miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment
Abstract
1. Introduction
2. Biogenesis and Regulatory Functions
3. miRNA: A Factor in Multifactorial Diseases
4. miRNAs and Their Role in the Tumor Microenvironment
5. Extracellular miRNAs
6. miRNAs-Based Therapy and Clinical Trials
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Robinson, V.L. Rethinking the central dogma: Noncoding RNAs are biologically relevant. Urol. Oncol. Semin. Ori. 2009, 27, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C-Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Caygill, E.E.; Johnston, L.A. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef]
- Sun, X.; Liu, J.; Xu, C.W.; Tang, S.C.; Ren, H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J. Cell Mol. Med. 2016, 20, 1779–1788. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Flynt, A.S.; Lai, E.C. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008, 9, 831–842. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant. Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Bio. 2008, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Hobert, O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb. Sym. 2006, 71, 181–188. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Mladinov, D.; Liu, Y.; Mattson, D.L.; Liang, M.Y. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase beta 1. Nucleic Acids Res. 2013, 41, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Ultimo, S.; Martelli, A.M.; Zauli, G.; Vitale, M.; Calin, G.A.; Neri, L.M. Roles and clinical implications of microRNAs in acute lymphoblastic leukemia. J. Cell. Physiol. 2018, 233, 5642–5654. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Komatsu, S.; Ichikawa, D.; Tsujiura, M.; Takeshita, H.; Hirajima, S.; Miyamae, M.; Okajima, W.; Ohashi, T.; Imamura, T.; et al. Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. Int. J. Mol. Sci. 2016, 17, 1459. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.J.; Qin, Z.L.; Xue, L.X. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin. Epigenetics 2018, 10. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef]
- Yang, N.; Zhu, S.; Lv, X.P.; Qiao, Y.; Liu, Y.J.; Chen, J.T. MicroRNAs: Pleiotropic Regulators in the Tumor Microenvironment. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Chou, J.; Shahi, P.; Werb, Z. microRNA-mediated regulation of the tumor microenvironment. Cell Cycle 2013, 12, 3262–3271. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: stepwise processing and subcellular localization. Embo. J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Song, M.S.; Rossi, J.J. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.C.; Iwasaki, S.; Liu, Q.H.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias (vol 115, pg 209, 2003). Cell 2003, 115, 505. [Google Scholar] [CrossRef]
- Hu, S.; Zhao, X.Y.; Mao, G.P.; Zhang, Z.J.; Wen, X.Z.; Zhang, C.Y.; Liao, W.M.; Zhang, Z.Q. MicroRNA-455-3p promotes TGF-beta signaling and inhibits osteoarthritis development by directly targeting PAK2. Exp. Mol. Med. 2019, 51. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.S.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Izaurralde, E. The Role of GW182 Proteins in miRNA-Mediated Gene Silencing. Ten Years Prog. Gw/P Body Res. 2013, 768, 147–163. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Xie, M.Y.; Li, M.F.; Vilborg, A.; Lee, N.; Shu, M.D.; Yartseva, V.; Sestan, N.; Steitz, J.A. Mammalian 5 ‘-Capped MicroRNA Precursors that Generate a SingleMicroRNA. Cell 2013, 155, 1568–1580. [Google Scholar] [CrossRef]
- Rorbach, G.; Unold, O.; Konopka, B.M. Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods. Sci. Rep.-UK 2018, 8. [Google Scholar] [CrossRef]
- Yang, J.S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes - Drosha, DGCR8, Dicer and Ago proteins. Febs. Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef]
- Knuckles, P.; Vogt, M.A.; Lugert, S.; Milo, M.; Chong, M.M.W.; Hautbergue, G.M.; Wilson, S.A.; Littman, D.R.; Taylor, V. Drosha regulates neurogenesis by controlling Neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012, 15, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Swahari, V.; Nakamura, A.; Baran-Gale, J.; Garcia, I.; Crowther, A.J.; Sons, R.; Gershon, T.R.; Hammond, S.; Sethupathy, P.; Deshmukh, M. Essential Function of Dicer in Resolving DNA Damage in the Rapidly Dividing Cells of the Developing and Malignant Cerebellum. Cell. Rep. 2016, 14, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.H.; Shin, S.; Jung, S.R.; Kim, E.; Song, J.J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell. 2015, 59, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Boland, A.; Huntzinger, E.; Weichenrieder, O.; Izaurralde, E. Structure of the PAN3 Pseudokinase Reveals the Basis for Interactions with the PAN2 Deadenylase and the GW182 Proteins. Mol. Cell 2013, 51, 360–373. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Peters, L.; Meister, G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell. 2007, 26, 611–623. [Google Scholar] [CrossRef]
- Hutvagner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. MRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Gene Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global Analyses of the Effect of Different Cellular Contexts on MicroRNA Targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Blazie, S.M.; Geissel, H.C.; Wilky, H.; Joshi, R.; Newbern, J.; Mangone, M. Alternative Polyadenylation Directs Tissue-Specific miRNA Targeting in Caenorhabditis elegans Somatic Tissues. Genetics 2017, 206, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Ramchandran, R.; Chaluvally-Raghavan, P. miRNA-Mediated RNA Activation in Mammalian Cells. Adv. Exp. Med. Biol. 2017, 983, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Vaschetto, L.M. miRNA activation is an endogenous gene expression pathway. RNA Biol. 2018, 15, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.S.; Tao, Y.G. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.Z.; Xi, Y.P.; Zhang, L.; Ding, C.; Luo, H.B.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 464, 126. [Google Scholar] [CrossRef]
- Yuan, K.; Ai, W.B.; Wan, L.Y.; Tan, X.; Wu, J.F. The miR-290-295 cluster as multi-faceted players in mouse embryonic stem cells. Cell. Biosci. 2017, 7. [Google Scholar] [CrossRef]
- Melton, C.; Blelloch, R. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv. Exp. Med. Biol. 2010, 695, 105–117. [Google Scholar] [CrossRef]
- Kosik, K.S.; Krichevsky, A.M. The elegance of the microRNAs: A neuronal perspective. Neuron 2005, 47, 779–782. [Google Scholar] [CrossRef]
- Jovicic, A.; Roshan, R.; Moisoi, N.; Pradervand, S.; Moser, R.; Pillai, B.; Luthi-Carter, R. Comprehensive Expression Analyses of Neural Cell-Type-Specific miRNAs Identify New Determinants of the Specification and Maintenance of Neuronal Phenotypes. J. Neurosci. 2013, 33, 5127–5137. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Johnston, R.J.; Frokjaer-Jensen, C.; Lockery, S.; Hobert, O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 2004, 430, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.J.; Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 2003, 426, 845–849. [Google Scholar] [CrossRef]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: from master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Yu, C.Y.; Wang, Y.; Liu, L.; Zhang, K.; Fang, C.; Liu, F.F.; Bian, G.L.; Song, B.; Yang, A.G.; et al. miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3 beta pathway by targeting Rap2a. Sci. Rep.-UK 2016, 6. [Google Scholar] [CrossRef]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.L.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development (vol 439, pg 283, 2006). Nature 2006, 441. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Zhao, Y.H.; Feng, Z.H. MicroRNA Control of p53. J. Cell Biochem. 2017, 118, 7–14. [Google Scholar] [CrossRef]
- Hoffman, Y.; Pilpel, Y.; Oren, M. microRNAs and Alu elements in the p53-Mdm2-Mdm4 regulatory network. J. Mol. Cell Biol. 2014, 6, 192–197. [Google Scholar] [CrossRef]
- Hermeking, H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat. Rev. Cancer 2012, 12, 613–626. [Google Scholar] [CrossRef]
- Tarasov, V.; Jung, P.; Verdoodt, B.; Lodygin, D.; Epanchintsev, A.; Menssen, A.; Meister, G.; Hermeking, H. Differential regulation of microRNAs by p53 revealed by massively parallel Sequencing - miR-34a is a p53 target that induces apoptosis and G(1)-arrest. Cell Cycle 2007, 6, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.Y.; Lim, L.P.; De Stanchina, E.; Xuan, Z.Y.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Zhu, S.M.; Wu, F.T.; Wu, H.L.; Walia, V.; Kumar, S.; Elble, R.; Watabe, K.; Mo, Y.Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA 2009, 106, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Yao, G.D.; Yin, M.M.; Lu, M.R.; Tian, H.; Liu, L.; Lian, J.; Huang, X.X.; Sun, F. Transcriptional cooperation between p53 and NF-kappa B p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Mol. Cell. Endocrinol. 2013, 370, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.L.; Xue, G.; Mei, Q.; Wang, Y.Z.; Ding, F.X.; Liu, M.F.; Lu, M.H.; Tang, Y.; Yu, H.Y.; Sun, S.H. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. Embo. J. 2009, 28, 2719–2732. [Google Scholar] [CrossRef]
- Duarte, F.V.; Palmeira, C.M.; Rolo, A.P. The Emerging Role of MitomiRs in the Pathophysiology of Human Disease. Adv. Exp. Med. Biol. 2015, 888, 123–154. [Google Scholar] [CrossRef]
- Duarte, F.V.; Amorim, J.A.; Palmeira, C.M.; Rolo, A.P. Regulation of Mitochondrial Function and its Impact in Metabolic Stress. Curr. Med. Chem. 2015, 22, 2468–2479. [Google Scholar] [CrossRef]
- Shepherd, D.L.; Hathaway, Q.A.; Pinti, M.V.; Nichols, C.E.; Durr, A.J.; Sreekumar, S.; Hughes, K.M.; Stine, S.M.; Martinez, I.; Hollander, J.M. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J. Mol. Cell. Cardiol. 2017, 110, 15–25. [Google Scholar] [CrossRef]
- Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Stricker, J.C.; Croston, T.L.; Baseler, W.A.; Lewis, S.E.; Martinez, I.; Hollander, J.M. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ. Cardiovasc. Genet. 2015, 8, 785–802. [Google Scholar] [CrossRef]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 2019, 79, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.I.; Goergen, D.; Zheng, J.F.; Song, Y.T.; Schuttler, C.G.; Fehr, C.; Junemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo. J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Arend, K.C.; Li, Y.; Yamane, D.; McGivern, D.R.; Kato, T.; Wakita, T.; Moorman, N.J.; Lemon, S.M. miR-122 Stimulates Hepatitis C Virus RNA Synthesis by Altering the Balance of Viral RNAs Engaged in Replication versus Translation. Cell Host Microbe 2015, 17, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Sedano, C.D.; Sarnow, P. Hepatitis C Virus Subverts Liver-Specific miR-122 to Protect the Viral Genome from Exoribonuclease Xrn2. Cell Host Microbe 2014, 16, 257–264. [Google Scholar] [CrossRef]
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.M.; Feray, C.; Trabucchi, M. Viruses and miRNAs: More Friends than Foes. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Grundhoff, A.; Sullivan, C.S. Virus-encoded microRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef]
- Hook, L.M.; Grey, F.; Grabski, R.; Tirabassi, R.; Doyle, T.; Hancock, M.; Landais, I.; Jeng, S.; McWeeney, S.; Britt, W.; et al. Cytomegalovirus miRNAs Target Secretory Pathway Genes to Facilitate Formation of the Virion Assembly Compartment and Reduce Cytokine Secretion. Cell Host Microbe 2014, 15, 363–373. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Sullivan, C.S. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathogens 2012, 8. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Singha, R.K.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Wang, M.L.; Qin, L.X.; Tang, B.S. MicroRNAs in Alzheimer’s Disease. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Lei, X.F.; Lei, L.J.; Zhang, Z.L.; Zhang, Z.Q.; Cheng, Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int J. Clin. Exp. Pathol. 2015, 8, 1565–1574. [Google Scholar]
- Ulrich, J.D.; Holtzman, D.M. TREM2 Function in Alzheimer’s Disease and Neurodegeneration. ACS Chem. Neurosci. 2016, 7, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Tiribuzi, R.; Crispoltoni, L.; Porcellati, S.; Di Lullo, M.; Florenzano, F.; Pirro, M.; Bagaglia, F.; Kawarai, T.; Zampolini, M.; Orlacchio, A.; et al. miR128 up-regulation correlates with impaired amyloid beta(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol. Aging 2014, 35, 345–356. [Google Scholar] [CrossRef]
- Gupta, A.; Iadecola, C. Impaired A beta clearance: a potential link between atherosclerosis and Alzheimer’s disease. Front. Aging Neurosci. 2015, 7. [Google Scholar] [CrossRef]
- Ma, X.H.; Liu, L.L.; Meng, J. MicroRNA-125b promotes neurons cell apoptosis and Tau phosphorylation in Alzheimer’s disease. Neurosci. Lett. 2017, 661, 57–62. [Google Scholar] [CrossRef]
- Wang, G.; Huang, Y.; Wang, L.L.; Zhang, Y.F.; Xu, J.; Zhou, Y.; Lourenco, G.F.; Zhang, B.; Wang, Y.; Ren, R.J.; et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep.-UK 2016, 6. [Google Scholar] [CrossRef]
- Perez Ortiz, J.M.; Swerdlow, R.H. Mitochondrial dysfunction in Alzheimer’s disease: Role in pathogenesis and novel therapeutic opportunities. Br. J. Pharmacol. 2019, 176, 3489–3507. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Z.F.; Li, W.; Hong, H.; Chen, J.; Tian, Y.; Liu, Z.Y. Protective effects of microRNA-330 on amyloid -protein production, oxidative stress, and mitochondrial dysfunction in Alzheimer’s disease by targeting VAV1 via the MAPK signaling pathway. J. Cell. Biochem. 2018, 119, 5437–5448. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.T.; Liu, Q.Y.; Tan, M.S.; Zhang, W.; Hu, N.; Wang, Y.L.; Sun, L.; Jiang, T.; Tan, L. Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 2014, 336, 52–56. [Google Scholar] [CrossRef]
- Bluher, M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Landrier, J.F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef] [PubMed]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; deLemos, A.S.; Black, J.C.; Ramirez, C.M.; Li, Y.X.; Tewhey, R.; et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Singh, A.K.; Rotllan, N.; Price, N.; Fernandez-Hernando, C. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef]
- Giralt, M.; Villarroya, F. White, Brown, Beige/Brite: Different Adipose Cells for Different Functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef]
- Schulz, T.J.; Huang, T.L.; Tran, T.T.; Zhang, H.B.; Townsend, K.L.; Shadrach, J.L.; Cerletti, M.; McDougall, L.E.; Giorgadze, N.; Tchkonia, T.; et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. USA 2011, 108, 143–148. [Google Scholar] [CrossRef]
- Rosenwald, M.; Perdikari, A.; Rulicke, T.; Wolfrum, C. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 2013, 15, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Ahmed, K.; Esau, C.C.; Stoffel, M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 2012, 14, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.C.; Yu, J.; Bi, J.H.; Qi, H.M.; Di, W.J.; Wu, L.; Wang, L.; Zha, J.M.; Lv, S.; Zhang, F.; et al. Glucocorticoids Transcriptionally Regulate miR-27b Expression Promoting Body Fat Accumulation Via Suppressing the Browning of White Adipose Tissue. Diabetes 2015, 64, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Duarte, F.V.; Palmeira, C.M.; Rolo, A.P. The Role of microRNAs in Mitochondria: Small Players Acting Wide. Genes (Basel) 2014, 5, 865–886. [Google Scholar] [CrossRef]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mossenbock, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 Family Is Required for Human Adipogenesis and Drives Characteristics of Brown Adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef]
- Vargas-Castillo, A.; Fuentes-Romero, R.; Rodriguez-Lopez, L.A.; Torres, N.; Tovar, A.R. Understanding the Biology of Thermogenic Fat: Is Browning A New Approach to the Treatment of Obesity? Arch. Med. Res. 2017, 48, 401–413. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer; Garland Science: New York, NY, USA, 2007. [Google Scholar]
- Simioni, C.; Martelli, A.M.; Zauli, G.; Melloni, E.; Neri, L.M. Targeting mTOR in Acute Lymphoblastic Leukemia. Cells 2019, 8, 190. [Google Scholar] [CrossRef]
- Bazzichetto, C.; Conciatori, F.; Pallocca, M.; Falcone, I.; Fanciulli, M.; Cognetti, F.; Milella, M.; Ciuffreda, L. PTEN as a Prognostic/Predictive Biomarker in Cancer: An Unfulfilled Promise? Cancers 2019, 11, 435. [Google Scholar] [CrossRef]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell. Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef]
- Williams, A.; Henao-Mejia, J.; Harman, C.C.; Flavell, R.A. miR-181 and metabolic regulation in the immune system. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 223–230. [Google Scholar] [CrossRef]
- Henao-Mejia, J.; Williams, A.; Goff, L.A.; Staron, M.; Licona-Limon, P.; Kaech, S.M.; Nakayama, M.; Rinn, J.L.; Flavell, R.A. The MicroRNA miR-181 Is a Critical Cellular Metabolic Rheostat Essential for NKT Cell Ontogenesis and Lymphocyte Development and Homeostasis. Immunity 2013, 38, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Croce, C.M. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2008, 15, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Luo, X.Q.; Zhang, P.; Huang, L.B.; Zheng, Y.S.; Wu, J.; Zhou, H.; Qu, L.H.; Xu, L.; Chen, Y.Q. MicroRNA Patterns Associated with Clinical Prognostic Parameters and CNS Relapse Prediction in Pediatric Acute Leukemia. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, J.H.; Umezu, T.; Kobayashi, C.; Hamamura, R.S.; Tanaka, M.; Kuroda, M.; Ohyashiki, K. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: in vivo assessment of cell to plasma ratio of miR-92a. BMC Res. Notes 2010, 3, 347. [Google Scholar] [CrossRef]
- Nemes, K.; Csoka, M.; Nagy, N.; Mark, A.; Varadi, Z.; Danko, T.; Kovacs, G.; Kopper, L.; Sebestyen, A. Expression of Certain Leukemia/Lymphoma Related microRNAs and its Correlation with Prognosis in Childhood Acute Lymphoblastic Leukemia. Pathol. Oncol. Res. 2015, 21, 597–604. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. BCL2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef]
- Singh, P.K.; Purohit, V.; Chaika, N.; Johnson, K.R.; Hollingsworth, M.A. Regulation of glucose metabolism by HIF in pancreatic cancer. Cancer Res. 2011, 71. [Google Scholar] [CrossRef]
- Ahmad, A.; Aboukameel, A.; Kong, D.J.; Wang, Z.W.; Sethi, S.; Chen, W.; Sarkar, F.H.; Raz, A. Phosphoglucose Isomerase/Autocrine Motility Factor Mediates Epithelial-Mesenchymal Transition Regulated by miR-200 in Breast Cancer Cells. Cancer Res. 2011, 71, 3400–3409. [Google Scholar] [CrossRef]
- Karamitopoulou, E.; Haemmig, S.; Baumgartner, U.; Schlup, C.; Wartenberg, M.; Vassella, E. MicroRNA dysregulation in the tumor microenvironment influences the phenotype of pancreatic cancer. Modern Pathol. 2017, 30, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Nocchi, L.; Staffolani, S.; Manzella, N.; Amati, M.; Goodwin, J.; Kluckova, K.; Nguyen, M.; Strafella, E.; Bajzikova, M.; et al. MicroRNA-126 suppresses mesothelioma malignancy by targeting IRS1 and interfering with the mitochondrial function. Antioxid. Redox Signal. 2014, 21, 2109–2125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, C.; Zhang, X. Mitochondrial Damage Mediated by miR-1 Overexpression in Cancer Stem Cells. Mol. Ther. Nucleic Acids 2019, 18, 938–953. [Google Scholar] [CrossRef] [PubMed]
- Taddei, M.L.; Giannoni, E.; Comito, G.; Chiarugi, P. Microenvironment and tumor cell plasticity: An easy way out. Cancer Lett. 2013, 341, 80–96. [Google Scholar] [CrossRef]
- Psaila, B.; Lyden, D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, H.; Powathil, G.; Kim, H.; Trucu, D.; Lee, W.; Lawler, S.; Chaplain, M. Role of extracellular matrix and microenvironment in regulation of tumor growth and LAR-mediated invasion in glioblastoma. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Ogawa, D.; Ansari, K.; Nowicki, M.O.; Salinska, E.; Bronisz, A.; Godlewski, J. MicroRNA-451 Inhibits Migration of Glioblastoma while Making It More Susceptible to Conventional Therapy. Non-Coding RNA 2019, 5, 25. [Google Scholar] [CrossRef]
- Sahay, D.; Leblanc, R.; Grunewald, T.G.P.; Ambatipudi, S.; Ribeiro, J.; Clezardin, P.; Peyruchaud, O. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget 2015, 6, 20604–20620. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.H.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.A.; Xiang, R.; et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef]
- Yu, T.; Zuo, Q.F.; Gong, L.; Wang, L.N.; Zou, Q.M.; Xiao, B. MicroRNA-491 regulates the proliferation and apoptosis of CD8(+) T cells. Sci. Rep.-UK 2016, 6. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, L.H.; Chen, Q.Y.; Song, Y.J.; Xu, S.; Ma, F.; Wang, X.J.; Wang, J.L.; Yu, H.; Cao, X.T.; et al. MicroRNA-494 Is Required for the Accumulation and Functions of Tumor-Expanded Myeloid-Derived Suppressor Cells via Targeting of PTEN. J. Immunol. 2012, 188, 5500–5510. [Google Scholar] [CrossRef] [PubMed]
- Bullock, M.D.; Pickard, K.M.; Nielsen, B.S.; Sayan, A.E.; Jenei, V.; Mellone, M.; Mitter, R.; Primrose, J.N.; Thomas, G.J.; Packham, G.K.; et al. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef]
- Naito, Y.; Sakamoto, N.; Oue, N.; Yashiro, M.; Sentani, K.; Yanagihara, K.; Hirakawa, K.; Yasui, W. MicroRNA-143 regulates collagen type III expression in stromal fibroblasts of scirrhous type gastric cancer. Cancer Sci. 2014, 105, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.Y.; Xin, J.X.; Liu, Y.; Zhang, Y.; Liang, X.; Su, X.M.; Yan, H.; Huang, Y.G.; Yang, R.C. MicroRNA-200c Promotes Suppressive Potential of Myeloid-Derived Suppressor Cells by Modulating PTEN and FOG2 Expression. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Ingenito, F.; Roscigno, G.; Affinito, A.; Nuzzo, S.; Scognamiglio, I.; Quintavalle, C.; Condorelli, G. The Role of Exo-miRNAs in Cancer: A Focus on Therapeutic and Diagnostic Applications. Int. J. Mol. Sci. 2019, 20, 4687. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Bauer, A.; ElSharawy, A.; Haas, J.; Backes, C.; Wendschlag, A.; Giese, N.; Tjaden, C.; Ott, K.; et al. Toward the blood-borne miRNome of human diseases. Nat. Methods 2011, 8, 841–843. [Google Scholar] [CrossRef]
- Bell, E.; Taylor, M.A. Functional Roles for Exosomal MicroRNAs in the Tumour Microenvironment. Comput. Struct. Biotec. 2017, 15, 8–13. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs Modulate MicroRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell. Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef]
- Hergenreider, E.; Heydt, S.; Treguer, K.; Boettger, T.; Horrevoets, A.J.G.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.K.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell. Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lasser, C.; Szabo, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzas, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Than, U.T.T.; Guanzon, D.; Broadbent, J.A.; Leavesley, D.I.; Salomon, C.; Parker, T.J. Differential Expression of Keratinocyte-Derived Extracellular Vesicle Mirnas Discriminate Exosomes From Apoptotic Bodies and Microvesicles. Front. Endocrinol. (Lausanne) 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Turiak, L.; Misjak, P.; Szabo, T.G.; Aradi, B.; Paloczi, K.; Ozohanics, O.; Drahos, L.; Kittel, A.; Falus, A.; Buzas, E.I.; et al. Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. J. Proteomics 2011, 74, 2025–2033. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell. Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Hobor, F.; Dallmann, A.; Ball, N.J.; Cicchini, C.; Battistelli, C.; Ogrodowicz, R.W.; Christodoulou, E.; Martin, S.R.; Castello, A.; Tripodi, M.; et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Torres, L.F.C.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.B.; Li, Z.L.; Luo, D.H.; Huang, B.J.; Chen, Y.S.; Zhang, X.S.; Cui, J.; Zeng, Y.X.; Li, J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 2014, 5, 5439–5452. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.W.; Zhang, Y.J.; Li, J.; Wang, Z.Y.; Chen, X.L.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF- and miR23a transfer. Oncoimmunology 2016, 5. [Google Scholar] [CrossRef]
- Ding, G.P.; Zhou, L.J.; Qian, Y.M.; Fu, M.N.; Chen, J.; Chen, J.H.; Xiang, J.Y.; Wu, Z.R.; Jiang, G.X.; Cao, L.P. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.H.; Zhou, L.J.; Chen, W.C.; Ding, G.P.; Cao, L.P. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated Polarization of Tumor-Associated Macrophages by miR-145 via Colorectal Cancer-Derived Extracellular Vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.Q.; Su, F.; Yu, B.; Su, F.X.; Lin, L.; Liu, Y.J.; Huang, J.D.; Song, E.W. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10. [Google Scholar] [CrossRef]
- Lin, X.J.; Fang, J.H.; Yang, X.J.; Zhang, C.; Yuan, Y.F.; Zheng, L.M.; Zhuang, S.M. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol. Ther.-Nucl. Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, F.; Wang, B.R.; Li, H.Q.; Xu, Y.; Liu, X.L.; Shi, L.; Lu, X.L.; Xu, W.C.; Lu, L.; et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016, 370, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sun, S.; Li, Z.Y.; Yang, Q.; Li, B.; Zhu, S.; Wang, L.J.; Wu, J.; Yuan, J.P.; Yang, C.H.; et al. Tumour-originated exosomal miR-155 triggers cancer-associated cachexia to promote tumour progression. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Li, Z.; Sun, S.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Zhang, Y.; Wang, C. Exosomes from the tumour-adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J. Exp. Clin. Canc. Res. 2019, 38, 223. [Google Scholar] [CrossRef] [PubMed]
- Bandini, E.; Rossi, T.; Gallerani, G.; Fabbri, F. Adipocytes and microRNAs Crosstalk: A Key Tile in the Mosaic of Breast Cancer Microenvironment. Cancers 2019, 11, 1451. [Google Scholar] [CrossRef] [PubMed]
- Takahara, K.; Ii, M.; Inamoto, T.; Nakagawa, T.; Ibuki, N.; Yoshikawa, Y.; Tsujino, T.; Uchimoto, T.; Saito, K.; Takai, T.; et al. microRNA-145 Mediates the Inhibitory Effect of Adipose Tissue-Derived Stromal Cells on Prostate Cancer. Stem Cells Dev. 2016, 25, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Kim, S.M.; Woo, E.Y.; Han, K.C.; Park, E.J.; Ko, S.; Choi, E.W.; Jang, M. Stemness-Attenuating miR-503-3p as a Paracrine Factor to Regulate Growth of Cancer Stem Cells. Stem Cells Int. 2018. [Google Scholar] [CrossRef]
- Gernapudi, R.; Yao, Y.; Zhang, Y.S.; Wolfson, B.; Roy, S.; Duru, N.; Eades, G.; Yang, P.X.; Zhou, Q. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res. Treat. 2015, 150, 685–695. [Google Scholar] [CrossRef]
- Chan, B.; Manley, J.; Lee, J.; Singh, S.R. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015, 356, 301–308. [Google Scholar] [CrossRef]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef]
- Schmittgen, T.D. Exosomal miRNA Cargo as Mediator of Immune Escape Mechanisms in Neuroblastoma. Cancer Res. 2019, 79, 1293–1294. [Google Scholar] [CrossRef]
- Park, J.K.; Kogure, T.; Nuovo, G.J.; Jiang, J.M.; He, L.; Kim, J.H.; Phelps, M.A.; Papenfuss, T.L.; Croce, C.M.; Patel, T.; et al. miR-221 Silencing Blocks Hepatocellular Carcinoma and Promotes Survival. Cancer Res. 2011, 71, 7608–7616. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.C.; Valencia, T.; Allerson, C.; Schairer, A.; Flaten, A.; Yheskel, M.; Kersjes, K.; Li, J.; Gatto, S.; Takhar, M.; et al. Discovery and preclinical evaluation of anti-miR-17 oligonucleotide RGLS4326 for the treatment of polycystic kidney disease. Nat. Commun. 2019, 10, 4148. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.; Wang, S.; Li, P.; Zheng, C.; Zhou, X.; Tao, Y.; Chen, X.; Sun, L.; Wang, A.; et al. Blockage of transferred exosome-shuttled miR-494 inhibits melanoma growth and metastasis. J. Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.J.; Tu, C.G.; Zhang, J.W.; Wang, J.H. Inhibition of multiple myeloma-derived exosomes uptake suppresses the functional response in bone marrow stromal cell. Int. J. Oncol. 2019, 54, 1061–1070. [Google Scholar] [CrossRef]
- Hanna, J.; Hossein, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; Rebhan, M.A.E.; Crivelli, S.E.M.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014, 42, 609–621. [Google Scholar] [CrossRef]
- Andriani, F.; Majorini, M.T.; Mano, M.; Landoni, E.; Miceli, R.; Facchinetti, F.; Mensah, M.; Fontanella, E.; Dugo, M.; Giacca, M.; et al. MiR-16 regulates the pro-tumorigenic potential of lung fibroblasts through the inhibition of HGF production in an FGFR-1- and MEK1-dependent manner. J. Hematol. Oncol. 2018, 11, 45. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef]
- Singh, S.; Narang, A.S.; Mahato, R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011, 28, 2996–3015. [Google Scholar] [CrossRef]
- Lin, X.Y.; Ruan, X.; Anderson, M.G.; McDowell, J.A.; Kroeger, P.E.; Fesik, S.W.; Shen, Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005, 33, 4527–4535. [Google Scholar] [CrossRef]
- Nielsen, C.B.; Shomron, N.; Sandberg, R.; Hornstein, E.; Kitzman, J.; Burge, C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. Rna 2007, 13, 1894–1910. [Google Scholar] [CrossRef] [PubMed]
- Judge, A.; Maclachlan, I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008, 19, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Betel, D.; Miller, M.L.; Sander, C.; Leslie, C.S.; Marks, D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009, 27, 671. [Google Scholar] [CrossRef]
- Bhome, R.; Del Vecchio, F.; Lee, G.H.; Bullock, M.D.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018, 420, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Gahan, P.B. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Non-Coding RNA 2019, 5, 28. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
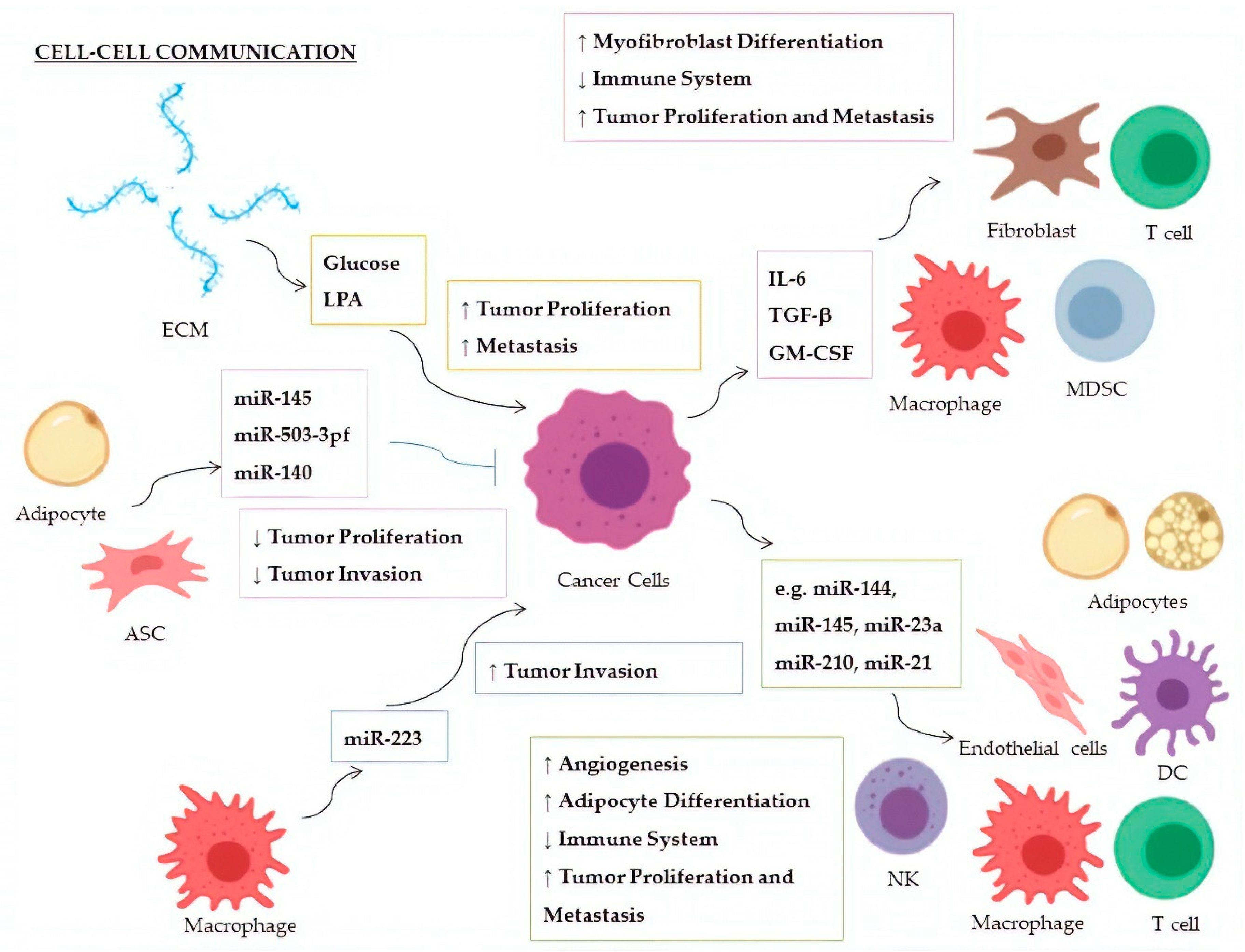
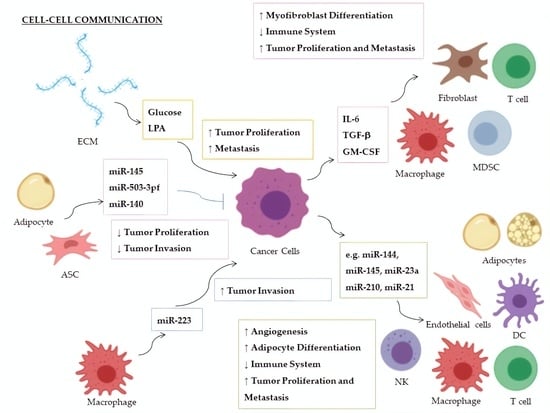
| Factor | miRNA | Donor | Recipient | Targeted Molecule | Target Expression | Effect | Ref. |
|---|---|---|---|---|---|---|---|
| Glucose | miR-451 | CSPGs (ECM) | Glioblastoma | AMPK | ↓ | Increased cellular proliferation; decreased tumor migration | [147,148] |
| LPA | miR-21 | ECM | Breast Cancer | PTEN | ↓ | Promotion of metastasis | [149] |
| IL-6 | miR-19a-3p | Breast Cancer | TAMs | Fra-1 | ↑ | Increased expression of VEGF and STAT3; M2-polarization of TAMs; increased tumor metastasis and migration | [150] |
| TGF-β | miR-491 | Colorectal Cancer | CD8+ T cells | CDK4, TCF-1, Bcl-xL | ↓ | Decreased cell proliferation and IFN-γ production; increased apoptosis | [151] |
| TGF-β1 | miR-494 | Breast Cancer | MDSCs | PTEN | ↓ | Accumulation of MDSCs in tumor tissues; Akt activation; tumor growth and metastasis | [152] |
| TGF-β | miR-21 | Colorectal Cancer | CAFs | α−SMAD | ↑ | Differentiation to myofibroblasts; increased tumor cell proliferation | [153] |
| TGF-β | miR-143 | Gastric Cancer | CAFs | Collagen type III | ↑ | Promotion of tumorigenesis | [154] |
| GM-CSF | miR-200c | Bone marrow exposed to several cancers | MDSCs | PTEN, FOG2 | ↓ | Expansion of MDSCs in tumor tissues and suppression of immune system; PI3K/Akt, STAT3 activation | [155] |
| miRNA | Donor | Recipient | Targeted Molecule | Target Expression | Effect | Ref. |
|---|---|---|---|---|---|---|
| miR-24-3p, miR-891a, miR-106a-5p, miR-20a-5p, miR-1908 | NPCs | Th1 Cells | MARK1 | ↑ | Increase of the pro-inflammatory cytokines IL-1β, IL-6, and IL-10; reduction of INF-γ, IL-2, and IL-17; inhibition of T-cell proliferation and Th1, and Th17 differentiation; promotion of Treg proliferation | [174] |
| miR-214 | Lung Cancer | Treg Cells | PTEN | ↓ | Tregs expansion; increase of IL-10; tumor growth | [175] |
| miR-23a | Leukemia, Lung Cancer | NKs | INF-γ, CD107a | ↓ | Decrease of immunosuppressive action by NK cells | [176] |
| miR-212-3p | Pancreatic Cancer | DCs | RFXAP | ↓ | Decreased expression of MHC-II; induction of DCs immune tolerance | [177] |
| miR-203 | Pancreatic Cancer | DCs | TLR4 | ↓ | Decrease of TNF-α and IL-12 production | [178] |
| miR-145 | Colorectal Cancer | TAMs | HDAC11, TLR4 | ↓ | Promotion of TMAs polarization to M2-like macrophages; promotion of tumor progression and enlargement of tumor volume | [179] |
| miR-223 | TAMs | Breast Cancer | Mef2c-β-catenin | ↑ | Promotion of tumor invasion and aggressiveness | [180] |
| miR-210-3p (miR-210) | Liver Cancer | CAEs | SMAD4, STAT6 | ↓ | Enhanced angiogenesis; tubulogenesis promotion of endothelial cells | [181] |
| miR-21 | Lung Cancer | Endothelial Cells | STAT3 | ↑ | Enhanced VEGF expression and angiogenesis | [182] |
| miR-155 | Breast Cancer | Adipocytes | PPARγ | ↓ | Promotion of beige/brown adipocyte differentiation and remodelling of metabolism | [183] |
| miR-126a | Breast Cancer | Adipocytes | IRS1 | ↓ | Increased catabolism and release of metabolites and HIF1α expression in adipocytes | [184] |
| miR-144 | Breast Cancer | Adipocytes | MAP3K8/ ERK1/2/ PPARγ | ↓ | Promotion of beige/brown adipocyte differentiation | [184] |
| miR-145 | ASCs | Prostate Cancer | Bcl-xL | ↓ | Promotion of apoptosis; decrease of tumor growth | [186] |
| miR-503-3pf | ASCs | Liver Cancer | Pluripotency genes (e.g., Nanog) | ↓ | Smaller tumor volume and cancer stemness-attenuation | [187] |
| miR-140 | Adipocytes | DCIS | SOX9 | ↓ | Inhibition of tumor invasion following antitumor Shikonin treatment | [188] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conti, I.; Varano, G.; Simioni, C.; Laface, I.; Milani, D.; Rimondi, E.; Neri, L.M. miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells 2020, 9, 220. https://doi.org/10.3390/cells9010220
Conti I, Varano G, Simioni C, Laface I, Milani D, Rimondi E, Neri LM. miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells. 2020; 9(1):220. https://doi.org/10.3390/cells9010220
Chicago/Turabian StyleConti, Ilaria, Gabriele Varano, Carolina Simioni, Ilaria Laface, Daniela Milani, Erika Rimondi, and Luca M. Neri. 2020. "miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment" Cells 9, no. 1: 220. https://doi.org/10.3390/cells9010220
APA StyleConti, I., Varano, G., Simioni, C., Laface, I., Milani, D., Rimondi, E., & Neri, L. M. (2020). miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells, 9(1), 220. https://doi.org/10.3390/cells9010220