The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis Via Regulation of a Specific mRNA Set
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks and Genetics
2.2. Immunofluorescence Staining and Confocal Microscopy
2.3. Antibodies
2.4. RNA Extraction, Reverse Transcription, and Real-Time Quantitative PCR
2.5. Fertility Tests
2.6. Electron Microscopy
2.7. CLIP-seq Analysis
2.8. Computational Analysis of CLIP Libraries
3. Results
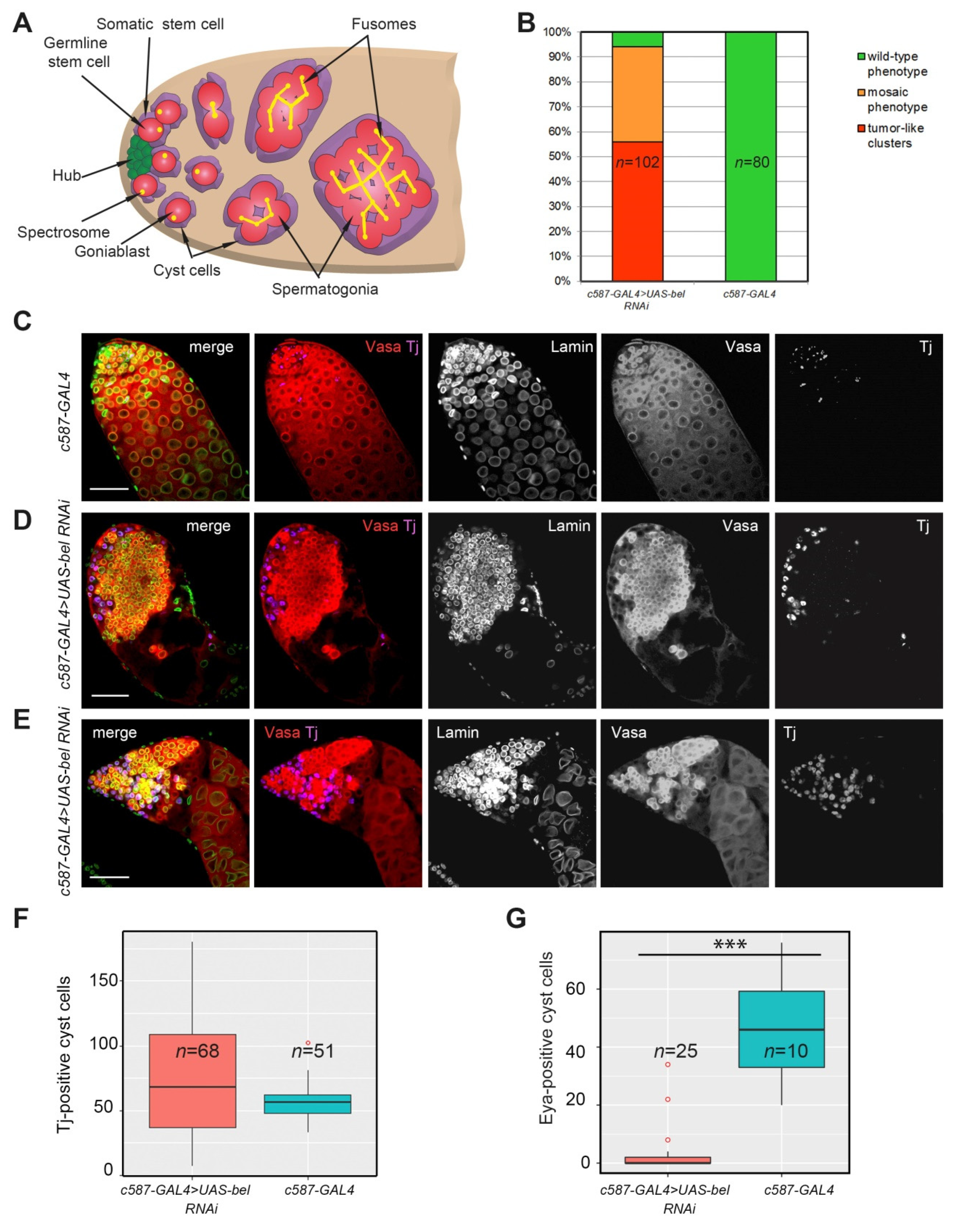
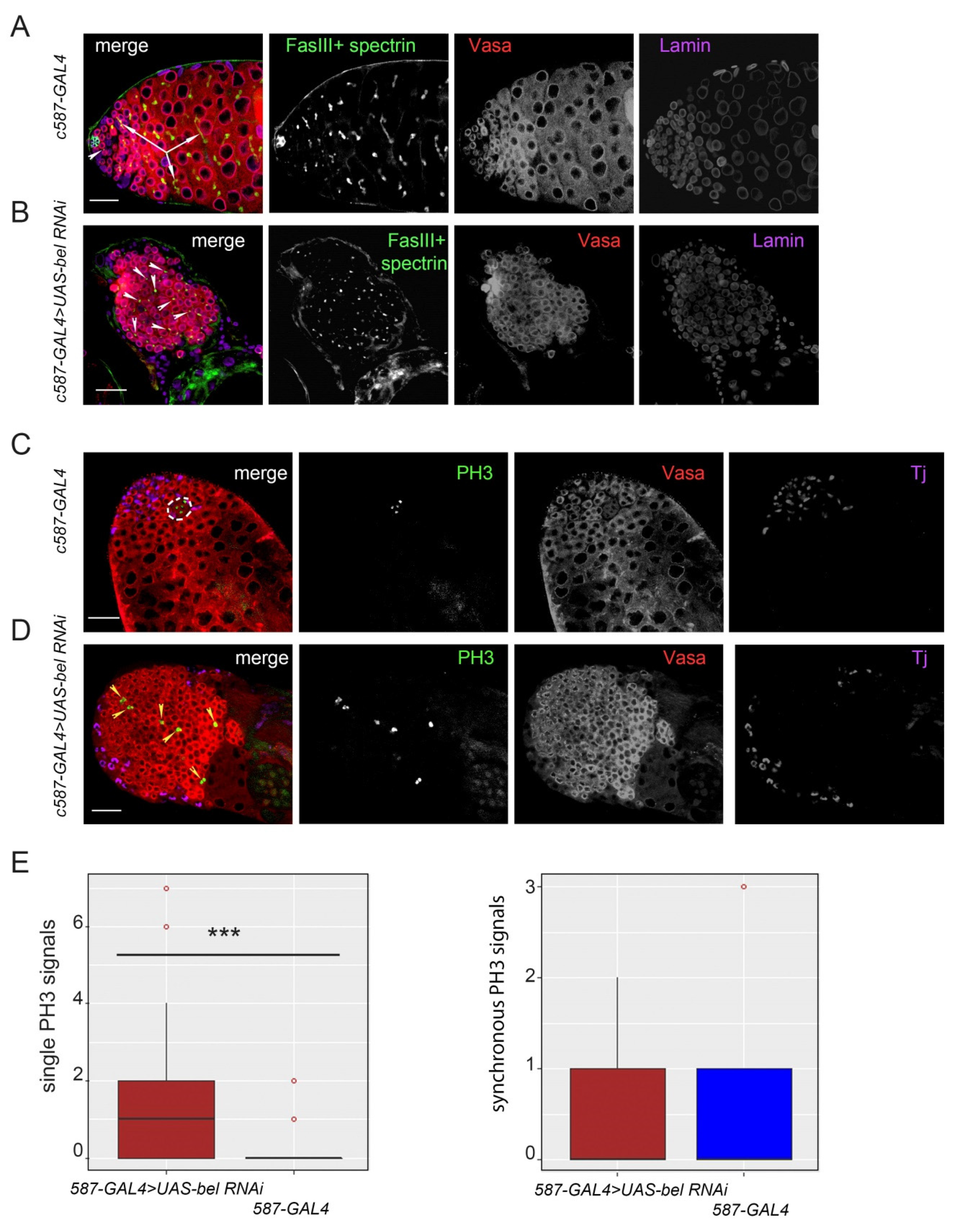
3.1. Functioning of Belle in Cyst Cells is Required for Differentiation of Early Germ Cells
3.2. Proliferation Activity of Germ Cells in the Clusters
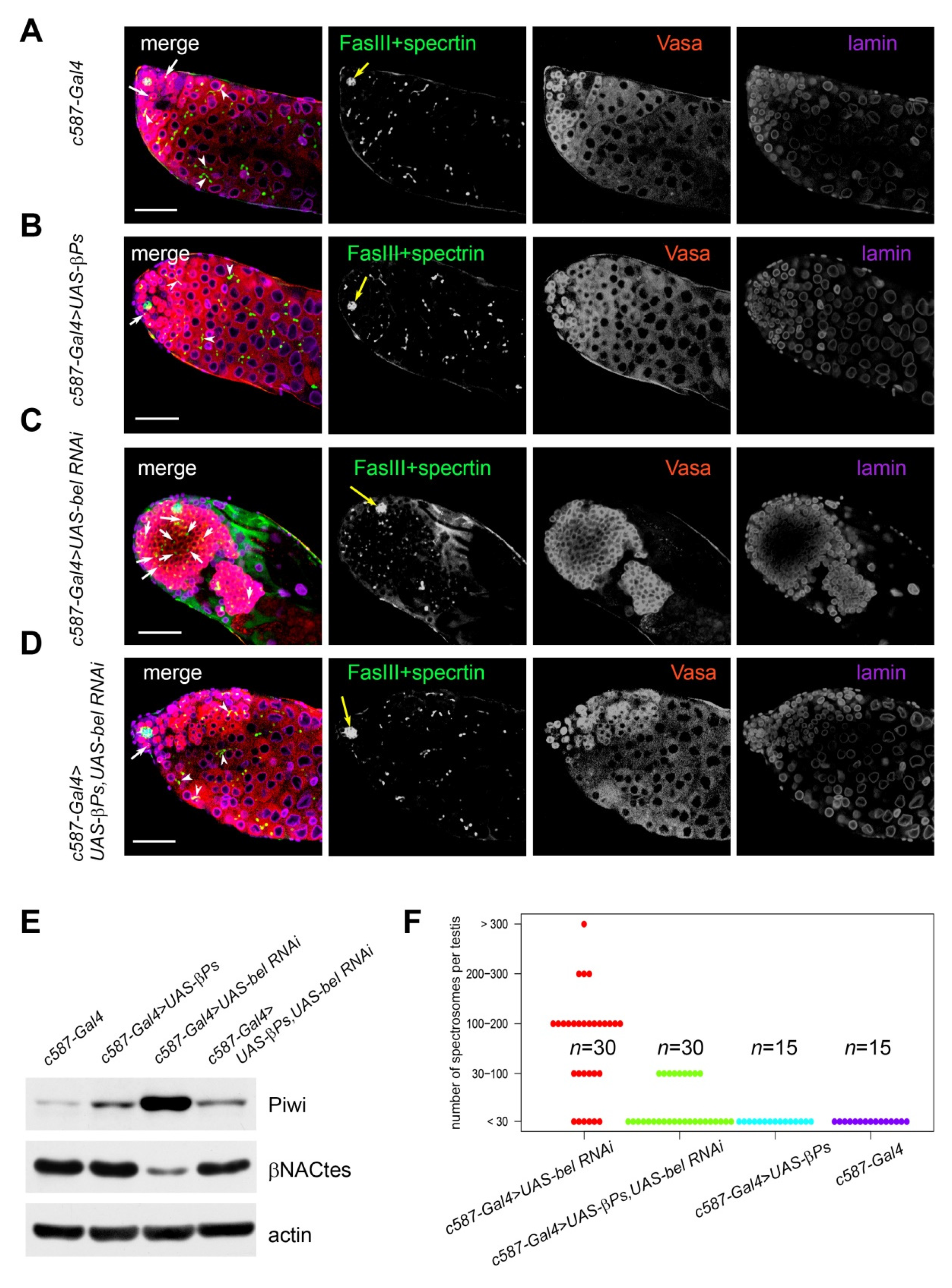
3.3. Ectopic Overexpression of A Transgenic Arms10 COPY in Cyst Cells Did Not Restore the Belle Knockdown Phenotype in the Testes
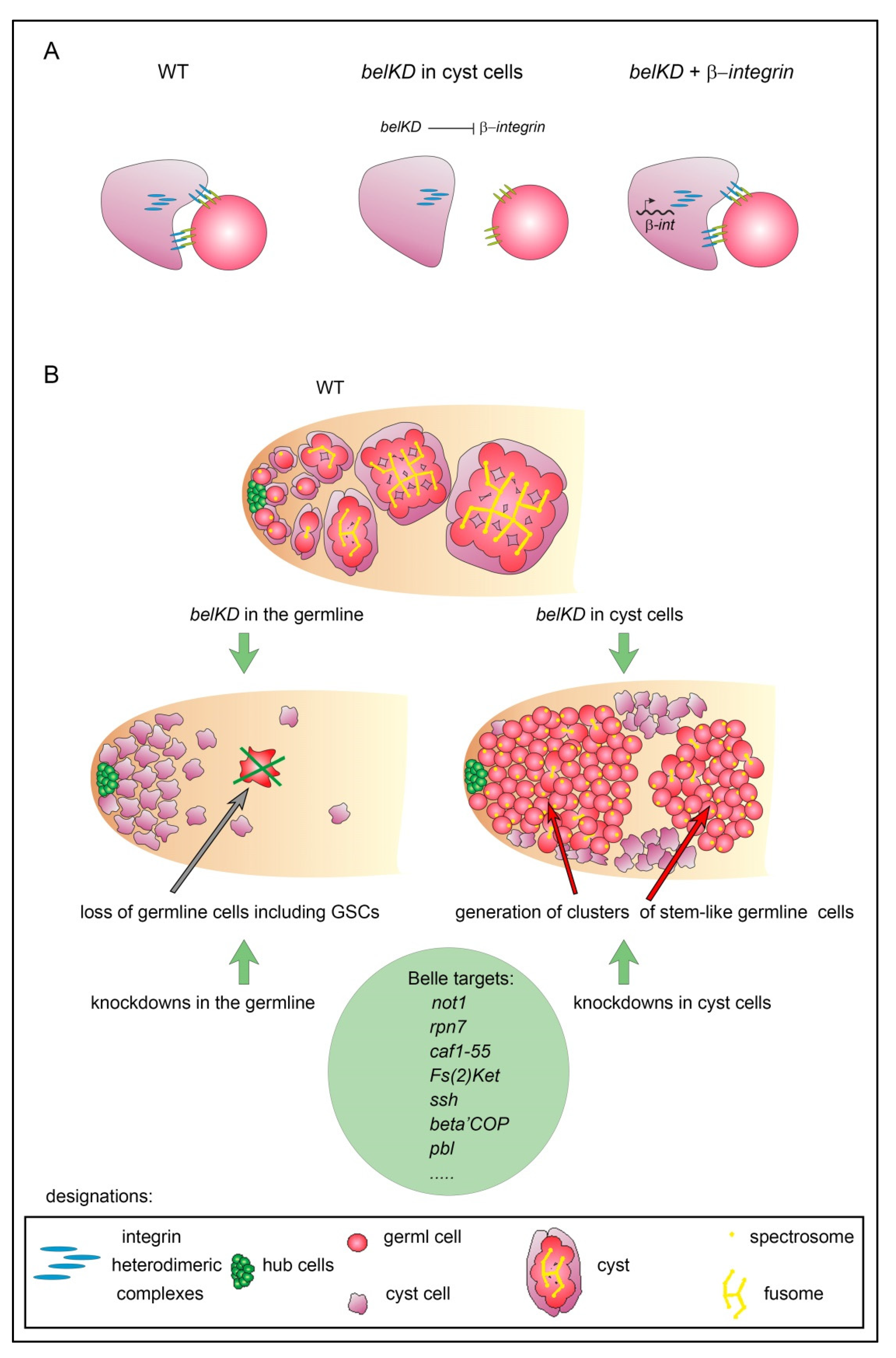
3.4. Ectopic Expression of β-Integrin Rescued Early Stages of Spermatogenesis in RNAi belKD Testes
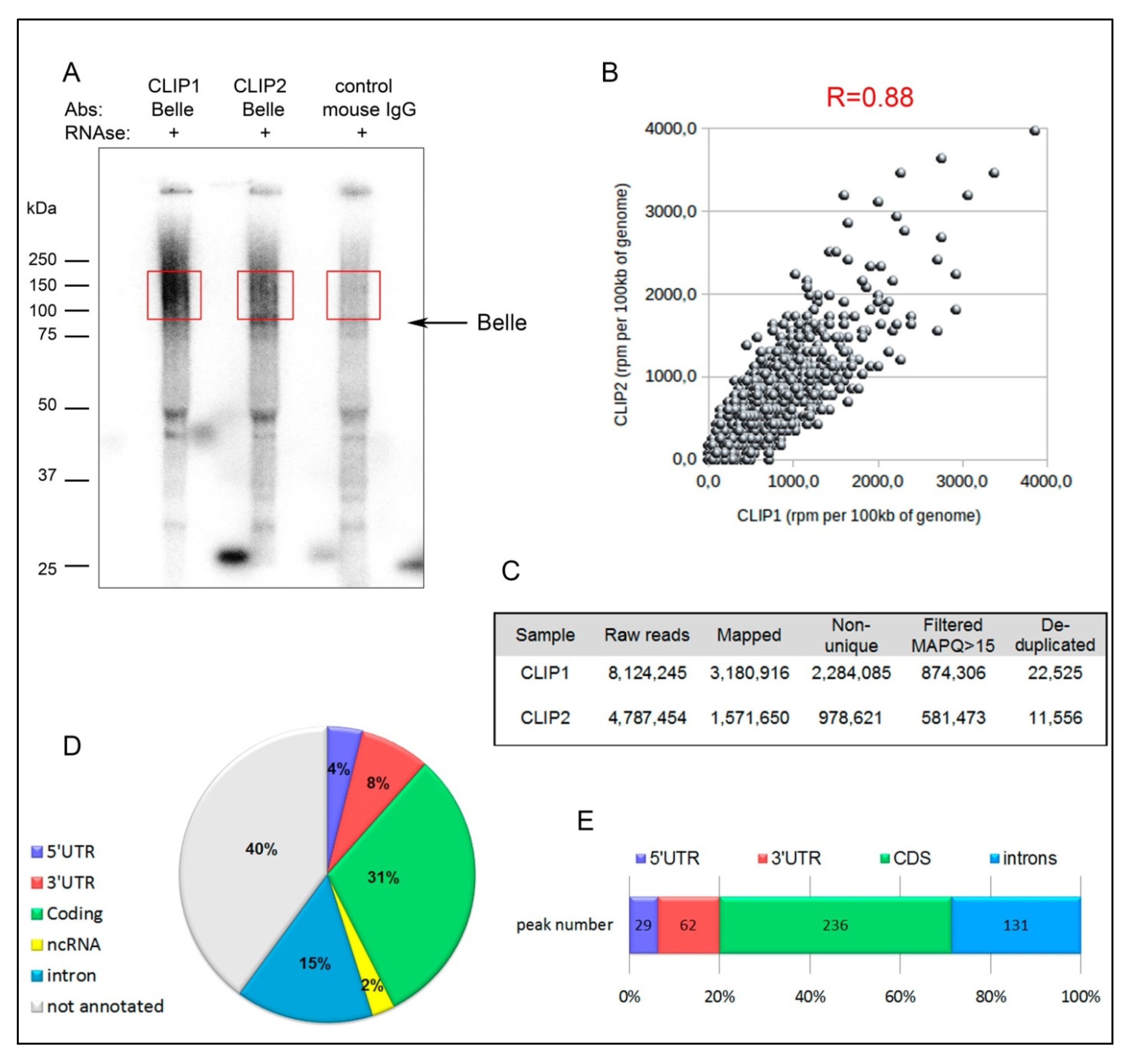
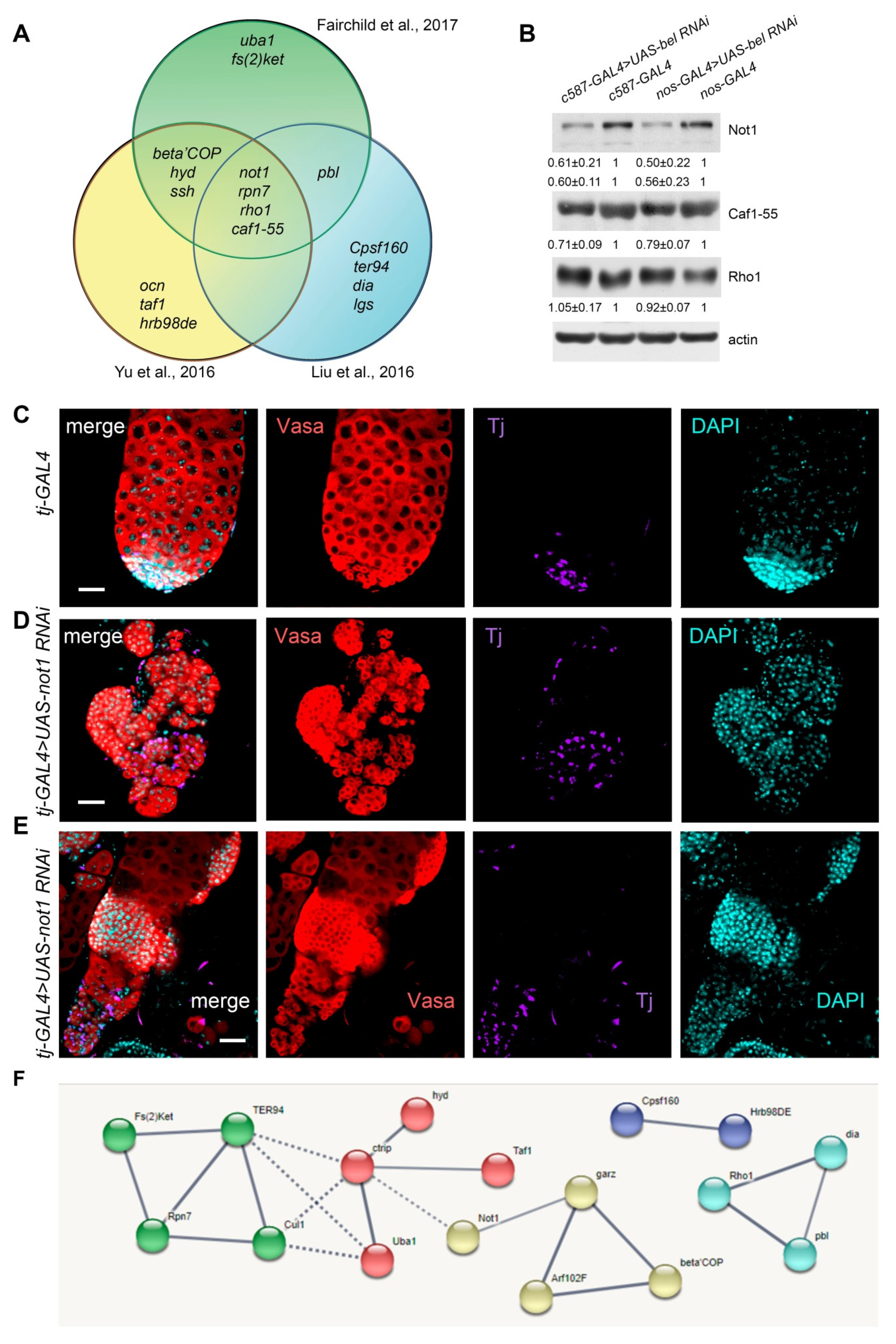
3.5. CLIP-seq Analysis for Identification of Belle mRNA Targets
3.6. Belle is Involved in Translational Regulation of Multiple mRNA Targets
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Linder, P.; Fuller-Pace, F.V. Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta. 2013, 1829, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Linder, P.; Jankowsky, E. From unwinding to clamping – the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Kotov, A.A.; Akulenko, N.V.; Kibanov, M.V.; Olenina, L.V. Dead-box RNA helicases in animal gametogenesis. Mol. Biol. (Mosk). 2014, 48, 16–28. [Google Scholar] [CrossRef]
- Lahn, B.T.; Page, D.C. Functional coherence of the human Y chromosome. Science 1997, 278, 675–680. [Google Scholar] [CrossRef]
- Ditton, H.J.; Zimmer, J.; Kamp, C.; Rajpert-De Meyts, E.; Vogt, P.H. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 2004, 13, 2333–2341. [Google Scholar] [CrossRef]
- Ramathal, C.; Angulo, B.; Sukhwani, M.; Cui, J.; Durruthy-Durruthy, J.; Fang, F.; Schanes, P.; Turek, P.J.; Orwig, K.E.; Reijo Pera, R. DDX3Y gene rescue of a Y chromosome AZFa deletion restores germ cell formation and transcriptional programs. Sci. Rep. 2015, 5, 15041. [Google Scholar] [CrossRef]
- Kamp, C.; Huellen, K.; Fernandes, S.; Sousa, M.; Schlegel, P.N.; Mielnik, A.; Kleiman, S.; Yavetz, H.; Krause, W.; Küpker, W.; et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol. Hum. Reprod. 2001, 7, 987–994. [Google Scholar] [CrossRef]
- Foresta, C.; Ferlin, A.; Moro, E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum. Mol. Genet. 2000, 9, 1161–1169. [Google Scholar] [CrossRef]
- Lardone, M.C.; Parodi, D.A.; Valdevenito, R.; Ebensperger, M.; Piottante, A.; Madariaga, M.; Smith, R.; Pommer, R.; Zambrano, N.; Castro, A. Quantification of DDX3Y, RBMY1, DAZ and TSPY mRNAs in testes of patients with severe impairment of spermatogenesis. Mol. Hum. Reprod. 2007, 13, 705–712. [Google Scholar] [CrossRef]
- Gueler, B.; Sonne, S.B.; Zimmer, J.; Hilscher, B.; Hilscher, W.; Græm, N.; Rajpert-De Meyts, E.; Vogt, P.H. AZFa protein DDX3Y is differentially expressed in human male germ cells during development and in testicular tumours: new evidence for phenotypic plasticity of germ cells. Hum. Reprod. 2012, 27, 1547–1555. [Google Scholar] [CrossRef]
- Botlagunta, M.; Vesuna, F.; Mironchik, Y.; Raman, A.; Lisok, A.; Winnard, P., Jr.; Mukadam, S.; Van Diest, P.; Chen, J.H.; Farabaugh, P.; et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 2008, 27, 3912–3922. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front. Genet. 2014, 5, 423. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Y.; Zhou, J.; Zhao, Y.; Cao, Y.; Chen, X. Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am. J. Cancer Res. 2016, 6, 387–402. [Google Scholar] [PubMed]
- Chen, H.H.; Tarn, W.Y. DDX3-mediated translational activation drives β-catenin signaling and cancer cell motility. Cancer Cell & Microenvironment 2014, 1, e392. [Google Scholar] [CrossRef][Green Version]
- Chen, H.H.; Yu, H.I.; Cho, W.C.; Tarn, W.Y. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene 2015, 34, 2790–2800. [Google Scholar] [CrossRef]
- Cruciat, C.M.; Dolde, C.; de Groot, R.E.; Ohkawara, B.; Reinhard, C.; Korswagen, H.C.; Niehrs, C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science 2013, 339, 1436–1441. [Google Scholar] [CrossRef]
- Johnstone, O.; Deuring, R.; Bock, R.; Linder, P.; Fuller, M.T.; Lasko, P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 2005, 277, 92–101. [Google Scholar] [CrossRef]
- Ihry, R.J.; Sapiro, A.L.; Bashirullah, A. Translational control by the DEAD Box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 2012, 8, e1003085. [Google Scholar] [CrossRef]
- Ulvila, J.; Parikka, M.; Kleino, A.; Sormunen, R.; Ezekowitz, R.A.; Kocks, C.; Rämet, M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 2006, 281, 14370–14375. [Google Scholar] [CrossRef]
- Zhou, R.; Hotta, I.; Denli, A.M.; Hong, P.; Perrimon, N.; Hannon, G.J. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell. 2008, 32, 592–599. [Google Scholar] [CrossRef]
- Poulton, J.S.; Huang, Y.C.; Smith, L.; Sun, J.; Leake, N.; Schleede, J.; Stevens, L.M.; Deng, W.M. The microRNA pathway regulates the temporal pattern of Notch signaling in Drosophila follicle cells. Development 2011, 138, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Pek, J.W.; Kai, T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA 2011, 108, 12007–12012. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, P.; Damulewicz, M.; Carbognin, E.; Caccin, L.; Puricella, A.; Specchia, V.; Bozzetti, M.P.; Costa, R.; Mazzotta, G.M. The RNA Helicase BELLE Is Involved in Circadian Rhythmicity and in Transposons Regulation in Drosophila melanogaster. Front. Physiol. 2019, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.M.; Frolov, M.V. Mutation of the DEAD-box helicase belle downregulates the cyclin-dependent kinase inhibitor Dacapo. Cell Cycle 2010, 9, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Kotov, A.A.; Olenkina, O.M.; Kibanov, M.V.; Olenina, L.V. RNA helicase Belle (DDX3) is essential for male germline stem cell maintenance and division in Drosophila. Biochim. Biophys. Acta. 2016, 1863, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.E.; Kandasamy, S.K.; Zhu, L.; Fukunaga, R. DEAD-box RNA helicase Belle posttranscriptionally promotes gene expression in an ATPase activity-dependent manner. RNA 2019, 25, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Yarunin, A.; Harris, R.E.; Ashe, M.P.; Ashe, H.L. Patterning of the Drosophila oocyte by a sequential translation repression program involving the d4EHP and Belle translational repressors. RNA Biol. 2011, 8, 904–912. [Google Scholar] [CrossRef]
- Götze, M.; Dufourt, J.; Ihling, C.; Rammelt, C.; Pierson, S.; Sambrani, N.; Temme, C.; Sinz, A.; Simonelig, M.; Wahle, E. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 2017, 23, 1552–1568. [Google Scholar] [CrossRef]
- Kotov, A.A.; Olenkina, O.M.; Godneeva, B.K.; Adashev, V.E.; Olenina, L.V. Progress in understanding the molecular functions of DDX3Y (DBY) in male germ cell development and maintenance. Biosci. Trends 2017, 11, 46–53. [Google Scholar] [CrossRef]
- Fabrizio, J.J.; Boyle, M.; DiNardo, S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev. Biol. 2003, 258, 117–128. [Google Scholar] [CrossRef]
- Kiger, A.A.; Jones, D.L.; Schulz, C.; Rogers, M.B.; Fuller, M.T. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 2001, 294, 2542–2545. [Google Scholar] [CrossRef]
- Lim, J.G.; Fuller, M.T. Somatic cell lineage is required for differentiation and not maintenance of germline stem cells in Drosophila testes. Proc. Natl. Acad. Sci. USA 2012, 109, 18477–18481. [Google Scholar] [CrossRef] [PubMed]
- White-Cooper, H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 2010, 139, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Spradling, A.; Fuller, M.T.; Braun, R.E.; Yoshida, S. Germline stem cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002642. [Google Scholar] [CrossRef] [PubMed]
- Beumer, K.J.; Rohrbough, J.; Prokop, A.; Broadie, K. A role for PS integrins in morphological growth and synaptic function at the postembryonic neuromuscular junction of Drosophila. Development 1999, 126, 5833–5846. [Google Scholar]
- Kibanov, M.V.; Kotov, A.A.; Olenina, L.V. Multicolor fluorescence imaging of whole-mount Drosophila testes for studying spermatogenesis. Anal. Biochem. 2013, 436, 55–64. [Google Scholar] [CrossRef]
- Osouda, S.; Nakamura, Y.; de Saint Phalle, B.; McConnell, M.; Horigome, T.; Sugiyama, S.; Fisher, P.A.; Furukawa, K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev. Biol. 2005, 284, 219–232. [Google Scholar] [CrossRef]
- Li, M.A.; Alls, J.D.; Avancini, R.M.; Koo, K.; Godt, D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell. Biol. 2003, 5, 994–1000. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Temme, C.; Zhang, L.; Kremmer, E.; Ihling, C.; Chartier, A.; Sinz, A.; Simonelig, M.; Wahle, E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 2010, 16, 1356–1370. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Uren, P.J.; Bahrami-Samani, E.; Burns, S.C.; Qiao, M.; Karginov, F.V.; Hodges, E.; Hannon, G.J.; Sanford, J.R.; Penalva, L.O.; Smith, A.D. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 2012, 28, 3013–3020. [Google Scholar] [CrossRef]
- Huang, d.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Voog, J.; D’Alterio, C.; Jones, D.L. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 2008, 454, 1132–1136. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 2000, 127, 503–514. [Google Scholar]
- Saito, K.; Nishida, K.M.; Mori, T.; Kawamura, Y.; Miyoshi, K.; Nagami, T.; Siomi, H.; Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006, 20, 2214–2222. [Google Scholar] [CrossRef]
- Pai, L.M.; Orsulic, S.; Bejsovec, A.; Peifer, M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 1997, 124, 2255–2266. [Google Scholar]
- Singh, S.R.; Liu, Y.; Zhao, J.; Zeng, X.; Hou, S.X. The novel tumour suppressor Madm regulates stem cell competition in the Drosophila testis. Nat. Commun. 2016, 7, 10473. [Google Scholar] [CrossRef] [PubMed]
- Kogan, G.L.; Akulenko, N.V.; Abramov, Y.A.; Sokolova, O.A.; Fefelova, E.A.; Gvozdev, V.A. Nascent polypeptide-associated complex as tissue-specific cofactor during germinal cell differentiation in Drosophila testes. Mol. Biol. (Mosk.) 2017, 51, 596–601. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, Q.; Chan, B.; Liu, H.; Singh, S.R.; Manley, J.; Lee, J.; Weideman, A.M.; Hou, G.; Hou, S.X. Whole-animal genome-wide RNAi screen identifies networks regulating male germline stem cells in Drosophila. Nat. Commun. 2016, 7, 12149. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, M.J.; Islam, F.; Tanentzapf, G. Identification of genetic networks that act in the somatic cells of the testis to mediate the developmental program of spermatogenesis. PLoS Genet. 2017, 13, e1007026. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lan, X.; Chen, X.; Yu, C.; Xu, Y.; Liu, Y.; Xu, L.; Fan, H.Y.; Tong, C. Protein synthesis and degradation are essential to regulate germline stem cell homeostasis in Drosophila testes. Development 2016, 143, 2930–2945. [Google Scholar] [CrossRef]
- Tulina, N.; Matunis, E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 2001, 294, 2546–2549. [Google Scholar] [CrossRef]
- Leatherman, J.L.; Dinardo, S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008, 3, 44–54. [Google Scholar] [CrossRef]
- Leatherman, J.L.; Dinardo, S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell. Biol. 2010, 12, 806–811. [Google Scholar] [CrossRef]
- Amoyel, M.; Sanny, J.; Burel, M.; Bach, E.A. Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development 2013, 140, 56–65. [Google Scholar] [CrossRef]
- Kiger, A.A.; White-Cooper, H.; Fuller, M.T. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 2000, 407, 750–754. [Google Scholar] [CrossRef]
- Tran, J.; Brenner, T.J.; DiNardo, S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature 2000, 407, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Parikh, N.; Hearn, S.A.; Fuller, M.T.; Tazuke, S.I.; Schulz, C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr. Biol. 2007, 17, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Kawase, E.; Wong, M.D.; Ding, B.C.; Xie, T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 2004, 131, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Shivdasani, A.A.; Ingham, P.W. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr. Biol. 2003, 13, 2065–2072. [Google Scholar] [CrossRef]
- Lee, S.; Zhou, L.; Kim, J.; Kalbfleisch, S.; Schöck, F. Lasp anchors the Drosophila male stem cell niche and mediates spermatid individualization. Mech. Dev. 2008, 125, 768–776. [Google Scholar] [CrossRef]
- Tanentzapf, G.; Devenport, D.; Godt, D.; Brown, N.H. Integrin-dependent anchoring of a stem-cell niche. Nat. Cell. Biol. 2007, 9, 1413–1418. [Google Scholar] [CrossRef]
- Issigonis, M.; Tulina, N.; de Cuevas, M.; Brawley, C.; Sandler, L.; Matunis, E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 2009, 326, 153–156. [Google Scholar] [CrossRef]
- Singh, S.R.; Zheng, Z.; Wang, H.; Oh, S.W.; Chen, X.; Hou, S.X. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J. Cell. Physiol. 2010, 223, 500–510. [Google Scholar] [CrossRef]
- Kim, C.; Ye, F.; Hu, X.; Ginsberg, M.H. Talin activates integrins by altering the topology of the β transmembrane domain. J. Cell. Biol. 2012, 197, 605–611. [Google Scholar] [CrossRef]
- Campbell, I.D.; Humphries, M.J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Brown, N.H.; Gregory, S.L.; Rickoll, W.L.; Fessler, L.I.; Prout, M.; White, R.A.; Fristrom, J.W. Talin is essential for integrin function in Drosophila. Dev. Cell 2002, 3, 569–579. [Google Scholar] [CrossRef]
- Lai, M.C.; Chang, W.C.; Shieh, S.Y.; Tarn, W.Y. DDX3 regulates cell growth through translational control of cyclin E1. Mol. Cell. Biol. 2010, 30, 5444–5453. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Sun, H.S.; Wang, S.W.; Tarn, W.Y. DDX3 functions in antiviral innate immunity through translational control of PACT. FEBS J. 2016, 283, 88–101. [Google Scholar] [CrossRef]
- Valentin-Vega, Y.A.; Wang, Y.D.; Parker, M.; Patmore, D.M.; Kanagaraj, A.; Moore, J.; Rusch, M.; Finkelstein, D.; Ellison, D.W.; Gilbertson, R.J.; et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016, 6, 25996. [Google Scholar] [CrossRef]
- Ku, Y.C.; Lai, M.H.; Lo, C.C.; Cheng, Y.C.; Qiu, J.T.; Tarn, W.Y.; Lai, M.C. DDX3 Participates in Translational Control of Inflammation Induced by Infections and Injuries. Mol. Cell. Biol. 2018, 39, e00285. [Google Scholar] [CrossRef]
- Temme, C.; Simonelig, M.; Wahle, E. Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front. Genet. 2014, 5, 143. [Google Scholar] [CrossRef]
- Wen, P.; Quan, Z.; Xi, R. The biological function of the WD40 repeat-containing protein p55/Caf1 in Drosophila. Dev. Dyn. 2012, 241, 455–464. [Google Scholar] [CrossRef]
- Angulo, B.; Srinivasan, S.; Bolival, B.J.; Olivares, G.H.; Spence, A.C.; Fuller, M.T. DREF Genetically Counteracts Mi-2 and Caf1 to Regulate Adult Stem Cell Maintenance. PLoS Genet. 2019, 15, e1008187. [Google Scholar] [CrossRef]
- Lippai, M.; Tirián, L.; Boros, I.; Mihály, J.; Erdélyi, M.; Belecz, I.; Máthé, E.; Pósfai, J.; Nagy, A.; Udvardy, A.; et al. The Ketel gene encodes a Drosophila homologue of importin-beta. Genetics 2000, 156, 1889–1900. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotov, A.A.; Godneeva, B.K.; Olenkina, O.M.; Adashev, V.E.; Trostnikov, M.V.; Olenina, L.V. The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis Via Regulation of a Specific mRNA Set. Cells 2020, 9, 550. https://doi.org/10.3390/cells9030550
Kotov AA, Godneeva BK, Olenkina OM, Adashev VE, Trostnikov MV, Olenina LV. The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis Via Regulation of a Specific mRNA Set. Cells. 2020; 9(3):550. https://doi.org/10.3390/cells9030550
Chicago/Turabian StyleKotov, Alexei A., Baira K. Godneeva, Oxana M. Olenkina, Vladimir E. Adashev, Mikhail V. Trostnikov, and Ludmila V. Olenina. 2020. "The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis Via Regulation of a Specific mRNA Set" Cells 9, no. 3: 550. https://doi.org/10.3390/cells9030550
APA StyleKotov, A. A., Godneeva, B. K., Olenkina, O. M., Adashev, V. E., Trostnikov, M. V., & Olenina, L. V. (2020). The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis Via Regulation of a Specific mRNA Set. Cells, 9(3), 550. https://doi.org/10.3390/cells9030550




