Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging
Abstract
1. Introduction
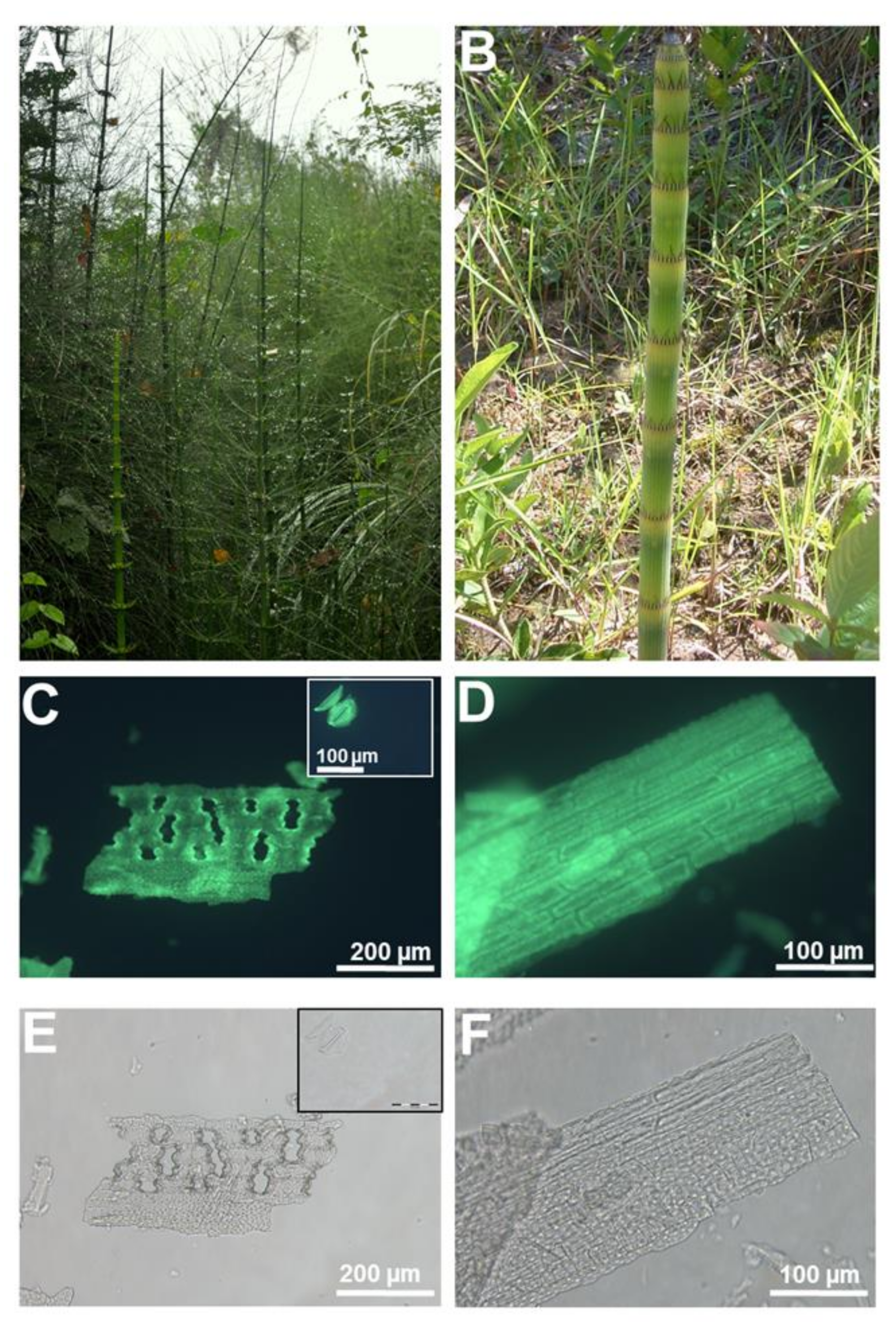
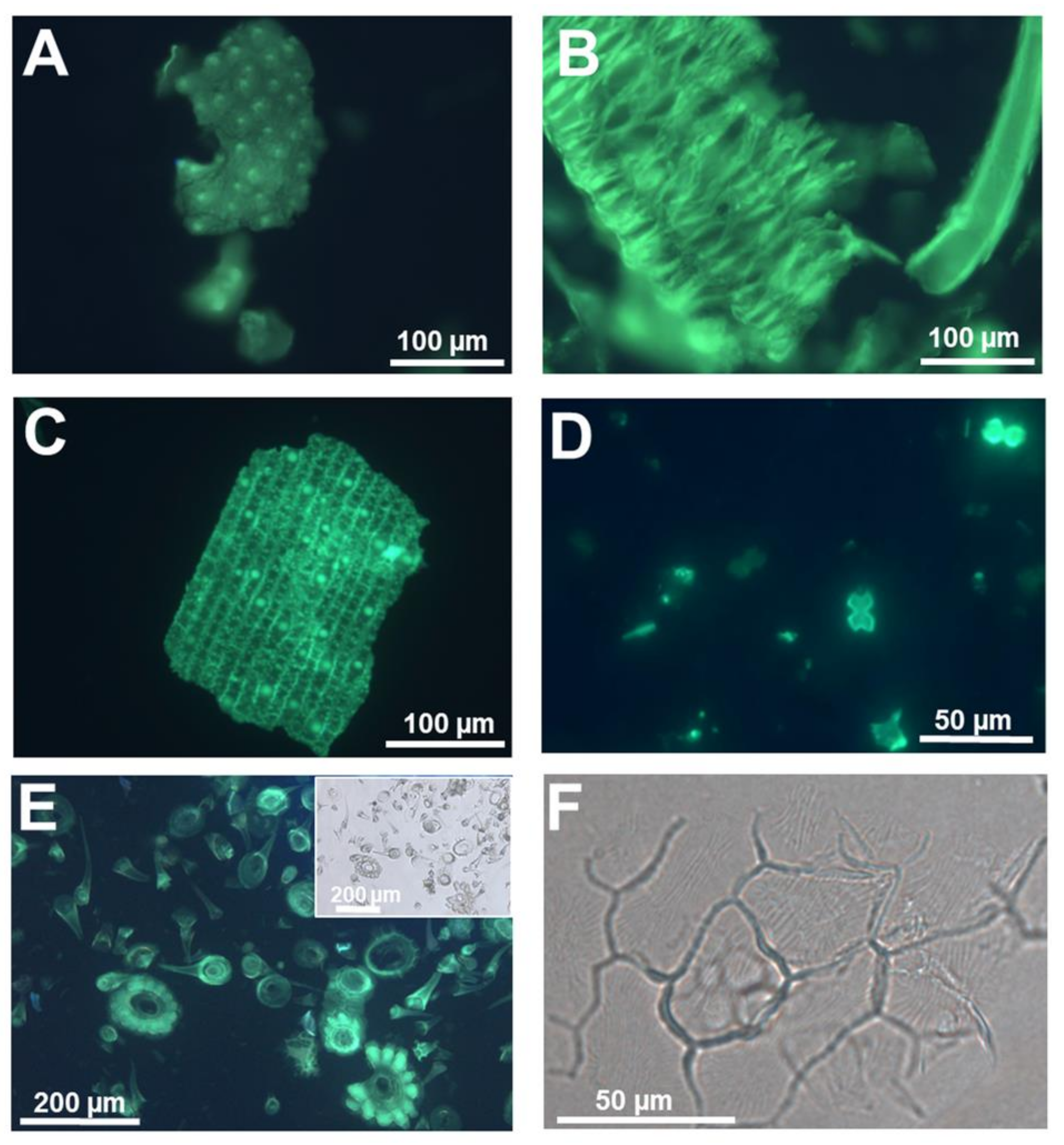
2. Dyes and Fluorophores to Visualize Plant Silica
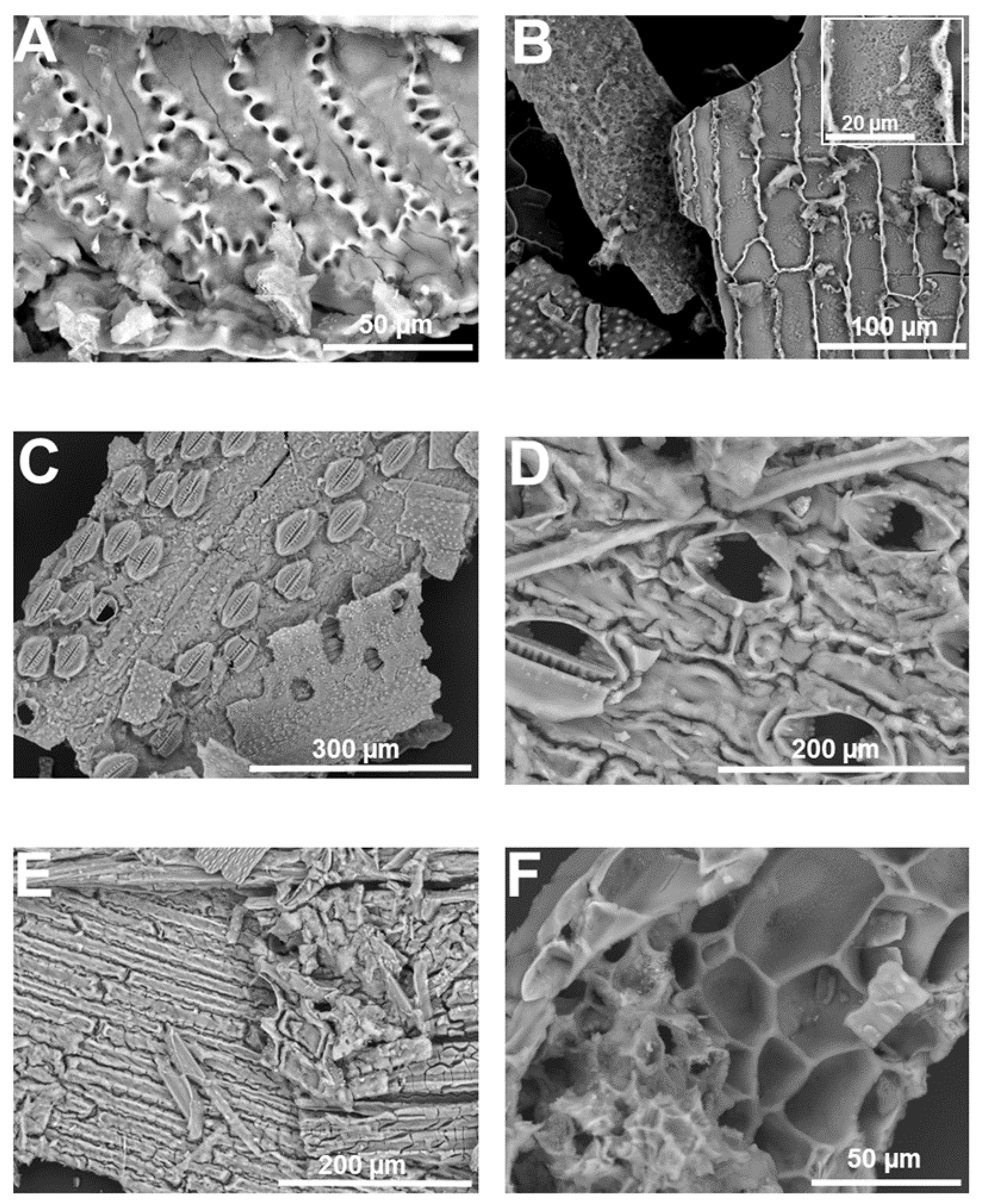
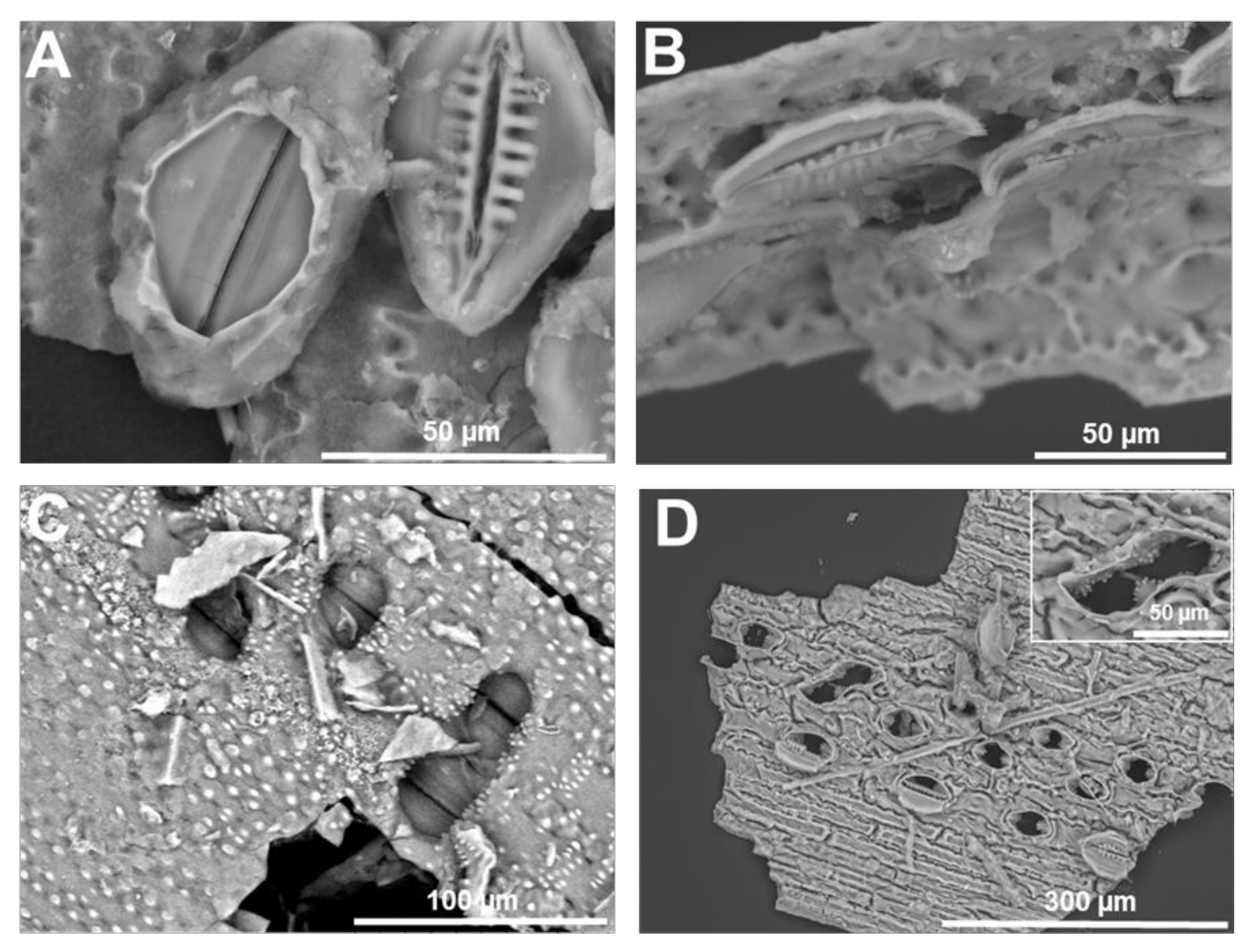
3. Micromorphology: Scanning Electron Microscopy of Extracted Plant Silica
4. Imaging Plant Silica Through Particle Induced X-ray Emission with Focused Beam and Micro-X-Ray Fluorescence Spectrometry
5. Mass Spectrometry Imaging of Si: SIMS Nano-Analysis
6. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Epstein, E. The anomaly of silicon in plant biology. Proc. Natl. Acad. Sci. USA 1994, 91, 11–17. [Google Scholar] [CrossRef]
- Guerriero, G.; Hausman, J.-F.; Legay, S. Silicon and the plant extracellular matrix. Front. Plant Sci. 2016, 7, 463. [Google Scholar] [CrossRef]
- Luyckx, M.; Hausman, J.-F.; Lutts, S.; Guerriero, G. Silicon and plants: Current knowledge and technological perspectives. Front. Plant Sci. 2017, 8, 411. [Google Scholar] [CrossRef]
- Exley, C. A possible mechanism of biological silicification in plants. Front. Plant Sci. 2015, 6, 853. [Google Scholar] [CrossRef]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef]
- Chiba, Y.; Mitani, N.; Yamaji, N.; Ma, J.F. HvLsi1 is a silicon influx transporter in barley. Plant J. 2009, 57, 810–818. [Google Scholar] [CrossRef]
- Grégoire, C.; Rémus-Borel, W.; Vivancos, J.; Labbé, C.; Belzile, F.; Bélanger, R.R. Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 2012, 72, 320–330. [Google Scholar] [CrossRef]
- Shivaraj, S.M.; Deshmukh, R.K.; Rai, R.; Bélanger, R.; Agrawal, P.K.; Dash, P.K. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 2017, 7, 46137. [Google Scholar] [CrossRef]
- Guerriero, G.; Deshmukh, R.; Sonah, H.; Sergeant, K.; Hausman, J.-F.; Lentzen, E.; Valle, N.; Siddiqui, K.S.; Exley, C. Identification of the aquaporin gene family in Cannabis sativa and evidence for the accumulation of silicon in its tissues. Plant Sci. 2019, 287, 110167. [Google Scholar] [CrossRef]
- Mitani, N.; Ma, J.F. Uptake system of silicon in different plant species. J. Exp. Bot. 2005, 56, 1255–1261. [Google Scholar] [CrossRef]
- Takahashi, E.; Ma, J.F.; Miyake, Y. The possibility of silicon as an essential element for higher plants. Comments Agric. Food Chem. 1990, 2, 99–102. [Google Scholar]
- Laane, H.-M. The effects of foliar sprays with different silicon compounds. Plants 2018, 7, 45. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, Y.; Yao, H.; Wu, J.; Sun, H.; Gong, H. Silicon improves seed germination and alleviates oxidative stress of bud seedlings in tomato under water deficit stress. Plant Physiol. Biochem. 2014, 78, 27–36. [Google Scholar] [CrossRef]
- Fry, S.C.; Nesselrode, B.H.W.A.; Miller, J.G.; Mewburn, B.R. Mixed-linkage (1→3,1→4)-β-d-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 2008, 179, 104–115. [Google Scholar] [CrossRef]
- He, C.; Wang, L.; Liu, J.; Liu, X.; Li, X.; Ma, J.; Lin, Y.; Xu, F. Evidence for “silicon” within the cell walls of suspension-cultured rice cells. New Phytol. 2013, 200, 700–709. [Google Scholar] [CrossRef]
- He, C.; Ma, J.; Wang, L. A hemicellulose-bound form of silicon with potential to improve the mechanical properties and regeneration of the cell wall of rice. New Phytol. 2015, 206, 1051–1062. [Google Scholar] [CrossRef]
- Kido, N.; Yokoyama, R.; Yamamoto, T.; Furukawa, J.; Iwai, H.; Satoh, S.; Nishitani, K. The matrix polysaccharide (1;3,1;4)-β-d-glucan is involved in silicon-dependent strengthening of rice cell wall. Plant Cell Physiol. 2015, 56, 268–276. [Google Scholar] [CrossRef]
- Law, C.; Exley, C. New insight into silica deposition in horsetail (Equisetum arvense). BMC Plant Biol. 2011, 11, 112. [Google Scholar] [CrossRef]
- Guerriero, G.; Law, C.; Stokes, I.; Moore, K.L.; Exley, C. Rough and tough. How does silicic acid protect horsetail from fungal infection? J. Trace Elem. Med. Biol. 2018, 47, 45–52. [Google Scholar] [CrossRef]
- Guerriero, G.; Stokes, I.; Exley, C. Is callose required for silicification in plants? Biol. Lett. 2018, 14, 20180338. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtíková, Z.; Sabol, P.; Ortmannová, J.; Neděla, V.; Tihlaříková, E.; Žárský, V. Exocyst subunit EXO70H4 has a specific role in callose synthase secretion and silica accumulation. Plant Physiol. 2018, 176, 2040–2051. [Google Scholar] [CrossRef]
- Brugiére, T.; Exley, C. Callose-associated silica deposition in Arabidopsis. J. Trace Elem. Med. Biol. 2017, 39, 86–90. [Google Scholar] [CrossRef]
- Kroukamp, E.M.; Wondimu, T.; Forbes, P.B.C. Metal and metalloid speciation in plants: Overview, instrumentation, approaches and commonly assessed elements. TrAC Trends Anal. Chem. 2016, 77, 87–99. [Google Scholar] [CrossRef]
- Tran, T.H.M.; Nguyen, K.G. Metal and metalloid concentrations in soil, surface water, and vegetables and the potential ecological and human health risks in the northeastern area of Hanoi, Vietnam. Environ. Monit. Assess. 2018, 190, 624. [Google Scholar] [CrossRef]
- Khan, Z.I.; Ugulu, I.; Ahmad, K.; Yasmeen, S.; Noorka, I.R.; Mehmood, N.; Sher, M. Assessment of trace metal and metalloid accumulation and human health risk from vegetables consumption through spinach and coriander specimens irrigated with wastewater. Bull. Environ. Contam. Toxicol. 2018, 101, 787–795. [Google Scholar] [CrossRef]
- Alam, M.Z.; Hoque, M.A.; Ahammed, G.J.; McGee, R.; Carpenter-Boggs, L. Arsenic accumulation in lentil (Lens culinaris) genotypes and risk associated with the consumption of grains. Sci. Rep. 2019, 9, 9431. [Google Scholar] [CrossRef]
- Lombi, E.; Scheckel, K.G.; Kempson, I.M. In situ analysis of metal(loid)s in plants: State of the art and artefacts. Environ. Exp. Bot. 2011, 72, 3–17. [Google Scholar] [CrossRef]
- Gierlinger, N.; Schwanninger, M. The potential of Raman microscopy and Raman imaging in plant research. Spectroscopy 2007, 21, 69–89. [Google Scholar] [CrossRef]
- Pierantoni, M.; Tenne, R.; Brumfeld, V.; Kiss, V.; Oron, D.; Addadi, L.; Weiner, S. Plants and light manipulation: The integrated mineral system in okra leaves. Adv. Sci. 2017, 4, 1600416. [Google Scholar] [CrossRef]
- Barron, A.; Turner, M.; Beeching, L.; Bellwood, P.; Piper, P.; Grono, E.; Jones, R.; Oxenham, M.; Kien, N.K.T.; Senden, T.; et al. MicroCT reveals domesticated rice (Oryza sativa) within pottery sherds from early Neolithic sites (4150–3265 cal BP) in Southeast Asia. Sci. Rep. 2017, 7, 1–5. [Google Scholar] [CrossRef]
- Cremer, C.; Masters, B.R. Resolution enhancement techniques in microscopy. Eur. Phys. J. H 2013, 38, 281–344. [Google Scholar] [CrossRef]
- Fouquet, C.; Gilles, J.-F.; Heck, N.; Dos Santos, M.; Schwartzmann, R.; Cannaya, V.; Morel, M.-P.; Davidson, R.S.; Trembleau, A.; Bolte, S. Improving axial resolution in confocal microscopy with new high refractive index mounting media. PLoS ONE 2015, 10, e0121096. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Kourousias, G.; Stolfa, A.; Kaulich, B. Recent developments at the TwinMic beamline at ELETTRA: An 8 SDD detector setup for low energy X-ray Fluorescence. J. Phys. Conf. Ser. 2013, 425, 182001. [Google Scholar] [CrossRef]
- Moore, K.L.; Chen, Y.; van de Meene, A.M.; Hughes, L.; Liu, W.; Geraki, T.; Mosselmans, F.; McGrath, S.P.; Grovenor, C.; Zhao, F.-J. Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 2014, 201, 104–115. [Google Scholar] [CrossRef]
- Sangely, L.; Boyer, B.; de Chambost, E.; Valle, N.; Audinot, J.-N.; Ireland, T.; Wiedenbeck, M.; Aléon, J.; Jungnickel, H.; Barnes, J.-P.; et al. CHAPTER 15: Secondary ion mass spectrometry. In Sector Field Mass Spectrometry for Elemental and Isotopic Analysis; Royal Society of Chemistry: London, UK, 2014; pp. 439–499. [Google Scholar]
- Dayanandan, P.; Kaufman, P.B.; Franklin, C.I. Detection of silica in plants. Am. J. Bot. 1983, 70, 1079–1084. [Google Scholar] [CrossRef]
- Nondek, L.; Buszewski, B.; Berek, D. Retention of pyridine and 2,6-dimethylpyridine on silanized silica: A simple test on residual silanols? J. Chromatogr. A 1986, 360, 241–246. [Google Scholar] [CrossRef]
- Blecher, I.C.; Seidel, R.; Thomann, R.; Speck, T. Comparison of different methods for the detection of silica inclusions in plant tissues. Int. J. Plant Sci. 2012, 173, 229–238. [Google Scholar] [CrossRef]
- Yokoyama, R.; Kido, N.; Yamamoto, T.; Furukawa, J.; Iwai, H.; Satoh, S.; Nishitani, K. Histochemical staining of silica body in rice leaf blades. Bio Protocol 2015, 5. [Google Scholar] [CrossRef]
- Isa, M.; Bai, S.; Yokoyama, T.; Ma, J.F.; Ishibashi, Y.; Yuasa, T.; Iwaya-Inoue, M. Silicon enhances growth independent of silica deposition in a low-silica rice mutant, lsi1. Plant Soil 2010, 331, 361–375. [Google Scholar] [CrossRef]
- Kao, T.-T.; Chen, S.-J.; Chiou, W.; Chuang, Y.-C.; Kuo-Huang, L.-L. Various microscopic methods for investigating the venuloid idioblasts of Pteris grevilleana wall. Taiwania 2008, 53, 394–400. [Google Scholar] [CrossRef]
- Raven, J.A. The transport and function of silicon in plants. Biol. Rev. 1983, 58, 179–207. [Google Scholar] [CrossRef]
- Strömberg, C.A.E.; Stilio, V.S.D.; Song, Z. Functions of phytoliths in vascular plants: An evolutionary perspective. Funct. Ecol. 2016, 30, 1286–1297. [Google Scholar] [CrossRef]
- Diwu, Z.; Chen, C.S.; Zhang, C.; Klaubert, D.H.; Haugland, R.P. A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem. Biol. 1999, 6, 411–418. [Google Scholar] [CrossRef]
- Shimizu, K.; Del Amo, Y.; Brzezinski, M.A.; Stucky, G.D.; Morse, D.E. A novel fluorescent silica tracer for biological silicification studies. Chem. Biol. 2001, 8, 1051–1060. [Google Scholar] [CrossRef]
- Perry, C.C.; Fraser, M.A. Silica deposition and ultrastructure in the cell wall of Equisetum arvense: The importance of cell wall structures and flow control in biosilicification? Philos. Trans. Biol. Sci. 1991, 334, 149–157. [Google Scholar]
- Cullen, E.; Rudall, P.J. The remarkable stomata of horsetails (Equisetum): Patterning, ultrastructure and development. Ann. Bot. 2016, 118, 207–218. [Google Scholar] [CrossRef]
- Jones, L.H.P.; Milne, A.A.; Wadham, S.M. Studies of silica in the oat plant. Plant Soil 1963, 18, 358–371. [Google Scholar] [CrossRef]
- Hayward, D.M.; Parry, D.W. Scanning electron microscopy of silica deposition in the leaves of barley (Hordeum sativum L.). Ann. Bot. 1975, 39, 1003–1009. [Google Scholar] [CrossRef]
- Vogel-Mikuš, K.; Pongrac, P.; Pelicon, P. Micro-PIXE elemental mapping for ionome studies of crop plants. Int. J. PIXE 2014, 24, 217–233. [Google Scholar] [CrossRef]
- Ortega, R.; Devès, G.; Carmona, A. Bio-metals imaging and speciation in cells using proton and synchrotron radiation X-ray microspectroscopy. J. R. Soc. Interface 2009, 6, S649–S658. [Google Scholar] [CrossRef][Green Version]
- Van der Ent, A.; Przybyłowicz, W.J.; de Jonge, M.D.; Harris, H.H.; Ryan, C.G.; Tylko, G.; Paterson, D.J.; Barnabas, A.D.; Kopittke, P.M.; Mesjasz-Przybyłowicz, J. X-ray elemental mapping techniques for elucidating the ecophysiology of hyperaccumulator plants. New Phytol. 2018, 218, 432–452. [Google Scholar] [CrossRef] [PubMed]
- Cloete, K.J.; Šmit, Ž.; Minnis-Ndimba, R.; Vavpetič, P.; du Plessis, A.; le Roux, S.G.; Pelicon, P. Physico-elemental analysis of roasted organic coffee beans from Ethiopia, Colombia, Honduras, and Mexico using X-ray micro-computed tomography and external beam particle induced X-ray emission. Food Chem. X 2019, 2, 100032. [Google Scholar] [CrossRef] [PubMed]
- Vavpetič, P.; Vogel-Mikuš, K.; Jeromel, L.; Ogrinc Potočnik, N.; Pongrac, P.; Drobne, D.; Pipan Tkalec, Ž.; Novak, S.; Kos, M.; Koren, Š.; et al. Elemental distribution and sample integrity comparison of freeze-dried and frozen-hydrated biological tissue samples with nuclear microprobe. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2015, 348, 147–151. [Google Scholar] [CrossRef]
- Vogel-Mikuš, K.; Pongrac, P.; Pelicon, P.; Vavpetič, P.; Povh, B.; Bothe, H.; Regvar, M. Micro-PIXE analysis for localization and quantification of elements in roots of mycorrhizal metal-tolerant plants. In Symbiotic Fungi: Principles and Practice; Varma, A., Kharkwal, A.C., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 227–242. ISBN 978-3-540-95894-9. [Google Scholar]
- Vogel-Mikus, K.; Regvar, M.; Mesjasz-Przybyłowicz, J.; Przybyłowicz, W.J.; Simcic, J.; Pelicon, P.; Budnar, M. Spatial distribution of cadmium in leaves of metal hyperaccumulating Thlaspi praecox using micro-PIXE. New Phytol. 2008, 179, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Vogel-Mikus, K.; Simcic, J.; Pelicon, P.; Budnar, M.; Kump, P.; Necemer, M.; Mesjasz-Przybyłowicz, J.; Przybyłowicz, W.J.; Regvar, M. Comparison of essential and non-essential element distribution in leaves of the Cd/Zn hyperaccumulator Thlaspi praecox as revealed by micro-PIXE. Plant Cell Environ. 2008, 31, 1484–1496. [Google Scholar] [CrossRef]
- Vogel-Mikuš, K.; Pongrac, P.; Kump, P.; Nečemer, M.; Simčič, J.; Pelicon, P.; Budnar, M.; Povh, B.; Regvar, M. Localisation and quantification of elements within seeds of Cd/Zn hyperaccumulator Thlaspi praecox by micro-PIXE. Environ. Pollut. 2007, 147, 50–59. [Google Scholar] [CrossRef]
- Weiersbye-Witkowski, I.M.; Przybylowicz, W.J.; Straker, C.J.; Mesjasz-Przybylowicz, J. Elemental micro-PIXE mapping of hypersensitive lesions in Lagenaria sphaerica (Cucurbitaceae) resistant to Sphaerotheca fuliginea (powdery mildew). Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 1997, 130, 388–395. [Google Scholar] [CrossRef]
- Stokes, I. Biosilicification in Oryza Sativa and Other Plants. Ph.D. Thesis, Keele University, Keele, UK, 2016. [Google Scholar]
- Dorairaj, D.; Ismail, M.R. Distribution of silicified microstructures, regulation of cinnamyl alcohol dehydrogenase and lodging resistance in silicon and paclobutrazol mediated Oryza sativa. Front. Physiol. 2017, 8, 491. [Google Scholar] [CrossRef]
- Kopittke, P.M.; Punshon, T.; Paterson, D.J.; Tappero, R.V.; Wang, P.; Blamey, F.P.C.; van der Ent, A.; Lombi, E. Synchrotron-based X-ray fluorescence microscopy as a technique for imaging of elements in plants. Plant Physiol. 2018, 178, 507–523. [Google Scholar] [CrossRef]
- Klančnik, K.; Vogel-Mikuš, K.; Gaberščik, A. Silicified structures affect leaf optical properties in grasses and sedge. J. Photochem. Photobiol. B Biol. 2014, 130, 1–10. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Vaccari, L.; Kourousias, G.; Cassese, D.; Bedolla, D.E.; Kenig, S.; Storici, P.; Lazzarino, M.; Kiskinova, M. Soft X-ray microscopy radiation damage on fixed cells investigated with synchrotron radiation FTIR microscopy. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tolrà, R.; Vogel-Mikuš, K.; Hajiboland, R.; Kump, P.; Pongrac, P.; Kaulich, B.; Gianoncelli, A.; Babin, V.; Barceló, J.; Regvar, M.; et al. Localization of aluminium in tea (Camellia sinensis) leaves using low energy X-ray fluorescence spectro-microscopy. J. Plant Res. 2011, 124, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M.; Gianoncelli, A.; Kourousias, G.; Green, K.; McKenna, B.A. Alleviation of Al toxicity by Si is associated with the formation of Al–Si complexes in root tissues of sorghum. Front. Plant Sci. 2017, 8, 2189. [Google Scholar] [CrossRef] [PubMed]
- Kump, P.; Vogel-Mikuš, K. Quantification of 2D elemental distribution maps of intermediate-thick biological sections by low energy synchrotron µ-X-ray fluorescence spectrometry. J. Instrum. 2018, 13, C05014. [Google Scholar] [CrossRef]
- Migeon, A.; Audinot, J.-N.; Eybe, T.; Richaud, P.; Damien, B.; Migeon, H.-N.; Chalot, M. Cadmium and zinc localization by SIMS in leaves of Populus deltoides (cv. Lena) grown in a metal polluted soil. Surf. Interface Anal. 2011, 43, 367–369. [Google Scholar] [CrossRef]
- Chandra, S.; Smith, D.R.; Morrison, G.H. Subcellular imaging by dynamic SIMS ion microscopy. Anal. Chem. 2000, 72, 104A–114A. [Google Scholar] [CrossRef]
- Moore, K.L.; Schröder, M.; Wu, Z.; Martin, B.G.H.; Hawes, C.R.; McGrath, S.P.; Hawkesford, M.J.; Ma, J.F.; Zhao, F.-J.; Grovenor, C.R.M. High-resolution secondary ion mass spectrometry reveals the contrasting subcellular distribution of arsenic and silicon in rice roots. Plant Physiol. 2011, 156, 913–924. [Google Scholar] [CrossRef]
- Sparks, J.P.; Chandra, S.; Derry, L.A.; Parthasarathy, M.V.; Daugherty, C.S.; Griffin, R. Subcellular localization of silicon and germanium in grass root and leaf tissues by SIMS: Evidence for differential and active transport. Biogeochemistry 2011, 104, 237–249. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Wu, C.; Xiong, L.; Chen, G.; Zhang, Q.; Wang, S. RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006, 34, D745–D748. [Google Scholar] [CrossRef]
- Jiao, Y.; Burke, J.; Chopra, R.; Burow, G.; Chen, J.; Wang, B.; Hayes, C.; Emendack, Y.; Ware, D.; Xin, Z. A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant Cell 2016, 28, 1551–1562. [Google Scholar] [CrossRef]
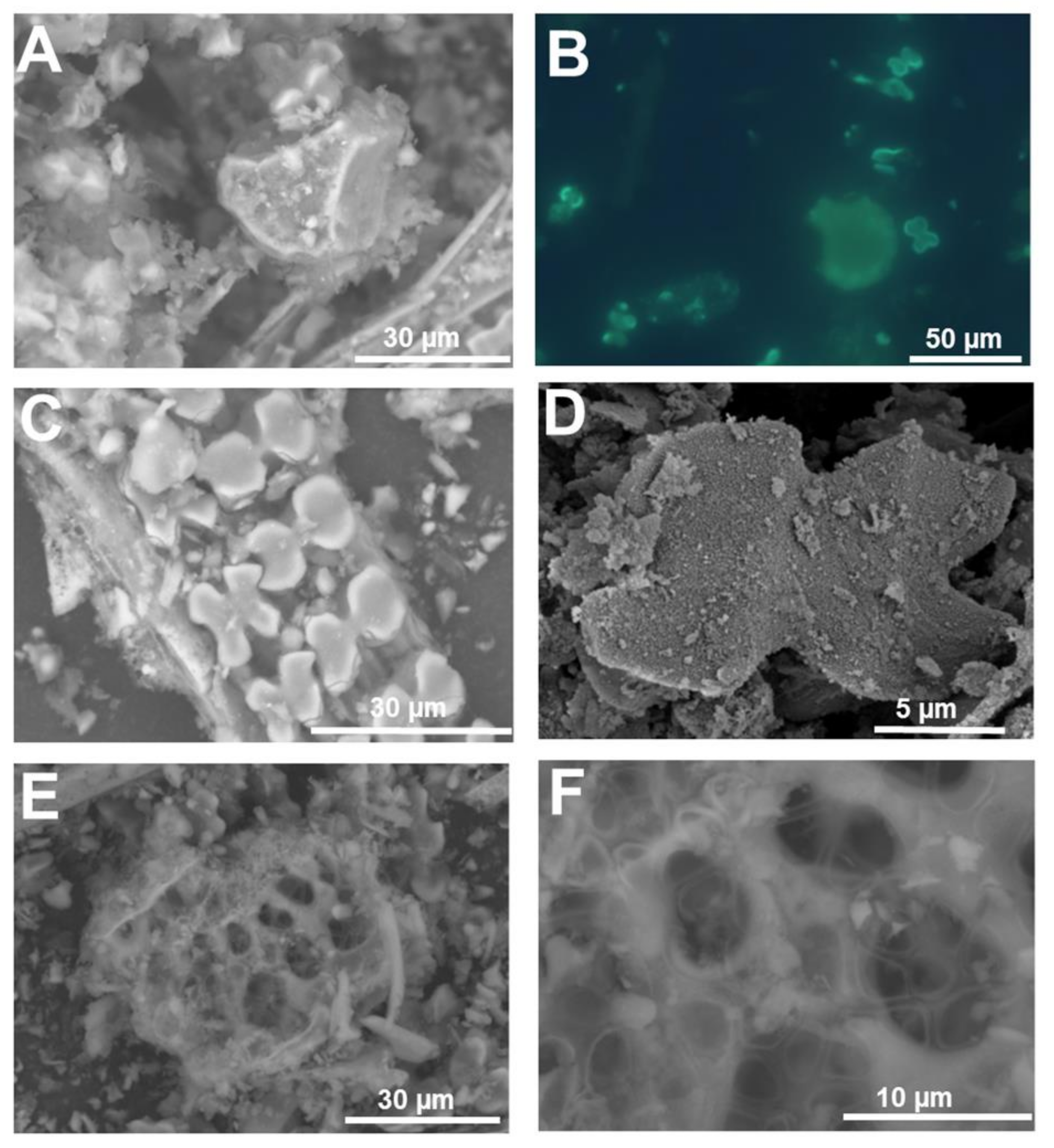
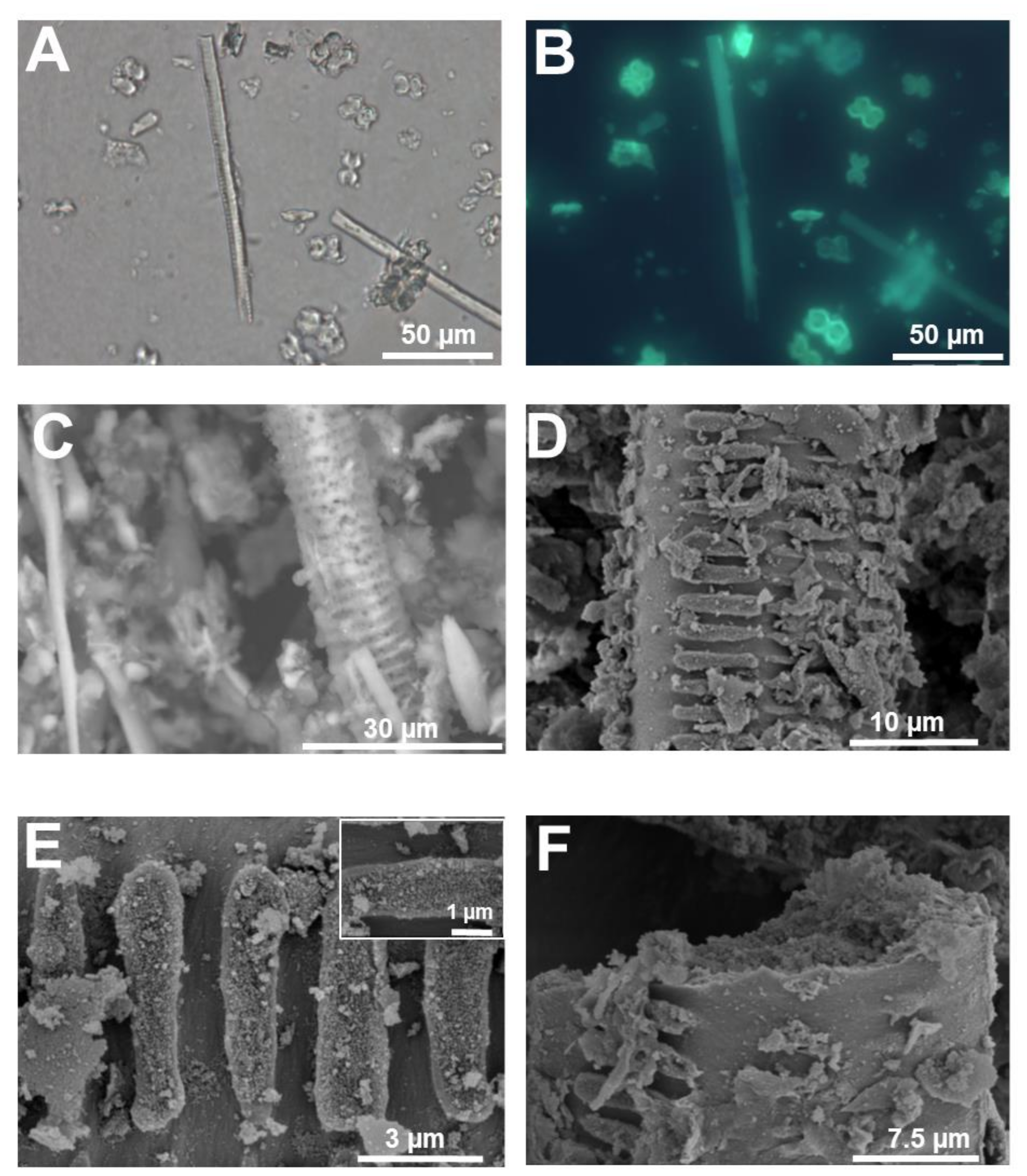
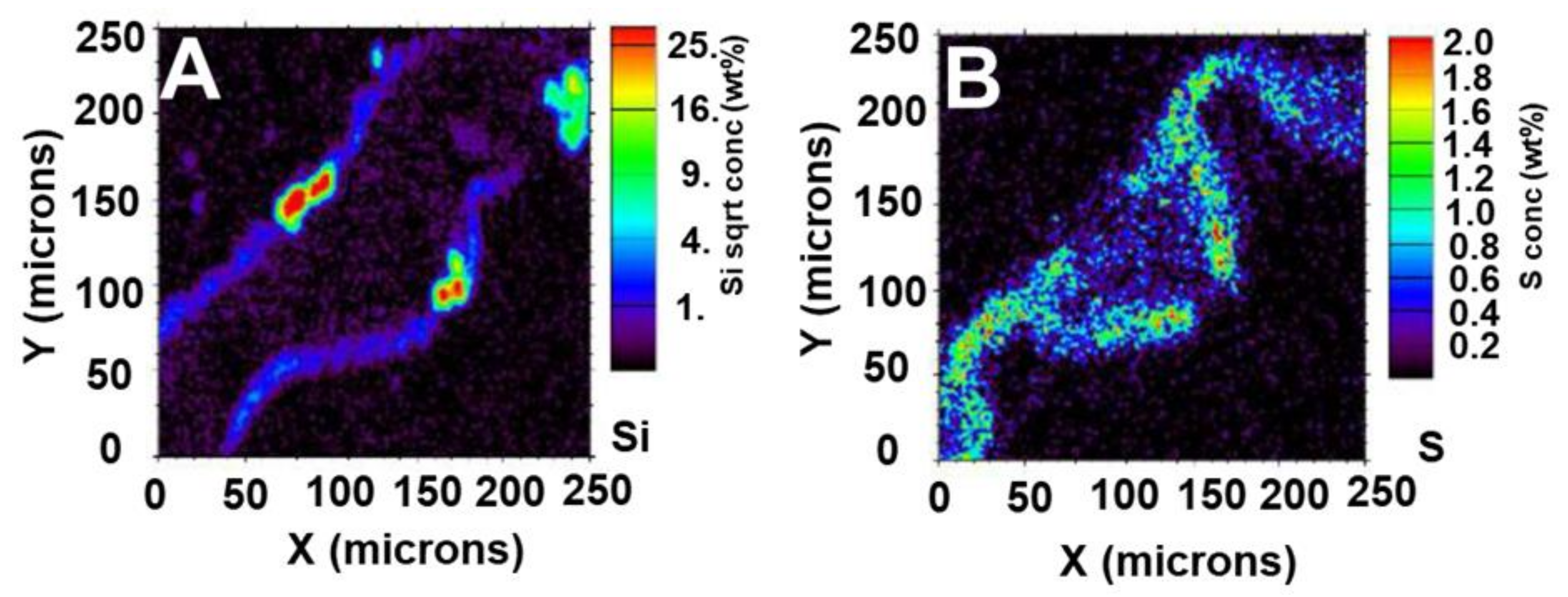

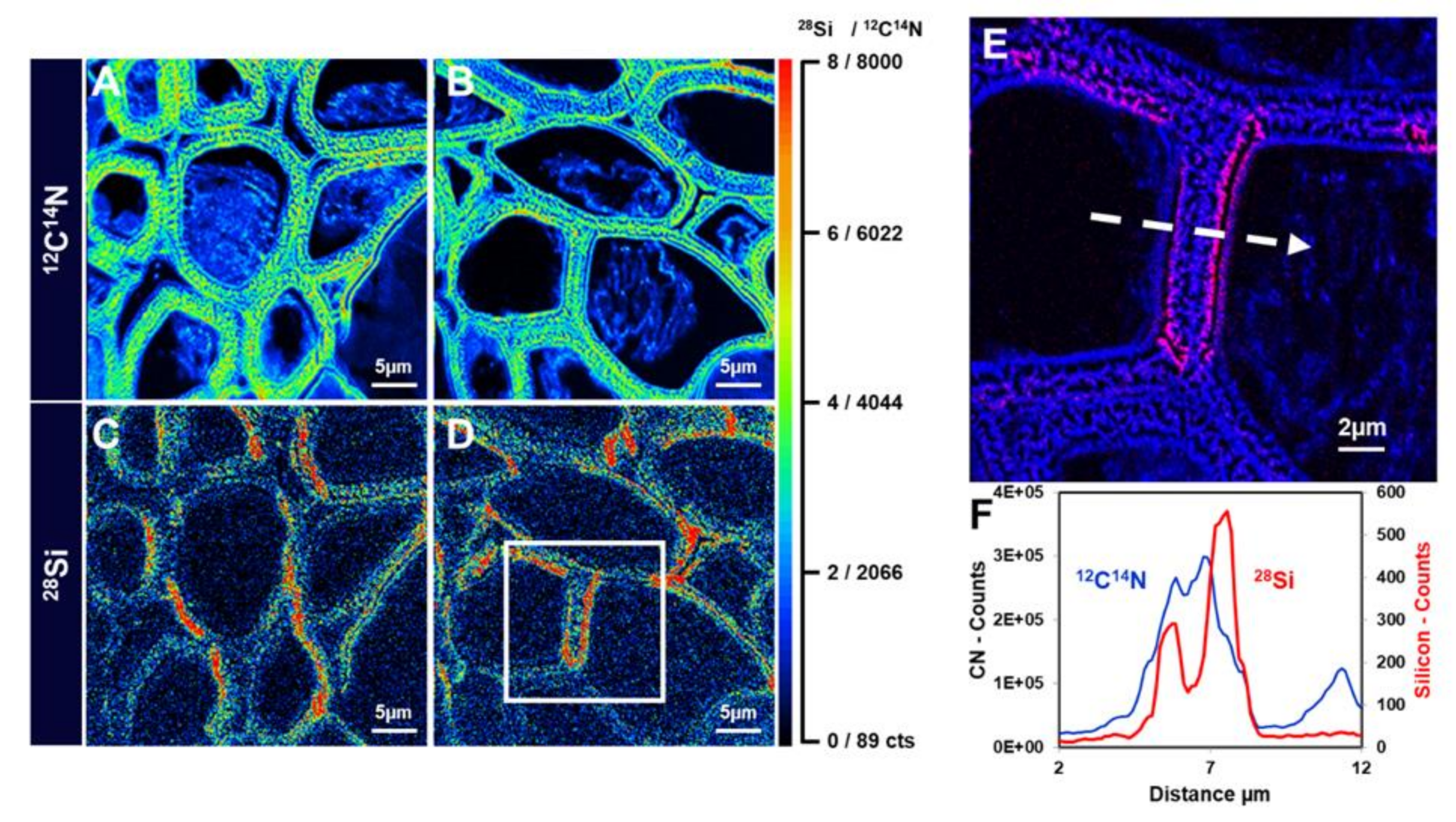
| Technique | Information | Advantages | Drawbacks | Spatial Resolution |
|---|---|---|---|---|
| Optical microscopy (dyes) | Tissue distribution | Instrument accessible in any laboratory | Invasive (tissue fixation) and slicing; etching; lack of discrimination between lignin and silica | 200 nm lateral 600 nm axial [31] |
| Confocal microscopy | Tissues distribution | Thick samples | Background fluorescence of chloroplasts | 180 nm lateral 500 nm axial [32] |
| Tabletop scanning electron microscopy (SEM) (on extracted silica) | Secondary electron images (SE): structure | Easy to use, rapidity;no sample preparation | Lower resolution than conventional SEM | 5–30 nm (depending on the model) |
| Conventional SEM (on extracted silica) | SE images: very thin structure | High resolution | Lengthy procedure for sample preparation with the use of hazardous chemicals | 1 nm |
| Micro-PIXE (microbeam particle-induced X-ray emission) | Quantitative elemental mapping | Quantification | Limit in the analysis of some light element (F, Li, B); time devoted to cryotechniques to preserve a state as close as possible to the native one | 1 µm |
| LEXRF (low-energy X-ray fluorescence) | Quantitative elemental mapping | Quantification | Limited access to synchrotron facilities; time devoted to cryotechniques for analysis of plant tissues | 100 nm [33] |
| NanoSIMS | Elemental mapping | High sensitivityHigh lateral resolution3D imaging | Sample preparation (removal of water without causing diffusion of element and damage of the sample structure) | 50 nm [34,35] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerriero, G.; Stokes, I.; Valle, N.; Hausman, J.-F.; Exley, C. Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging. Cells 2020, 9, 1066. https://doi.org/10.3390/cells9041066
Guerriero G, Stokes I, Valle N, Hausman J-F, Exley C. Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging. Cells. 2020; 9(4):1066. https://doi.org/10.3390/cells9041066
Chicago/Turabian StyleGuerriero, Gea, Ian Stokes, Nathalie Valle, Jean-Francois Hausman, and Christopher Exley. 2020. "Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging" Cells 9, no. 4: 1066. https://doi.org/10.3390/cells9041066
APA StyleGuerriero, G., Stokes, I., Valle, N., Hausman, J.-F., & Exley, C. (2020). Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging. Cells, 9(4), 1066. https://doi.org/10.3390/cells9041066







