2′O-Ribose Methylation of Ribosomal RNAs: Natural Diversity in Living Organisms, Biological Processes, and Diseases
Abstract
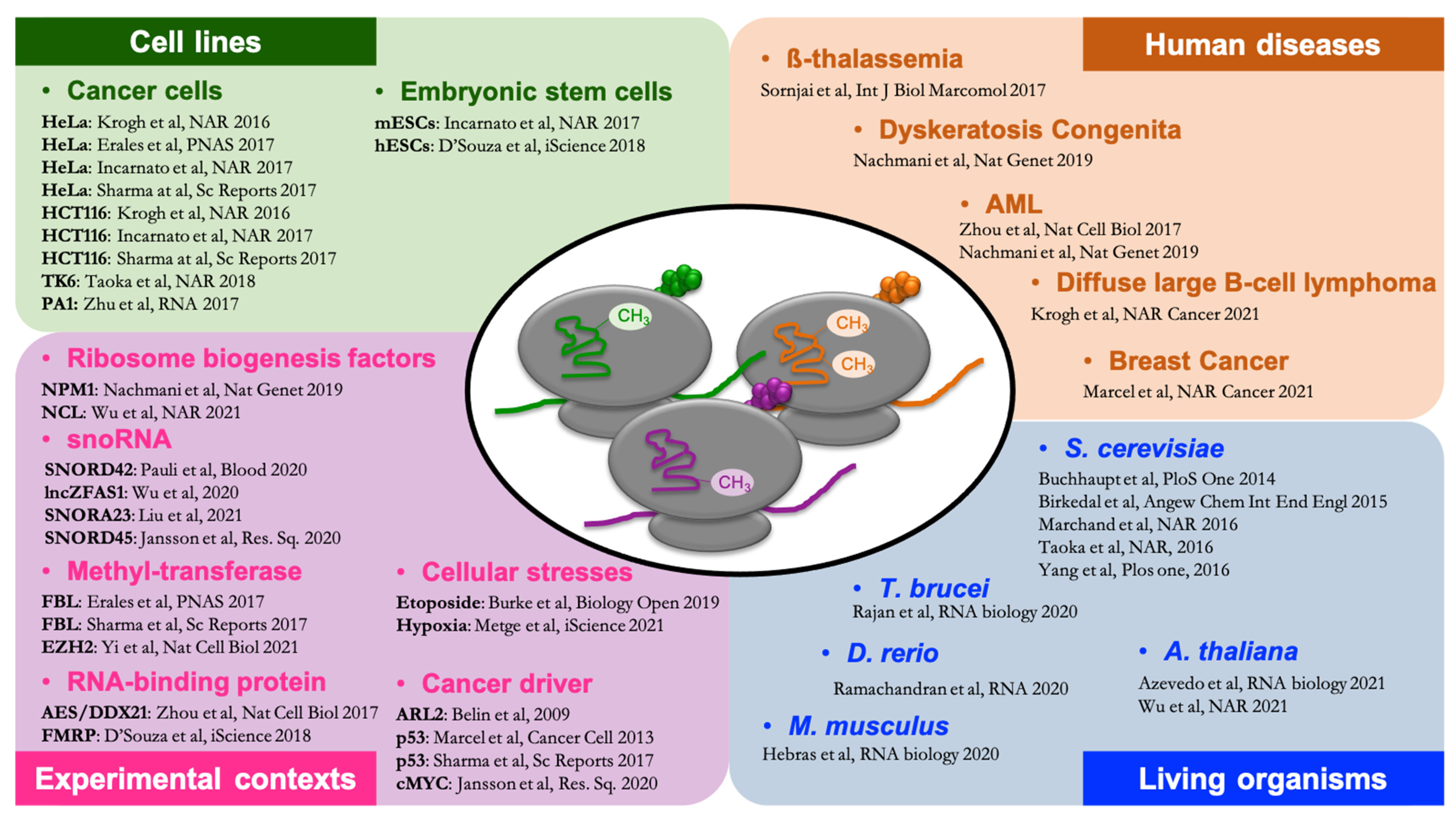
1. Introduction
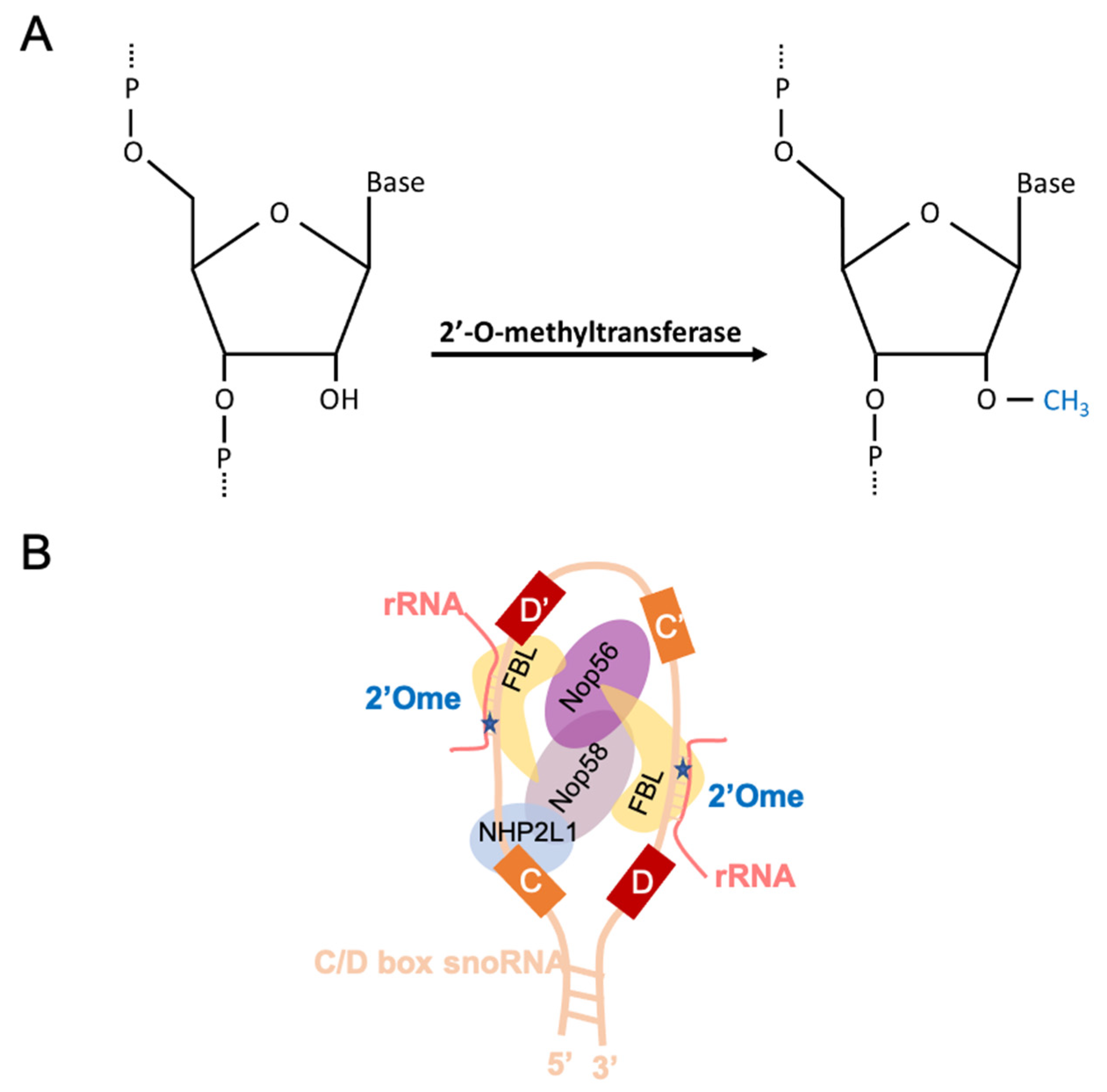
2. A Brief Overview of rRNA 2′Ome
3. Natural Variability of rRNA 2′Ome in Yeast
4. Variation of rRNA in Metazoan Cell Lines
4.1. Intra-Variability of rRNA 2′Ome in Human Cell Lines
4.2. Physiopathological Variability of rRNA 2′Ome in Cell Lines
4.2.1. rRNA 2′Ome Variability in Response to Modulations of Ribosome Biogenesis Factor (RBF) Expression and Activity
4.2.2. rRNA 2′Ome Variability in Physio-Pathological Contexts
5. Diversity of rRNA 2′Ome Profiles in Human Cancer Tissues
| Site 1 | Biological Context of Variability | Variation | Related snoRNA (Host Gene) | Method | Reference |
|---|---|---|---|---|---|
| 18S-Um116 | FBL depletion | Down | SNORD42A/B | RMS 2-Illumina | [44] |
| SNORD42A depletion | Down | (RPL23A) | RMS-Illumina + RTL-P | [54] | |
| EZH2 depletion | Down | RMS-Illumina | [50] | ||
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 18S-Cm174 | FMRP depletion | Up | SNORD45C | RMS-Illumina | [51] |
| FBL depletion | Down | (RABGGTB) | RMS-Illumina | [44] | |
| c-MYC overexpression | Up | RMS-Ion Torrent | [59] | ||
| EZH2 depletion | Down | RMS-Illumina | [50] | ||
| Cell lines | Up and down | RMS-Ion Torrent | [26] | ||
| 18S-Gm867 | FMRP depletion | Down | SNORD98 | RMS-Illumina | [51] |
| EZH2 depletion | Down | (CCAR1) | RMS-Illumina | [50] | |
| FBL depletion | Down | RMS-Illumina | [44] | ||
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 28S-Cm1327 | EZH2 depletion | Down | SNORD104 | RMS-Illumina | [50] |
| NPM1 depletion and loss-of-function, between AML patients | Down | (lncSNHG25) | RTL-P | [52] | |
| FBL depletion | Down | RMS-Illumina | [44] | ||
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 28S-Am1858 | Reduced Arl2 expression | Up | SNORD38A/B | RTL-P | [64] |
| FBL depletion | Down | (RPS8) | RMS-Illumina | [44] | |
| EZH2 depletion | Down | RMS-Illumina | [50] | ||
| DLBCL patients vs. healthy donor | Down | RMS-Ion Torrent | [70] | ||
| ß-thalassemic patients vs. healthy donor | Up | RTL-P | [69] | ||
| 28S-Um2402 | FMRP depletion | Down | SNORD143, SNORD144 | RMS-Illumina | [51] |
| FBL depletion | Down | (SEC31A) | RMS-Illumina | [44] | |
| c-MYC overexpression | Down | RMS-Ion Torrent | [59] | ||
| cell lines | Up and down | RMS-Ion Torrent | [26] | ||
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| DLBCL patients vs. healthy donor | Down | RMS-Ion Torrent | [70] | ||
| 28S-Cm2409 | EZH2 depletion | Down | SNORD5 | RMS-Illumina | [50] |
| FMRP depletion | Up | (TAF1D) | RMS-Illumina | [51] | |
| FBL depletion | Down | RMS-Illumina | [44] | ||
| cell lines | Up and down | RMS-Ion Torrent | [26] | ||
| cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 28S-Am2774 | FMRP depletion | Down | SNORD99 | RMS-Illumina | [51] |
| FBL depletion | Down | (SNHG12) | RMS-Illumina | [44] | |
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| Brain, liver and skeletal muscle | Down in Liver | RMS-Ion Torrent | [70] | ||
| 28S-Cm2848 | EZH2 depletion | Down | SNORD50A/B | RMS-Illumina | [50] |
| FBL depletion | Down | (SNHG5) | RMS-Illumina | [44] | |
| Cell lines | Up and down | RMS-Ion Torrent | [26] | ||
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 28S-Gm3923 | FMRP depletion | Down | SNORD111/B | RMS-Illumina | [51] |
| FBL depletion | Down | (SF3B3) | RMS-Illumina | [44] | |
| Cell lines | Up and down | RMS-Ion Torrent | [26] | ||
| cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| 28S-Cm4506 | FBL depletion | Down | SNORD35A (RPL13A) | RMS-Illumina | [44] |
| SNORA23 depletion/overexpression | Up/down | SNORD35B (RPS11) | RTL-P | [48] | |
| AES depletion | Down | RTL-P | [55] | ||
| ß-thalassemic patients vs. healthy donor | Up | RTL-P | [69] | ||
| 28S-Gm4593 | FBL depletion | Down | SNORD78 | RMS-Illumina | [44] |
| ZFAS1 depletion/overexpression | Up/down | (GAS5) | RTL-P/DBPST | [47] | |
| Cell lines, between breast cancer patients | Up and down | RMS-Illumina | [71] | ||
| DLBCL patients vs. healthy donor, brain, liver and skeletal muscle | Up in DLBCL, Up in brain and liver | RMS-Ion Torrent | [70] |
6. In Vivo Variability of rRNA 2′Ome in Model Organisms
6.1. Trypanosoma Brucei
6.2. Arabidopsis Thaliana
6.3. Danio Rerio
6.4. Mus Musculus
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Natchiar, S.K.; Myasnikov, A.G.; Hazemann, I.; Klaholz, B.P. Visualizing the Role of 2′-OH rRNA Methylations in the Human Ribosome Structure. Biomolecules 2018, 8, 125. [Google Scholar] [CrossRef]
- Melnikov, S.; Ben-Shem, A.; Garreau de Loubresse, N.; Jenner, L.; Yusupova, G.; Yusupov, M. One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef]
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351. [Google Scholar] [CrossRef]
- Monaco, P.L.; Marcel, V.; Diaz, J.J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.G.; Teysset, L.; Carre, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E. Methylation map of Xenopus laevis ribosomal RNA. Nature 1980, 288, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Schimmang, T.; Tollervey, D.; Kern, H.; Frank, R.; Hurt, E.C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989, 8, 4015–4024. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Petfalski, E.; Tollervey, D.; Caceres, J.F. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell. Biol. 2003, 23, 8519–8527. [Google Scholar] [CrossRef]
- Dalla Venezia, N.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.J. Emerging Role of Eukaryote Ribosomes in Translational Control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef]
- Gabut, M.; Bourdelais, F.; Durand, S. Ribosome and Translational Control in Stem Cells. Cells 2020, 9, 497. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Lafontaine, D.L.J. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef]
- Birkedal, U.; Christensen-Dalsgaard, M.; Krogh, N.; Sabarinathan, R.; Gorodkin, J.; Nielsen, H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. 2015, 127, 461–465. [Google Scholar] [CrossRef]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef]
- Gautier, T.; Berges, T.; Tollervey, D.; Hurt, E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997, 17, 7088–7098. [Google Scholar] [CrossRef]
- Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef]
- Bonnerot, C.; Pintard, L.; Lutfalla, G. Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell 2003, 12, 1309–1315. [Google Scholar] [CrossRef]
- Lapeyre, B.; Purushothaman, S.K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell 2004, 16, 663–669. [Google Scholar] [CrossRef]
- Ayadi, L.; Galvanin, A.; Pichot, F.; Marchand, V.; Motorin, Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 253–269. [Google Scholar] [CrossRef]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef]
- Krogh, N.; Jansson, M.D.; Hafner, S.J.; Tehler, D.; Birkedal, U.; Christensen-Dalsgaard, M.; Lund, A.H.; Nielsen, H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016, 44, 7884–7895. [Google Scholar] [CrossRef]
- Marchand, V.; Blanloeil-Oillo, F.; Helm, M.; Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016, 44, e135. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 2001, 25, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.W.; Shao, P.; Diao, L.T.; Zhou, H.; Yu, C.H.; Qu, L.H. RTL-P: A sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 2012, 40, e157. [Google Scholar] [CrossRef]
- Taoka, M.; Nobe, Y.; Hori, M.; Takeuchi, A.; Masaki, S.; Yamauchi, Y.; Nakayama, H.; Takahashi, N.; Isobe, T. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: The complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res. 2015, 43, e115. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Anselmi, F.; Morandi, E.; Neri, F.; Maldotti, M.; Rapelli, S.; Parlato, C.; Basile, G.; Oliviero, S. High-throughput single-base resolution mapping of RNA 2-O-methylated residues. Nucleic Acids Res. 2017, 45, 1433–1441. [Google Scholar] [CrossRef]
- Zhu, Y.; Pirnie, S.P.; Carmichael, G.G. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 2017, 23, 1303–1314. [Google Scholar] [CrossRef]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Yamauchi, Y.; Ishikawa, H.; Takahashi, N.; Nakayama, H.; Isobe, T. The complete chemical structure of Saccharomyces cerevisiae rRNA: Partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. 2016, 44, 8951–8961. [Google Scholar] [CrossRef]
- Yang, J.; Sharma, S.; Watzinger, P.; Hartmann, J.D.; Kotter, P.; Entian, K.D. Mapping of Complete Set of Ribose and Base Modifications of Yeast rRNA by RP-HPLC and Mung Bean Nuclease Assay. PLoS ONE 2016, 11, e0168873. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, D.; Lehtonen, H.; Jansen, R.; Kern, H.; Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993, 72, 443–457. [Google Scholar] [CrossRef]
- Tollervey, D.; Lehtonen, H.; Carmo-Fonseca, M.; Hurt, E.C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991, 10, 573–583. [Google Scholar] [CrossRef]
- Liang, X.H.; Liu, Q.; Fournier, M.J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 2007, 28, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef]
- Esguerra, J.; Warringer, J.; Blomberg, A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA 2008, 14, 649–656. [Google Scholar] [CrossRef]
- Buchhaupt, M.; Sharma, S.; Kellner, S.; Oswald, S.; Paetzold, M.; Peifer, C.; Watzinger, P.; Schrader, J.; Helm, M.; Entian, K.D. Partial methylation at Am100 in 18S rRNA of baker’s yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS ONE 2014, 9, e89640. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Straub, A.U.; Doebele, C.; Bohnsack, M.T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013, 10, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Galan, O.; Garcia-Gomez, J.J.; de la Cruz, J. Yeast and human RNA helicases involved in ribosome biogenesis: Current status and perspectives. Biochim. Biophys. Acta 2013, 1829, 775–790. [Google Scholar] [CrossRef]
- Aquino, G.R.R.; Krogh, N.; Hackert, P.; Martin, R.; Gallesio, J.D.; van Nues, R.W.; Schneider, C.; Watkins, N.J.; Nielsen, H.; Bohnsack, K.E.; et al. RNA helicase-mediated regulation of snoRNP dynamics on pre-ribosomes and rRNA 2′-O-methylation. Nucleic Acids Res. 2021, 49, 4066–4084. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforets, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, R.; Jedlinski, D.J.; Schmidt, A.; Gypas, F.; Martin, G.; Vina-Vilaseca, A.; Zavolan, M. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Nucleic Acids Res. 2017, 45, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Sato, K.; Ishikawa, H.; Izumikawa, K.; Yamauchi, Y.; Hirota, K.; Nakayama, H.; Takahashi, N.; et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018, 46, 9289–9298. [Google Scholar] [CrossRef]
- Wu, H.; Qin, W.; Lu, S.; Wang, X.; Zhang, J.; Sun, T.; Hu, X.; Li, Y.; Chen, Q.; Wang, Y.; et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol. Cancer 2020, 19, 95. [Google Scholar] [CrossRef]
- Liu, Z.; Pang, Y.; Jia, Y.; Qin, Q.; Wang, R.; Li, W.; Jing, J.; Liu, H.; Liu, S. SNORA23 inhibits HCC tumorigenesis by impairing the 2′-O-ribose methylation level of 28S rRNA. Cancer Biol. Med. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Sharma, S.; Marchand, V.; Motorin, Y.; Lafontaine, D.L.J. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017, 7, 11490. [Google Scholar] [CrossRef]
- Yi, Y.; Li, Y.; Meng, Q.; Li, Q.; Li, F.; Lu, B.; Shen, J.; Fazli, L.; Zhao, D.; Li, C.; et al. A PRC2-independent function for EZH2 in regulating rRNA 2′-O methylation and IRES-dependent translation. Nat. Cell Biol. 2021, 23, 341–354. [Google Scholar] [CrossRef]
- D’Souza, M.N.; Gowda, N.K.C.; Tiwari, V.; Babu, R.O.; Anand, P.; Dastidar, S.G.; Singh, R.; James, O.G.; Selvaraj, B.; Pal, R.; et al. FMRP Interacts with C/D Box snoRNA in the Nucleus and Regulates Ribosomal RNA Methylation. iScience 2018, 9, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Nachmani, D.; Bothmer, A.H.; Grisendi, S.; Mele, A.; Bothmer, D.; Lee, J.D.; Monteleone, E.; Cheng, K.; Zhang, Y.; Bester, A.C.; et al. Germline NPM1 mutations lead to altered rRNA 2′-O-methylation and cause dyskeratosis congenita. Nat. Genet. 2019, 51, 1518–1529. [Google Scholar] [CrossRef] [PubMed]
- Kunchala, P.; Kuravi, S.; Jensen, R.; McGuirk, J.; Balusu, R. When the good go bad: Mutant NPM1 in acute myeloid leukemia. Blood Rev. 2018, 32, 167–183. [Google Scholar] [CrossRef]
- Pauli, C.; Liu, Y.; Rohde, C.; Cui, C.; Fijalkowska, D.; Gerloff, D.; Walter, C.; Krijgsveld, J.; Dugas, M.; Edemir, B.; et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 2020, 135, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Kohn, M.; Misiak, D.; Baumer, N.; Cui, C.; Gollner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Mulloy, J.C.; Cammenga, J.; MacKenzie, K.L.; Berguido, F.J.; Moore, M.A.; Nimer, S.D. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 2002, 99, 15–23. [Google Scholar] [CrossRef]
- Steffen, B.; Knop, M.; Bergholz, U.; Vakhrusheva, O.; Rode, M.; Kohler, G.; Henrichs, M.P.; Bulk, E.; Hehn, S.; Stehling, M.; et al. AML1/ETO induces self-renewal in hematopoietic progenitor cells via the Groucho-related amino-terminal AES protein. Blood 2011, 117, 4328–4337. [Google Scholar] [CrossRef][Green Version]
- Tonks, A.; Tonks, A.J.; Pearn, L.; Pearce, L.; Hoy, T.; Couzens, S.; Fisher, J.; Burnett, A.K.; Darley, R.L. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia 2004, 18, 1238–1245. [Google Scholar] [CrossRef][Green Version]
- Jansson, M.D.; Häfner, S.J.; Altinel, K.; Tehler, D.; Krogh, N.; Jakobsen, E.; Andersen, J.V.; Andersen, K.L.; Schoof, E.M.; Ménard, P.; et al. Ribosomal RNA methylation induced by MYC regulates translation. Res. Sq. 2020. preprint. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The p53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem. 2016, 85, 375–404. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Dichtel-Danjoy, M.L.; Sagne, C.; Hafsi, H.; Ma, D.; Ortiz-Cuaran, S.; Olivier, M.; Hall, J.; Mollereau, B.; Hainaut, P.; et al. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011, 18, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell Biol. 2000, 20, 5930–5938. [Google Scholar] [CrossRef]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.P.; Solano-Gonzalez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef]
- Belin, S.; Beghin, A.; Solano-Gonzalez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.C.; Mertani, H.C.; Dumontet, C.; Diaz, J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef]
- Metge, B.J.; Kammerud, S.C.; Pruitt, H.C.; Shevde, L.A.; Samant, R.S. Hypoxia re-programs 2′-O-Me modifications on ribosomal RNA. iScience 2021, 24, 102010. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.F.; McLaurin, D.M.; Logan, M.K.; Hebert, M.D. Alteration of 28S rRNA 2′-O-methylation by etoposide correlates with decreased SMN phosphorylation and reduced Drosha levels. Biol. Open 2019, 8, bio041848. [Google Scholar] [CrossRef]
- Burke, M.F.; Logan, M.K.; Hebert, M.D. Identification of additional regulatory RNPs that impact rRNA and U6 snRNA methylation. Biol. Open 2018, 7, bio036095. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.R.; Vicino, I.; Adachi, H.; Yu, Y.T.; Hebert, M.D. Regulatory RNPs: A novel class of ribonucleoproteins that potentially contribute to ribosome heterogeneity. Biol. Open 2017, 6, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Sornjai, W.; Lithanatudom, P.; Erales, J.; Joly, P.; Francina, A.; Hacot, S.; Fucharoen, S.; Svasti, S.; Diaz, J.J.; Mertani, H.C.; et al. Hypermethylation of 28S ribosomal RNA in beta-thalassemia trait carriers. Int. J. Biol. Macromol. 2017, 94, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Krogh, N.; Asmar, F.; Côme, C.; Munch-Petersen, H.F.; Grønbæk, K.; Nielsen, H. Profiling of ribose methylations in ribosomal RNA from diffuse large B-cell lymphoma patients for evaluation of ribosomes as drug targets. NAR Cancer 2020, 2, zcaa035. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Kielbassa, J.; Marchand, V.; Kundhavai, S.N.; Paraqindes, H.; Van Long, F.N.; Ayadi, L.; Bourguignon-Igel, V.; Lo Monaco, P.; Monchiet, D.; et al. Ribosomal RNA 2′O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR Cancer 2020, 2, zcaa036. [Google Scholar] [CrossRef]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Baudin-Baillieu, A.; Fabret, C.; Liang, X.H.; Piekna-Przybylska, D.; Fournier, M.J.; Rousset, J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009, 37, 7665–7677. [Google Scholar] [CrossRef]
- Matthews, K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005, 118, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.S.; Zhu, Y.; Adler, K.; Doniger, T.; Cohen-Chalamish, S.; Srivastava, A.; Shalev-Benami, M.; Matzov, D.; Unger, R.; Tschudi, C.; et al. The large repertoire of 2′-O-methylation guided by C/D snoRNAs on Trypanosoma brucei rRNA. RNA Biol. 2020, 17, 1018–1039. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Wang, J.; Li, X.; Li, J.; Ye, K. Profiling of RNA ribose methylation in Arabidopsis thaliana. Nucleic Acids Res. 2021, 49, 4104–4119. [Google Scholar] [CrossRef]
- Azevedo-Favory, J.; Gaspin, C.; Ayadi, L.; Montacie, C.; Marchand, V.; Jobet, E.; Rompais, M.; Carapito, C.; Motorin, Y.; Saez-Vasquez, J. Mapping rRNA 2′-O-methylations and identification of C/D snoRNAs in Arabidopsis thaliana plants. RNA Biol. 2021, 1–18. [Google Scholar] [CrossRef]
- Locati, M.D.; Pagano, J.F.; Ensink, W.A.; van Olst, M.; van Leeuwen, S.; Nehrdich, U.; Zhu, K.; Spaink, H.P.; Girard, G.; Rauwerda, H.; et al. Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA 2017, 23, 446–456. [Google Scholar] [CrossRef]
- Locati, M.D.; Pagano, J.F.B.; Girard, G.; Ensink, W.A.; van Olst, M.; van Leeuwen, S.; Nehrdich, U.; Spaink, H.P.; Rauwerda, H.; Jonker, M.J.; et al. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 2017, 23, 1188–1199. [Google Scholar] [CrossRef]
- Tao, B.; Lo, L.J.; Peng, J.; He, J. rDNA subtypes and their transcriptional expression in zebrafish at different developmental stages. Biochem. Biophys. Res. Commun. 2020, 529, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Krogh, N.; Jorgensen, T.E.; Johansen, S.D.; Nielsen, H.; Babiak, I. The shift from early to late types of ribosomes in zebrafish development involves changes at a subset of rRNA 2′-O-Me sites. RNA 2020, 26, 1919–1934. [Google Scholar] [CrossRef] [PubMed]
- Hebras, J.; Krogh, N.; Marty, V.; Nielsen, H.; Cavaille, J. Developmental changes of rRNA ribose methylations in the mouse. RNA Biol. 2020, 17, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Begik, O.; Lucas, M.C.; Pryszcz, L.P.; Ramirez, J.M.; Medina, R.; Milenkovic, I.; Cruciani, S.; Liu, H.; Vieira, H.G.S.; Sas-Chen, A.; et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
| Method | Principle | 2′Ome Site | 2′Ome Detection | Reference |
|---|---|---|---|---|
| Primer Extension | The presence of a 2′Ome residue causes the Reverse Transcriptase to pause at low dNTP concentration, the pause being directly observable on a gel using a radioactive/fluorescent primer | site specific | qualitative | [28] |
| RTL-P | The presence of a 2′Ome residue decreases the reverse transcription efficiency at low dNTP concentration, the pause being quantified by regular or quantitative PCR | site specific | semi-quantitative | [29] |
| Mass Spectrometry | Detect rRNA fragments or nucleosides, the mass of which is shifted by specific modification including 2′Ome | global overview of all sites + site specific | quantitative (absolute) | [30] |
| RiboMeth-seq | The presence of a 2′Ome inhibits alkaline-mediated hydrolysis of the 3′-adjacent phosphodiester bond and is then detected by RNA-seq (Illumina or Ion Torrent sequencing) | global overview of known sites + site specific | quantitative (absolute) | [17] |
| 2′OMe-seq | The presence of a 2′Ome residue causes the Reverse Transcriptase to pause at low dNTP concentration and is then detected by RNA-seq (Illumina) | global overview of all sites + site specific | quantitative (relative) | [31] |
| RibOxi-seq | The 3′-terminal ribose of a 2′Ome residue is resistant to periodate cleavage. After RNA fragmentation using the endonuclease Benzonase and periodate oxidation, RNA fragments with a 2′Ome group at their 3′end are enriched and detected by RNA-seq | global overview of all sites | qualitative | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaafar, M.; Paraqindes, H.; Gabut, M.; Diaz, J.-J.; Marcel, V.; Durand, S. 2′O-Ribose Methylation of Ribosomal RNAs: Natural Diversity in Living Organisms, Biological Processes, and Diseases. Cells 2021, 10, 1948. https://doi.org/10.3390/cells10081948
Jaafar M, Paraqindes H, Gabut M, Diaz J-J, Marcel V, Durand S. 2′O-Ribose Methylation of Ribosomal RNAs: Natural Diversity in Living Organisms, Biological Processes, and Diseases. Cells. 2021; 10(8):1948. https://doi.org/10.3390/cells10081948
Chicago/Turabian StyleJaafar, Mariam, Hermes Paraqindes, Mathieu Gabut, Jean-Jacques Diaz, Virginie Marcel, and Sébastien Durand. 2021. "2′O-Ribose Methylation of Ribosomal RNAs: Natural Diversity in Living Organisms, Biological Processes, and Diseases" Cells 10, no. 8: 1948. https://doi.org/10.3390/cells10081948
APA StyleJaafar, M., Paraqindes, H., Gabut, M., Diaz, J.-J., Marcel, V., & Durand, S. (2021). 2′O-Ribose Methylation of Ribosomal RNAs: Natural Diversity in Living Organisms, Biological Processes, and Diseases. Cells, 10(8), 1948. https://doi.org/10.3390/cells10081948








