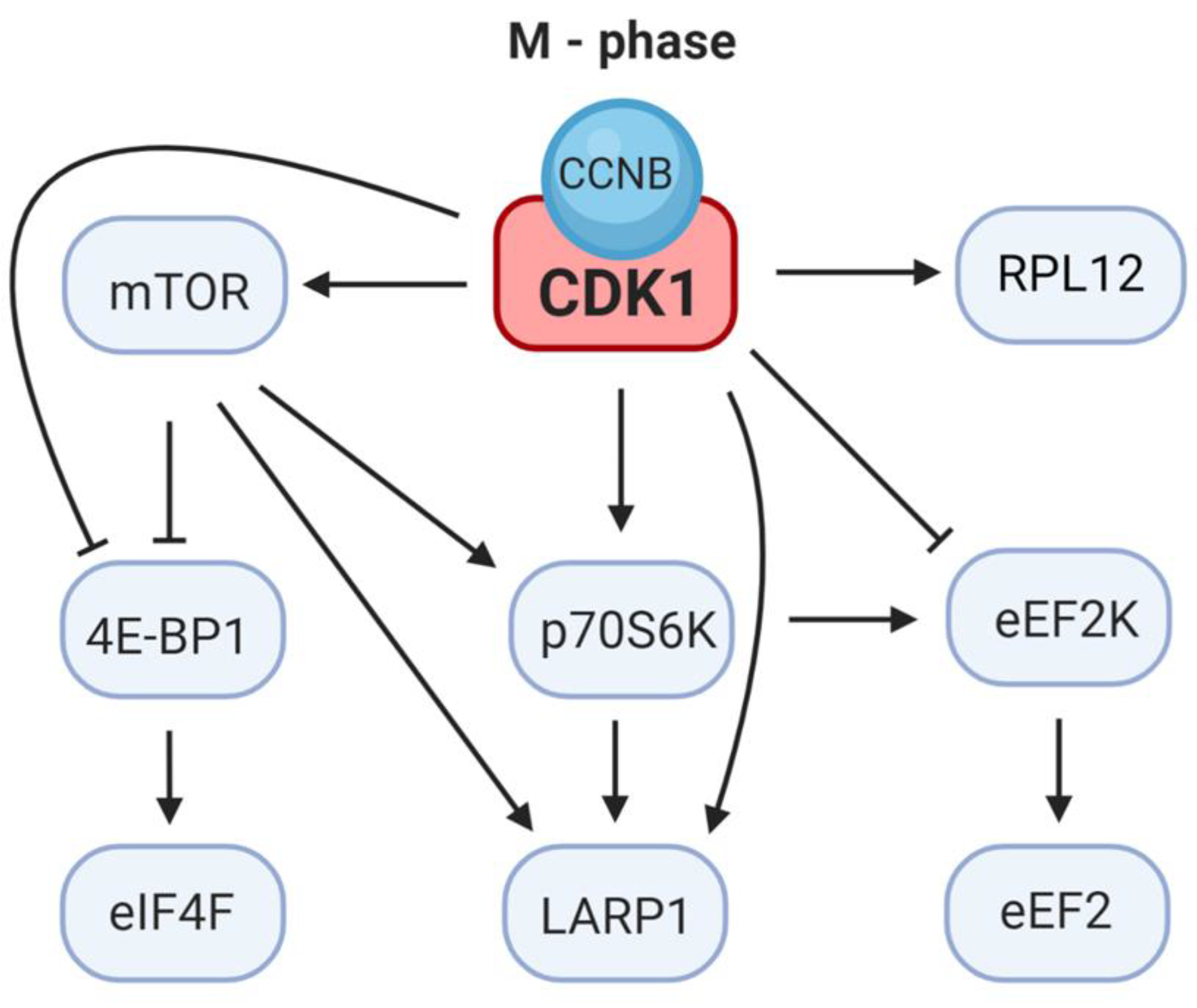
2. CDK1 Contributes to the Reprograming of Translation during M-Phase
3. CDK1 Activity and Localization in the Cell
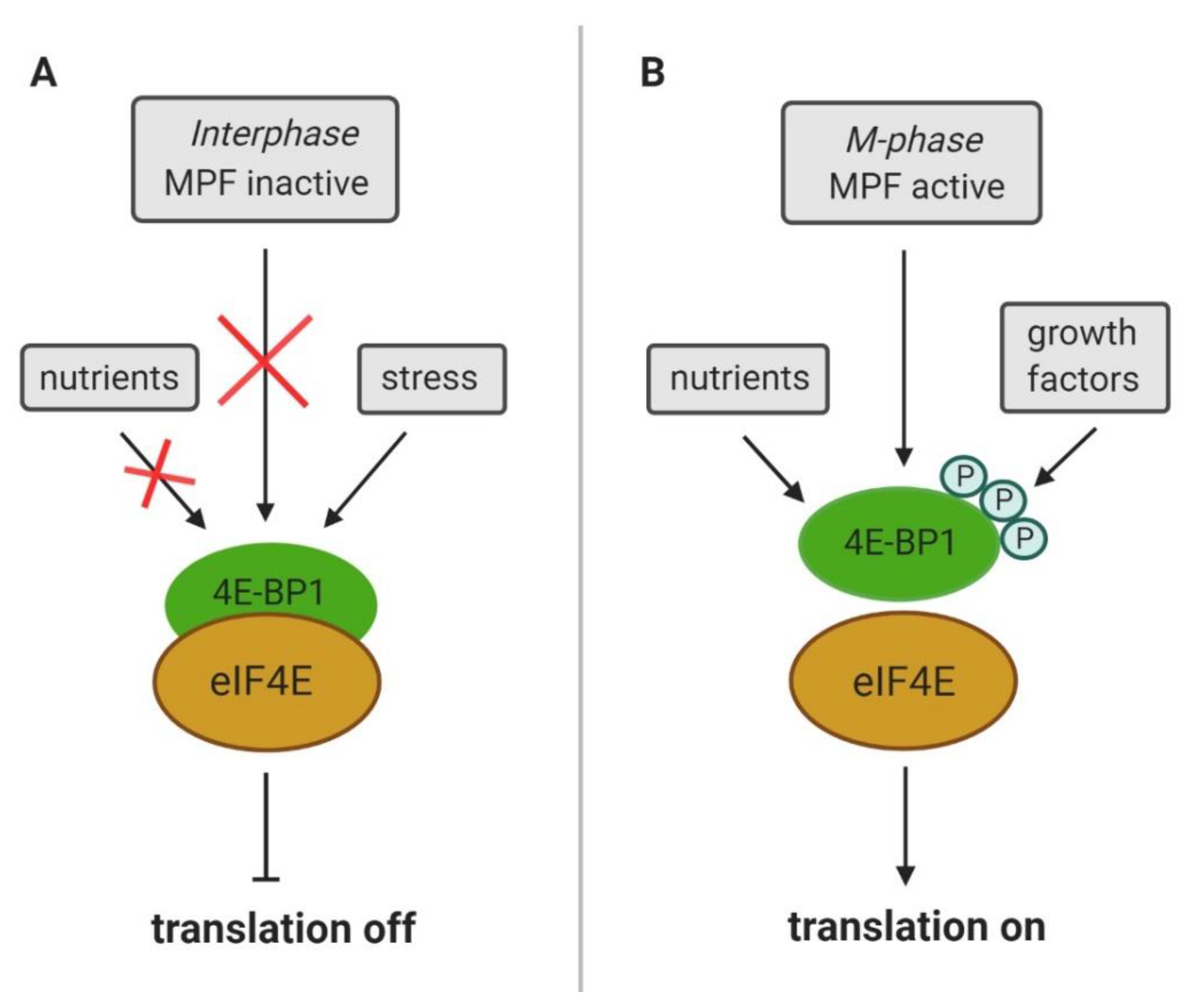
4. CDK1 Substitutes for mTOR Control of Cap-Dependent Translation
5. CDK1 Regulates the Elongation Step of Translation
6. CDK1 Modulates LARP1 Activity
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Adhikari, D.; Zheng, W.; Shen, Y.; Gorre, N.; Ning, Y.; Halet, G.; Kaldis, P.; Liu, K. Cdk1, but not Cdk2, is the sole Cdk that is essential and sufficient to drive resumption of meiosis in mouse oocytes. Hum. Mol. Genet. 2012, 21, 2476–2484. [Google Scholar] [CrossRef] [PubMed]
- Diril, M.K.; Ratnacaram, C.K.; Padmakumar, V.C.; Du, T.; Wasser, M.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 3826–3831. [Google Scholar] [CrossRef] [PubMed]
- Dorée, M.; Peaucellier, G.; Picard, A. Activity of the maturation-promoting factor and the extent of protein phosphorylation oscillate simultaneously during meiotic maturation of starfish oocytes. Dev. Biol. 1983, 99, 489–501. [Google Scholar] [CrossRef]
- Picard, A.; Labbe, J.C.; Doree, M. The cell cycle can occur in starfish oocytes and embryos without the production of transferable MPF (maturation-promoting factor). Dev. Biol. 1988, 128, 129–135. [Google Scholar] [CrossRef]
- Wasserman, W.; Masui, Y. Effects of cycloheximide on a cytoplasmic factor initiating meiotic maturation in Xenopus oocytes. Exp. Cell Res. 1975, 91, 381–388. [Google Scholar] [CrossRef]
- Enserink, J.M.; Kolodner, R.D. An overview of Cdk1-controlled targets and processes. Cell Div. 2010, 5, 1–41. [Google Scholar] [CrossRef]
- Jansova, D.; Koncicka, M.; Tetkova, A.; Cerna, R.; Malik, R.; del Llano, E.; Kubelka, M.; Susor, A. Regulation of 4E-BP1 activity in the mammalian oocyte. Cell Cycle 2017, 16, 927–939. [Google Scholar] [CrossRef]
- Velásquez, C.; Cheng, E.; Shuda, M.; Lee-Oesterreich, P.J.; Von Strandmann, L.P.; Gritsenko, M.A.; Jacobs, J.M.; Moore, P.S.; Chang, Y. Mitotic protein kinase CDK1 phosphorylation of mRNA translation regulator 4E-BP1 Ser83 may contribute to cell transformation. Proc. Natl. Acad. Sci. USA 2016, 113, 8466–8471. [Google Scholar] [CrossRef]
- Krek, W.; Nigg, E.A. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: Evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 1991, 10, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.J.; Glotzer, M.; Lee, T.H.; Philippe, M.; Kirschner, M.W. Cyclin activation of p34cdc2. Cell 1990, 63, 1013–1024. [Google Scholar] [CrossRef]
- Tachibana, K.; Ishiura, M.; Uchida, T.; Kishimoto, T. The starfish egg mRNA responsible for meiosis reinitiation encodes cyclin. Dev. Biol. 1990, 140, 241–252. [Google Scholar] [CrossRef]
- Li, J.; Tang, J.X.; Cheng, J.M.; Hu, B.; Wang, Y.Q.; Aalia, B.; Li, X.Y.; Jin, C.; Wang, X.X.; Deng, S.L.; et al. Cyclin B2 can compensate for Cyclin B1 in oocyte meiosis I. J. Cell Biol. 2018, 217, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Haneke, K.; Schott, J.; Lindner, D.; Hollensen, A.K.; Damgaard, C.K.; Mongis, C.; Knop, M.; Palm, W.; Ruggieri, A.; Stoecklin, G. CDK1 couples proliferation with protein synthesis. J. Cell Biol. 2020, 219, e201906147. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.; Zhang, L.; He, Z.; Feng, G.; Sun, H.; Wang, J.; Li, Z.; Liu, C.; Han, J.; et al. Cyclin b3 is required for metaphase to anaphase transition in oocyte meiosis I. J. Cell Biol. 2019, 218, 1553–1563. [Google Scholar] [CrossRef] [PubMed]
- Karasu, M.E.; Bouftas, N.; Keeney, S.; Wassmann, K. Cyclin B3 promotes anaphase i onset in oocyte meiosis. J. Cell Biol. 2019, 218, 1265–1281. [Google Scholar] [CrossRef]
- Kozak, M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 1978, 15, 1109–1123. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Shirokikh, N.E.; Preiss, T. Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip. Rev. RNA 2018, 9. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Imataka, H.; Gradi, A.; Sonenberg, N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998, 17, 7480–7489. [Google Scholar] [CrossRef]
- Sonenberg, N.; Morgan, M.A.; Merrick, W.C.; Shatkin, A.J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. USA 1978, 75, 4843–4847. [Google Scholar] [CrossRef] [PubMed]
- Sachs, A.B.; Davis, R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 1989, 58, 857–867. [Google Scholar] [CrossRef]
- Wells, S.E.; Hillner, P.E.; Vale, R.D.; Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 1998, 2, 135–140. [Google Scholar] [CrossRef]
- Sun, R.; Cheng, E.; Velásquez, C.; Chang, Y.; Moore, P.S. Mitosis-related phosphorylation of the eukaryotic translation suppressor 4E-BP1 and its interaction with eukaryotic translation initiation factor 4E (eIF4E). J. Biol. Chem. 2019, 294, 11840–11852. [Google Scholar] [CrossRef]
- Sengupta, C.; Peterson, T.R.; Sabatini, D.M.; Sengupta, S. Regulation of the mTOR Complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 2010, 40, 310–322. [Google Scholar] [CrossRef]
- Ellederova, Z.; Kovarova, H.; Melo-Sterza, F.; Livingstone, M.; Tomek, W.; Kubelka, M. Suppression of translation during in vitro maturation of pig oocytes despite enhanced formation of cap-binding protein complex eIF4F and 4E-BP1 hyperphosphorylation. Mol. Reprod. Dev. 2006, 73, 68–76. [Google Scholar] [CrossRef]
- Susor, A.; Jansova, D.; Cerna, R.; Danylevska, A.; Anger, M.; Toralova, T.; Malik, R.; Supolikova, J.; Cook, M.S.; Oh, J.S.; et al. Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat. Commun. 2015, 6, 6078. [Google Scholar] [CrossRef]
- Pyronnet, S.; Dostie, J.; Sonenberg, N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001, 15, 2083–2093. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Stern-Ginossar, N.; Weissman, J.S.; Vale, R.D. Regulation of mRNA translation during mitosis. eLife 2015, 4, e07957. [Google Scholar] [CrossRef]
- Fan, H.; Penman, S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 1970, 50, 655–670. [Google Scholar] [CrossRef]
- Tarnowka, M.A.; Baglioni, C. Regulation of protein synthesis in mitotic HeLa cells. J. Cell. Physiol. 1979, 99, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Coldwell, M.J.; Cowan, J.L.; Vlasak, M.; Mead, A.; Willett, M.; Perry, L.S.; Morley, S.J. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: A reappraisal of translation initiation during mitosis. Cell Cycle 2013, 12, 3615–3628. [Google Scholar] [CrossRef] [PubMed]
- Shuda, M.; Velásquez, C.; Cheng, E.; Cordek, D.G.; Kwun, H.J.; Chang, Y.; Moore, P.S. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci. USA 2015, 112, 5875–5882. [Google Scholar] [CrossRef] [PubMed]
- Anda, S.; Grallert, B. Cell-Cycle-Dependent Regulation of Translation: New interpretations of old observations in light of new approaches. BioEssays 2019, 41, e1900022. [Google Scholar] [CrossRef]
- Silva, R.C.; Dautel, M.; Di Genova, B.M.; Amberg, D.C.; Castilho, B.A.; Sattlegger, E. The Gcn2 Regulator Yih1 Interacts with the cyclin dependent kinase Cdc28 and promotes cell cycle progression through G2/M in budding yeast. PLoS ONE 2015, 10, e0131070. [Google Scholar] [CrossRef]
- Stonyte, V.; Boye, E.; Grallert, B. Regulation of global translation during the cell cycle. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Uppala, J.K.; Ghosh, C.; Sathe, L.; Dey, M. Phosphorylation of translation initiation factor eIF2α at Ser51 depends on site- and context-specific information. FEBS Lett. 2018, 592, 3116–3125. [Google Scholar] [CrossRef]
- Gordiyenko, Y.; Llácer, J.L.; Ramakrishnan, V. Structural basis for the inhibition of translation through eIF2α phosphorylation. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- De La Fuente, R.; Viveiros, M.M.; Burns, K.H.; Adashi, E.Y.; Matzuk, M.M.; Eppig, J.J. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev. Biol. 2004, 275, 447–458. [Google Scholar] [CrossRef]
- Eppig, J.J.; Schroeder, A.C. Capacity of Mouse Oocytes from Preantral Follicles to Undergo Embryogenesis and Development to Live Young after Growth, Maturation, and Fertilization in Vitro1. Biol. Reprod. 1989, 41, 268–276. [Google Scholar] [CrossRef]
- Šušor, A.; Jelínková, L.; Karabínová, P.; Torner, H.; Tomek, W.; Kovářová, H.; Kubelka, M. Regulation of cap-dependent translation initiation in the early stage porcine parthenotes. Mol. Reprod. Dev. 2008, 75, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Ellederová, Z.; Cais, O.; Šušor, A.; Uhlířová, K.; Kovářová, H.; Jelínková, L.; Tomek, W.; Kubelka, M. ERK1/2 map kinase metabolic pathway is responsible for phosphorylation of translation initiation factor eIF4E during in vitro maturation of pig oocytes. Mol. Reprod. Dev. 2008, 75, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Tomek, W.; Sterza, F.A.M.; Kubelka, M.; Wollenhaupt, K.; Torner, H.; Anger, M.; Kanitz, W. Regulation of Translation During In Vitro Maturation of Bovine Oocytes: The Role of MAP Kinase, eIF4E (Cap Binding Protein) Phosphorylation, and eIF4E-BP11. Biol. Reprod. 2002, 66, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Valle, F.; Badura, M.L.; Braunstein, S.; Narasimhan, M.; Schneider, R.J. Mitotic Raptor Promotes mTORC1 Activity, G2/M Cell Cycle Progression, and Internal Ribosome Entry Site-Mediated mRNA Translation. Mol. Cell. Biol. 2010, 30, 3151–3164. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Yoon, I.S.; Chung, J.H.; Hahm, S.H.; Park, M.J.; Lee, Y.R.; Ko, S.I.; Kang, L.W.; Kim, T.S.; Kim, J.; Han, Y.S. Ribosomal protein S3 is phosphorylated by Cdk1/cdc2 during G2/M phase. BMB Rep. 2011, 44, 529–534. [Google Scholar] [CrossRef]
- Susor, A.; Kubelka, M. Translational regulation in the mammalian oocyte. In Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2017; Volume 63, pp. 257–295. [Google Scholar]
- Jang, C.Y.; Kim, H.D.; Zhang, X.; Chang, J.S.; Kim, J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 2012, 429, 57–62. [Google Scholar] [CrossRef]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef]
- Imami, K.; Milek, M.; Bogdanow, B.; Yasuda, T.; Kastelic, N.; Zauber, H.; Ishihama, Y.; Landthaler, M.; Selbach, M. Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol. Cell 2018, 72, 84–98. [Google Scholar] [CrossRef]
- Odle, R.I.; Walker, S.A.; Oxley, D.; Kidger, A.M.; Balmanno, K.; Gilley, R.; Okkenhaug, H.; Florey, O.; Ktistakis, N.T.; Cook, S.J. An mTORC1-to-CDK1 Switch Maintains Autophagy Suppression during Mitosis. Mol. Cell 2020, 77, 228–240. [Google Scholar] [CrossRef]
- Berman, A.J.; Thoreen, C.C.; Dedeic, Z.; Chettle, J.; Roux, P.P.; Sarah, B.P. Controversies around the function of LARP1. RNA Biol. 2020, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dobrikov, M.I.; Shveygert, M.; Brown, M.C.; Gromeier, M. Mitotic Phosphorylation of Eukaryotic Initiation Factor 4G1 (eIF4G1) at Ser1232 by Cdk1:Cyclin B Inhibits eIF4A Helicase Complex Binding with RNA. Mol. Cell. Biol. 2014, 34, 439–451. [Google Scholar] [CrossRef]
- Sivan, G.; Aviner, R.; Elroy-Stein, O. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J. Biol. Chem. 2011, 286, 27927–27935. [Google Scholar] [CrossRef] [PubMed]
- Mulner-Lorillon, O.; Minella, O.; Cormier, P.; Capony, J.P.; Cavadore, J.C.; Morales, J.; Poulhe, R.; Bellé, R. Elongation factor EF-1 delta, a new target for maturation-promoting factor in Xenopus oocytes. J. Biol. Chem. 1994, 269, 20201–20207. [Google Scholar]
- Smith, E.M.; Proud, C.G. cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle and amino acid-dependent manner. EMBO J. 2008, 27, 1005–1016. [Google Scholar] [CrossRef]
- Gnad, F.; Gunawardena, J.; Mann, M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011, 39, D253–D260. [Google Scholar] [CrossRef]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics revealswidespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef]
- Murano, K.; Okuwaki, M.; Hisaoka, M.; Nagata, K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 2008, 28, 3114–3126. [Google Scholar] [CrossRef]
- Okuwaki, M.; Matsumoto, K.; Tsujimoto, M.; Nagata, K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001, 506, 272–276. [Google Scholar] [CrossRef]
- Yu, Y.; Maggi, L.B.; Brady, S.N.; Apicelli, A.J.; Dai, M.-S.; Lu, H.; Weber, J.D. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006, 26, 3798–3809. [Google Scholar] [CrossRef] [PubMed]
- Hisaoka, M.; Ueshima, S.; Murano, K.; Nagata, K.; Okuwaki, M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol. Cell. Biol. 2010, 30, 4952–4964. [Google Scholar] [CrossRef] [PubMed]
- Hagting, A.; Jackman, M.; Simpson, K.; Pines, J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 1999, 9, 680–689. [Google Scholar] [CrossRef]
- Lindqvist, A.; van Zon, W.; Karlsson Rosenthal, C.; Wolthuis, R.M.F. Cyclin B1–Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007, 5, e123. [Google Scholar] [CrossRef] [PubMed]
- Jackman, M.; Lindon, C.; Niggt, E.A.; Pines, J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef]
- Gavet, O.; Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 2010, 18, 533–543. [Google Scholar] [CrossRef]
- Karabinova, P.; Kubelka, M.; Susor, A. Proteasomal degradation of ubiquitinated proteins in oocyte meiosis and fertilization in mammals. Cell Tissue Res. 2011, 346, 1. [Google Scholar] [CrossRef]
- Pines, J.; Hunter, T. Cyclin-dependent kinases: A new cell cycle motif? Trends Cell Biol. 1991, 1, 117–121. [Google Scholar] [CrossRef]
- Koncicka, M.; Tetkova, A.; Jansova, D.; Del Llano, E.; Gahurova, L.; Kracmarova, J.; Prokesova, S.; Masek, T.; Pospisek, M.; Bruce, A.W.; et al. Increased expression of maturation promoting factor components speeds up meiosis in oocytes from aged females. Int. J. Mol. Sci. 2018, 19, 2841. [Google Scholar] [CrossRef]
- Schweizer, N.; Pawar, N.; Weiss, M.; Maiato, H. An organelle-exclusion envelope assists mitosis and underlies distinct molecular crowding in the spindle region. J. Cell Biol. 2015, 210, 695–704. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. mTORC1 Signaling: What We Still Don’t Know. J. Mol. Cell Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 2015, 1849, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Suzuki, Y.; Takeuchi, N.; Wakaguri, H.; Ueda, T.; Sugano, S.; Nakai, K. Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic Acids Res. 2008, 36, 3707–3715. [Google Scholar] [CrossRef] [PubMed]
- Fingar, D.C.; Richardson, C.J.; Tee, A.R.; Cheatham, L.; Tsou, C.; Blenis, J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/Eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004, 24, 200–216. [Google Scholar] [CrossRef]
- Miettinen, T.P.; Kang, J.H.; Yang, L.F.; Manalis, S.R. Mammalian cell growth dynamics in mitosis. eLife 2019, 8, e44700. [Google Scholar] [CrossRef]
- Livingstone, M.; Bidinosti, M. Rapamycin-insensitive mTORC1 activity controls eIF4E:4E-BP1 binding. F1000Research 2012, 1, 4. [Google Scholar] [CrossRef]
- Burnett, P.E.; Barrow, R.K.; Cohen, N.A.; Snyder, S.H.; Sabatini, D.M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 1998, 95, 1432–1437. [Google Scholar] [CrossRef]
- Romasko, E.J.; Amarnath, D.; Midic, U.; Latham, K.E. Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: Localized translational control supporting female meiosis in mammals. Genetics 2013, 195, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Papst, P.J.; Sugiyama, H.; Nagasawa, M.; Lucas, J.J.; Maller, J.L.; Terada, N. Cdc2-cyclin B phosphorylates p70 S6 kinase on Ser411 at mitosis. J. Biol. Chem. 1998, 273, 15077–15084. [Google Scholar] [CrossRef] [PubMed]
- Shah, O.J.; Ghosh, S.; Hunter, T. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by Cdc2. J. Biol. Chem. 2003, 278, 16433–16442. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Małecki, J.; Falnes, P. Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation. RNA Biol. 2018, 15, 314–319. [Google Scholar] [CrossRef]
- Sivan, G.; Elroy-Stein, O. Regulation of mRNA Translation during cellular division. Cell Cycle 2008, 7, 741–744. [Google Scholar] [CrossRef]
- Monnier, A.; Bellé, R.; Morales, J.; Cormier, P.; Boulben, S.; Mulner-Lorillon, O. Evidence for regulation of protein synthesis at the elongation step by CDK1/cyclin B phosphorylation. Nucleic Acids Res. 2001, 29, 1453–1457. [Google Scholar] [CrossRef][Green Version]
- Bellé, R.; Derancourt, J.; Poulhe, R.; Capony, J.P.; Ozon, R.; Mulner-Lorillon, O. A purified complex from Xenopus oocytes contains a p47 protein, an in vivo substrate of MPF, and a p30 protein respectively homologous to elongation factors EF-1γ and EF-1β. FEBS Lett. 1989, 255, 101–104. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kato, K.; Tanaka, M.; Kanamori, M.; Nishiyama, Y.; Yamanashi, Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1dekta. J. Virol. 2003, 77, 2359–2368. [Google Scholar] [CrossRef]
- Tcherkezian, J.; Cargnello, M.; Romeo, Y.; Huttlin, E.L.; Lavoie, G.; Gygi, S.P.; Roux, P.P. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014, 28, 357–371. [Google Scholar] [CrossRef]
- Aoki, K.; Adachi, S.; Homoto, M.; Kusano, H.; Koike, K.; Natsume, T. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013, 587, 2173–2178. [Google Scholar] [CrossRef]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yoon, S.O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Freeberg, M.A.; Han, T.; Kamath, A.; Yao, Y.; Fukuda, T.; Suzuki, T.; Kim, J.K.; Inoki, K. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife 2017, 6, e25237. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.; Hopkins, T.G.; Michael, T.; Abd-Latip, N.; Weir, J.; Aboagye, E.; Mauri, F.; Jameson, C.; Sturge, J.; Gabra, H.; et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 2015, 34, 5025–5036. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Huang, L.; Xie, S.; Xie, D.; Zhang, G.; Wang, P.; Peng, L.; Gao, Z. LARP1 predict the prognosis for early-stage and AFP-normal hepatocellular carcinoma. J. Transl. Med. 2013, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Lin, S.T.; Mi, Y.S.; Liu, Y.; Ma, Y.; Sun, H.M.; Peng, Z.H.; Fan, J.W. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumor Biol. 2016, 37, 14585–14594. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, J.; Lu, H.; Lin, B.; Cai, S.; Guo, J.; Zang, F.; Chen, R. LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol. Rep. 2017, 38, 3659–3667. [Google Scholar] [CrossRef]

| CDK1 Substrate | Ref. | Role of CDK1 Substrate | |
|---|---|---|---|
| CDK1 and mTOR | Raptor | [51] | mTOR phosphorylation and activation |
| mTOR (Ser2448) | [7] | 4E-BP1 and p70S6K phosphorylation | |
| p70S6K | [8,33] | Regulation of TOP mRNA translation | |
| LARP1 | [13,52] | Regulation of TOP mRNA translation | |
| CDK1 and 4E-BP1 | 4E-BP1 (Thr37/Thr46) | [33] | Thr37/Thr46 phosphorylation primes 4E-BP1 for Ser65 and Thr70 phosphorylation |
| 4E-BP1 (Thr70) | [7,8] | Thr70 phosphorylation regulates 4E-BP1 | |
| 4E-BP1 (Ser83) | [8] | The role of Ser83 phosphorylation is unknown | |
| CDK1 and ribo-some assembly | eIF2α | [13] | Regulation of eIF2 complex |
| eIF4G1 | [53] | mRNA recruitment to the ribosome | |
| RPL12 | [50] | Translational program | |
| CDK1 and elongation | eEF1B | [54] | Allows ribosome movement along RNA |
| eEF1D | [55] | Delivery of aminoacyl-tRNA to the ribosome | |
| eEF2K | [56] | Regulation the elongation step of protein synthesis |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalous, J.; Jansová, D.; Šušor, A. Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase. Cells 2020, 9, 1568. https://doi.org/10.3390/cells9071568
Kalous J, Jansová D, Šušor A. Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase. Cells. 2020; 9(7):1568. https://doi.org/10.3390/cells9071568
Chicago/Turabian StyleKalous, Jaroslav, Denisa Jansová, and Andrej Šušor. 2020. "Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase" Cells 9, no. 7: 1568. https://doi.org/10.3390/cells9071568
APA StyleKalous, J., Jansová, D., & Šušor, A. (2020). Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase. Cells, 9(7), 1568. https://doi.org/10.3390/cells9071568





