Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunofluorescence Image Data Sets of EpCAM-Enriched Cells and Extracellular Vesicles of 25 Healthy Individuals and 75 Metastatic Cancer Patients
2.2. Blood Samples of 10 Healthy Individuals
2.3. Immunofluorescence Imaging of Cells and Extracellular Vesicles in Whole Blood Samples
2.4. Automated Enumeration of Objects in Immunofluorescence Images Using the Open-Source ACCEPT Software
3. Results
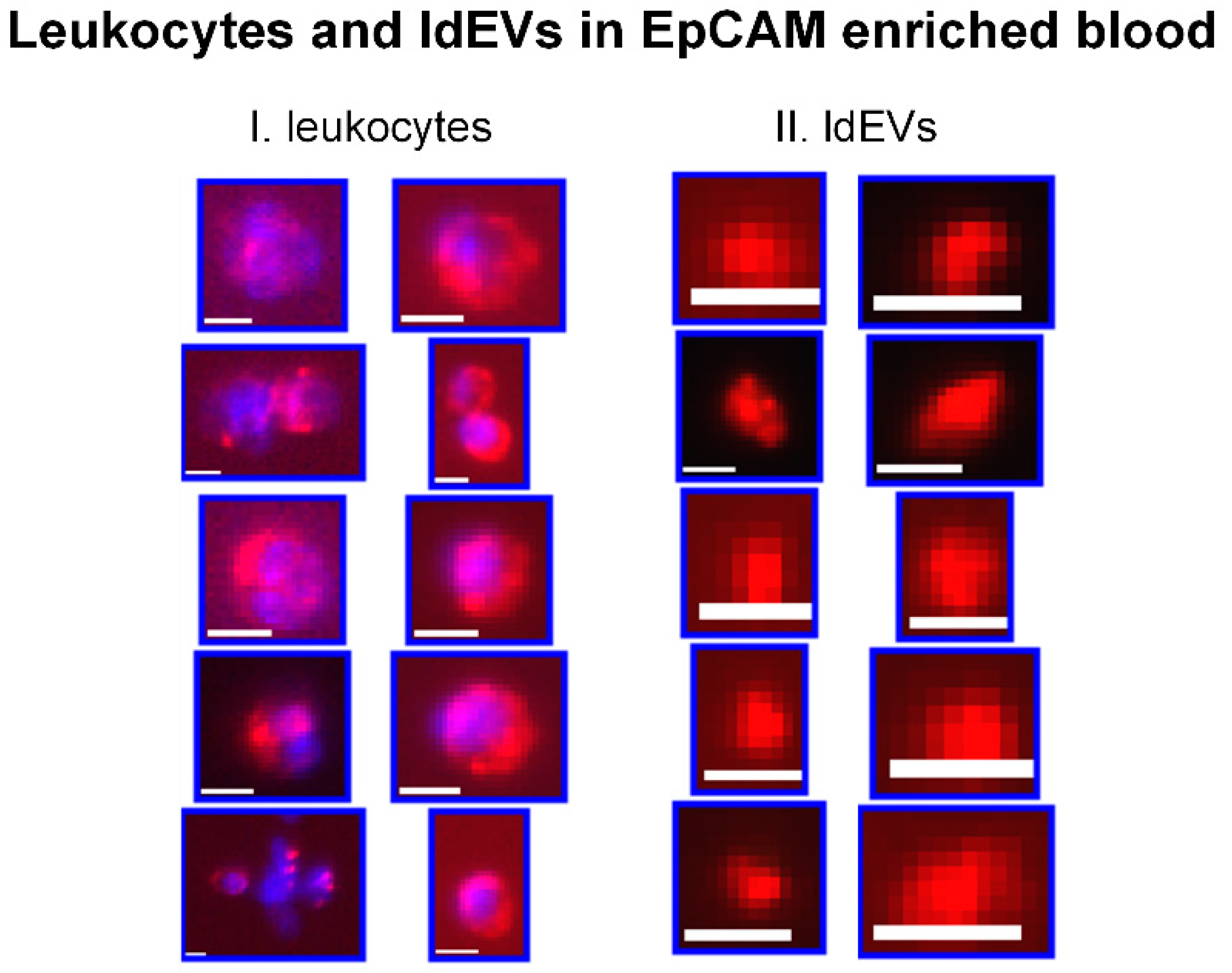
3.1. Detection of ldEVs in EpCAM-Enriched Blood Samples of Healthy Individuals and Metastatic Cancer Patients
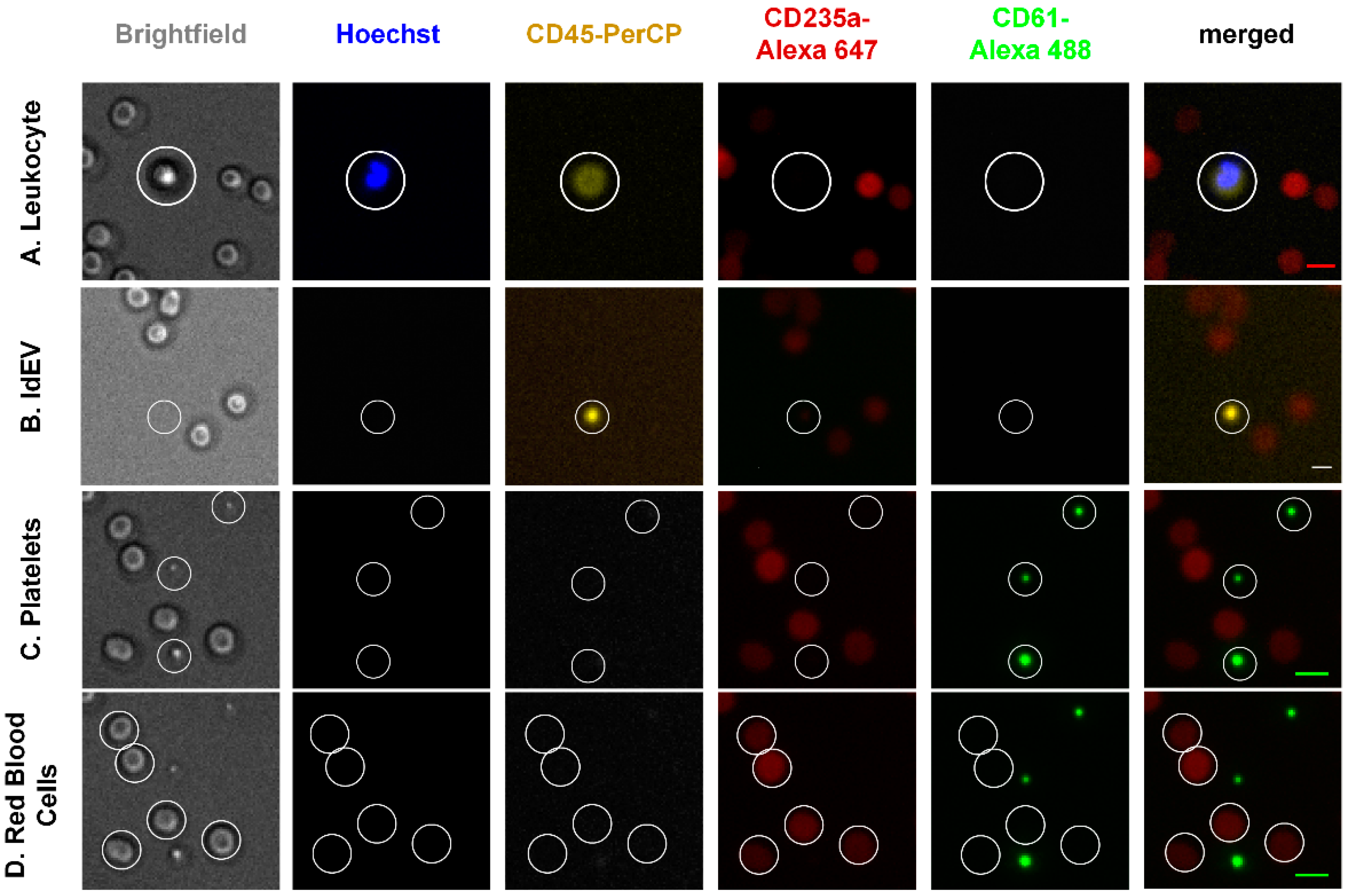
3.2. Detection of Cell and Extracellular Vesicle Classes in the Blood Of Healthy Individuals without EpCAM Enrichment
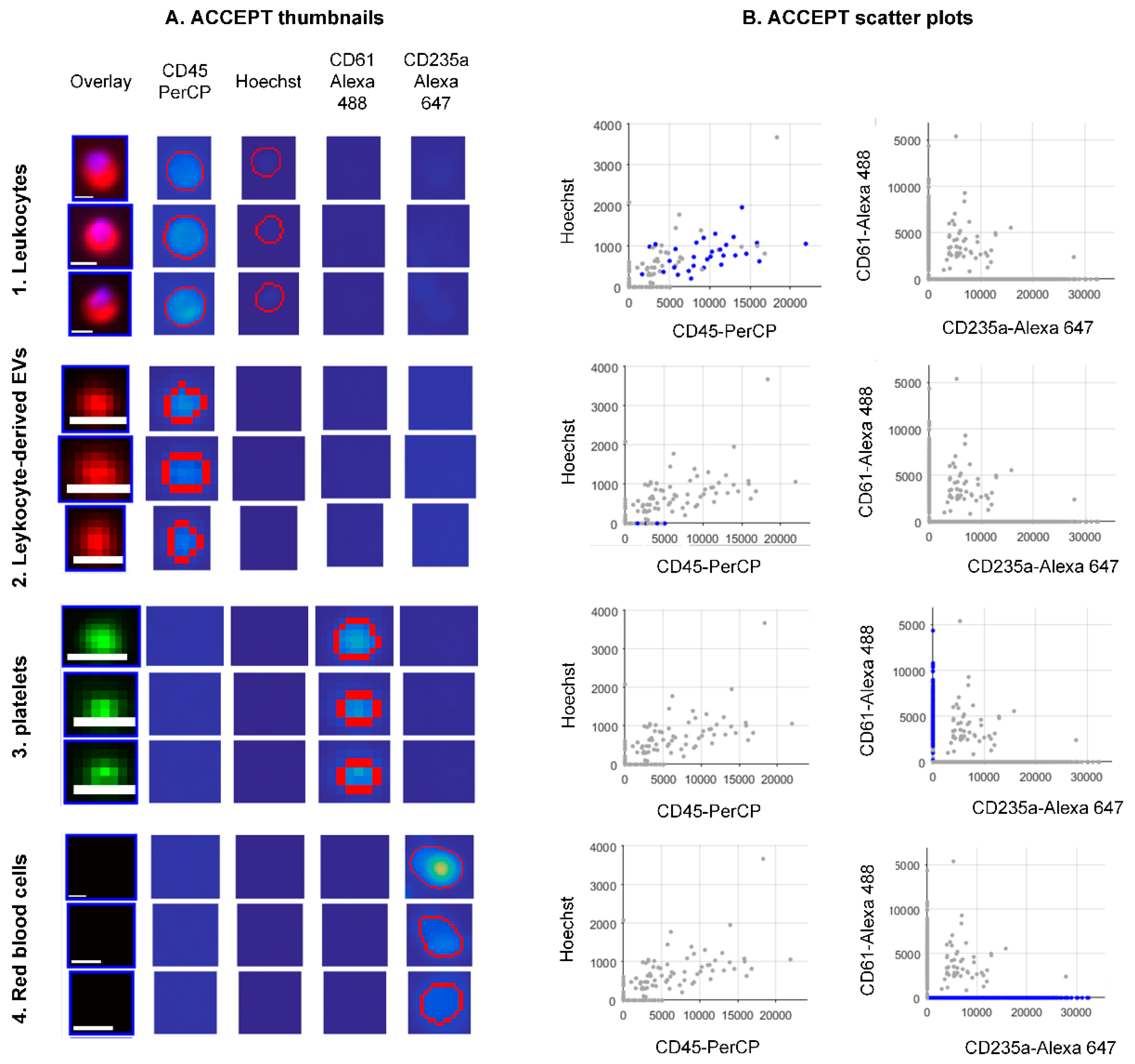
3.3. ACCEPT Gates for the Automated Enumeration of Different Classes in the Blood with and without EpCAM Enrichment.
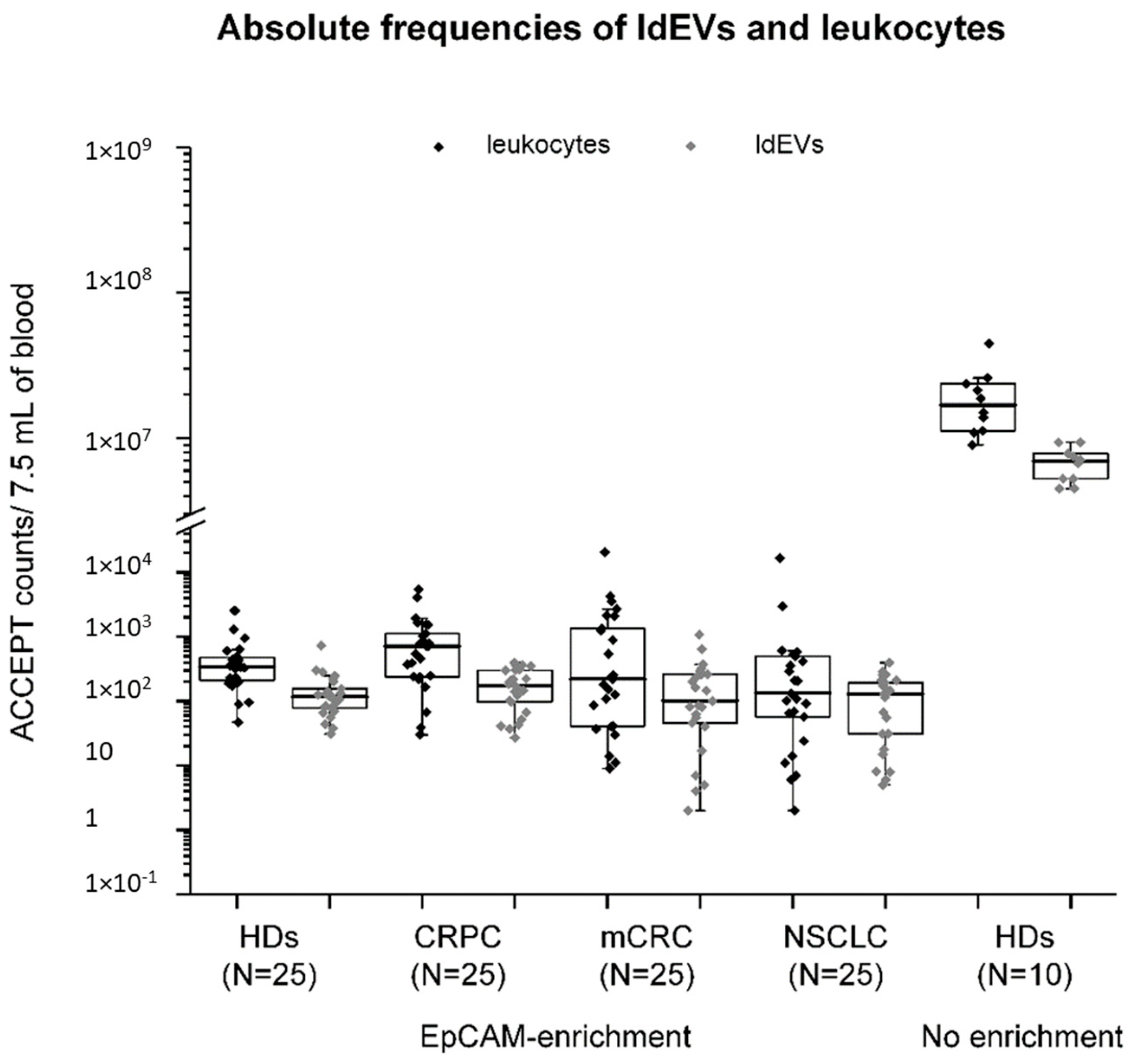
3.4. Absolute and Relative Frequencies of Leukocytes and ldEVs in 7.5 mL of EpCA- Enriched Blood of Healthy Individuals and Metastatic Cancer Patients.
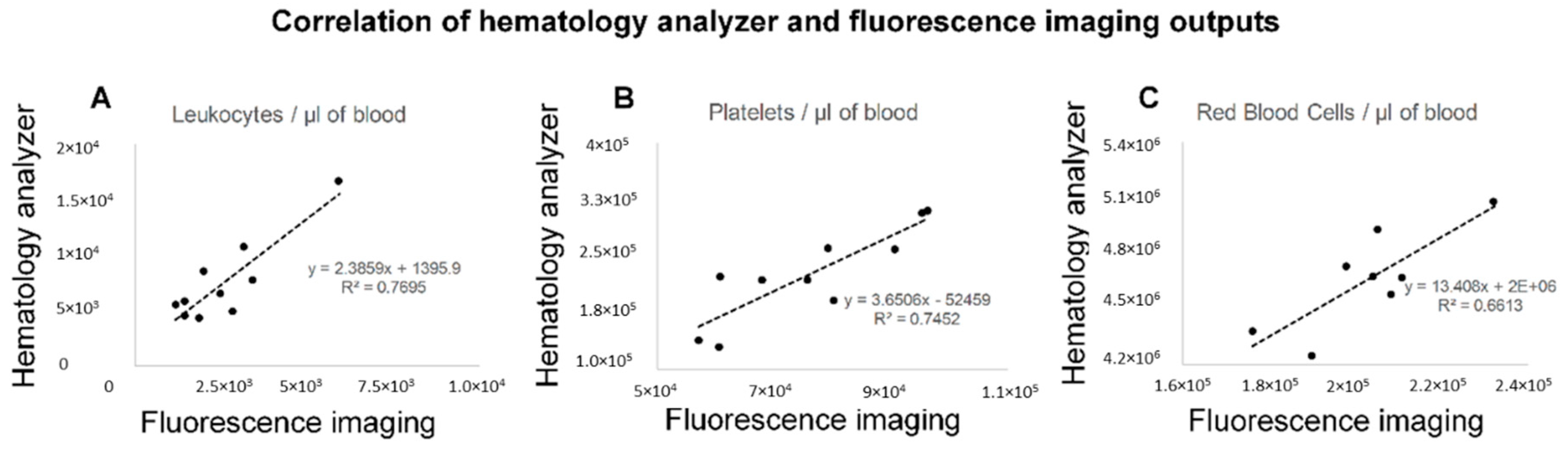
3.5. The Reproducibility of Measurements by Fluorescence Imaging and the Correlation with the Frequencies of Blood Cells by Hematology Analyzer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Niel, G.D.; Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell. Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Fais, S.O.; Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; Cordeiro da Silva, A.; Del Portillo, H.; El Andaloussi, S. Evidence-based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano 2016, 10, 3886–3899. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Jia, S.; Zhang, R.; Li, Z.; Li, J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 2017, 8, 55632–55645. [Google Scholar] [CrossRef] [Green Version]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 2143. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Yoshioka, Y.; Fujita, Y.; Ochiya, T. Versatile roles of extracellular vesicles in cancer. J. Clin. Investig. 2016, 126, 1163–1172. [Google Scholar] [CrossRef] [Green Version]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De S Posadas, E.M. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [Green Version]
- Ricklefs, F.L.; Maire, C.L.; Reimer, R.; Duhrsen, L.; Kolbe, K.; Holz, M.; Schneider, E.; Rissiek, A.; Babayan, A.; Hille, C.; et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J. Extracell. Vesicles. 2019, 8, 1588555. [Google Scholar] [CrossRef] [Green Version]
- Konig, L.; Kasimir-Bauer, S.; Bittner, A.K.; Hoffmann, O.; Wagner, B.; Santos Manvailer, L.F.; Kimmig, R.; Horn, P.A.; Rebmann, V. Elevated levels of extracellular vesicles are associated with therapy failure and disease progression in breast cancer patients undergoing neoadjuvant chemotherapy. Oncoimmunology 2017, 7, e1376153. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Slomka, A.; Urban, S.K.; Lukacs-Kornek, V.; Zekanowska, E.; Kornek, M. Large Extracellular Vesicles: Have We Found the Holy Grail of Inflammation? Front. Immunol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Padda, R.S.; Deng, F.K.; Brett, S.I.; Biggs, C.N.; Durfee, P.N.; Brinker, C.J.; Williams, K.C.; Leong, H.S. Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma. Prostate 2019, 79, 592–603. [Google Scholar] [CrossRef]
- Reategui, E.; van der Vos, K.E.; Lai, C.P.; Zeinali, M.; Atai, N.A.; Aldikacti, B.; Floyd, F.P.A.H.K., Jr.; Thapar, V.; Hochberg, F.H.; Sequist, L.V.; et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018, 9, 175. [Google Scholar] [CrossRef]
- Nanou, A.; Coumans, F.A.W.; van Dalum, G.; Zeune, L.L.; Dolling, D.; Onstenk, W.; Crespo, M.; Fontes, M.S.; Rescigno, P.; Fowler, G.; et al. Circulating tumor cells, tumor-derived extracellular vesicles and plasma cytokeratins in castration-resistant prostate cancer patients. Oncotarget 2018, 9, 19283–19293. [Google Scholar] [CrossRef] [Green Version]
- Nanou, A.; Zeune, L.L.; de Wit, S.; Miller, C.M.; Punt, C.J.A.; Groen, H.J.M.; Hayes, D.F.; de Bono, J.S.L.W.M.M.T. Tumor-Derived Extracellular Vesicles in Blood of Metastatic Breast, Colorectal, Prostate and Non-Small Cell Lung Cancer Patients Associate with Worse Survival; American Association for Cancer Research: Philadelphia, PA, USA, 2019. [Google Scholar]
- Zeune, L.; van Dalum, G.; Terstappen, L.W.M.M.; van Gils, S.A.; Brune, C. Multiscale Segmentation via Bregman Distances and Nonlinear Spectral Analysis. SIAM J. Imaging Sci. 2017, 10, 111–146. [Google Scholar] [CrossRef] [Green Version]
- Zeune, L. Automated CTC Classification, Enumeration and Pheno Typing: Where Marh Meets Biology. Medical Cell BioPhysics; University of Twente: Overijssel, The Netherland, 2019. [Google Scholar]
- Coumans, F.; Terstappen, L. Detection and Characterizati of Circulating Tumor Cells by the CellSearch Approach. Methods Mol. Biol. 2015, 1347, 263–278. [Google Scholar]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Tibbe, A.G.; de Grooth, B.G.; Greve, J.; Dolan, G.J.; Rao, C.; Terstappen, L.W. Magnetic field design for selecting and aligning immunomagnetic labeled cells. Cytometry 2002, 47, 163–172. [Google Scholar] [CrossRef]
- De Wit, S.; Zeune, L.L.; Hiltermann, T.J.N.; Groen, H.J.M.; Dalum, G.V.; Terstappen, L.W.M.M. Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis. Cancers 2018, 10, 377. [Google Scholar] [CrossRef]
- Zeune, L.L.; de Wit, S.; Berghuis, A.M.S.M.J.I.J.; Terstappen, L.; Brune, C. How to Agree on a CTC: Evaluating the Consensus in Circulating Tumor Cell Scoring. Cytom. A 2018, 93, 1202–1206. [Google Scholar] [CrossRef]
- De Wit, S.; Rossi, E.; Weber, S.; Tamminga, M.; Manicone, M.; Swennenhuis, J.F. Groothuis-Oudshoorn, C.G.M.; Vidotto, R.; Facchinetti, A.; Zeune, L.L.; et al. Single tube liquid biopsy for advanced non-small cell lung cancer. Int. J. Cancer 2019, 144, 3127–3137. [Google Scholar] [CrossRef]
- Van der Pol, E.; Boing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar]
- Pitanga, T.N.; de Aragao Franca, L.; Rocha, V.C.; Meirelles, T.; Borges, V.M.; Goncalves, M.S.; Pontes-de-Carvalho, L.C.; Noronha-Dutra, A.A.; dos-Santos, W.L. Neutrophil-derived microparticles induce myeloperoxidase-mediated damage of vascular endothelial cells. BMC Cell Biol. 2014, 15, 21. [Google Scholar] [CrossRef]
- Baka, Z.; Senolt, L.; Vencovsky, J.; Mann, H.; Simon, P.S.; Kittel, A.; Buzas, E.; Nagy, G. Increased serum concentration of immune cell derived microparticles in polymyositis/dermatomyositis. Immunol. Lett. 2010, 128, 124–130. [Google Scholar] [CrossRef]
- Canellini, G.; Rubin, O.; Delobel, J.; Crettaz, D.; Lion, N.; Tissot, J.D. Red blood cell microparticles and blood group antigens: An analysis by flow cytometry. Blood Transfus. 2012, 10, s39–s45. [Google Scholar]
- Wheway, J.; Latham, S.L.; Combes, V.; Grau, G.E.R. Endothelial microparticles interact with and support the proliferation of T cells. J. Immunol. 2014, 193, 3378–3387. [Google Scholar] [CrossRef]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Lo Iacono, M.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef]
- Ruhen, O.; Meehan, K. Tumor-Derived Extracellular Vesicles as a Novel Source of Protein Biomarkers for Cancer Diagnosis and Monitoring. Proteomics 2019, 19, e1800155. [Google Scholar] [CrossRef]
- Nanou, A.; Crespo, M.; Flohr, P.; De Bono, J.S.; Terstappen, L. Scanning Electron Microscopy of Circulating Tumor Cells and Tumor-Derived Extracellular Vesicles. Cancers 2018, 10, 416. [Google Scholar] [CrossRef]
- Rikkert, L.G.; van der Pol, E.; van Leeuwen, T.G.; Nieuwland, R.; Coumans, F.A.W. Centrifugation affects the purity of liquid biopsy-based tumor biomarkers. Cytometry A 2018, 93, 1207–1212. [Google Scholar] [CrossRef]
- Simak, J.; Holada, K.; Risitano, A.M.; Zivny, J.H.; Young, N.S.; Vostal, J.G. Elevated circulating endothelial membrane microparticles in paroxysmal nocturnal haemoglobinuria. Br. J. Haematol. 2004, 125, 804–813. [Google Scholar] [CrossRef]
- Lacroix, R.; Robert, S.; Poncelet, P.; Dignat-George, F. Overcoming limitations of microparticle measurement by flow cytometry. Semin. Thromb. Hemost. 2010, 36, 807–818. [Google Scholar] [CrossRef]
- Van der Pol, E.; Coumans, F.A.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef]
- Gool, E.L.; Stojanovic, I.; Schasfoort, R.B.M.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R.; Terstappen, L.; Coumans, F.A.W. Surface Plasmon Resonance is an Analytically Sensitive Method for Antigen Profiling of Extracellular Vesicles. Clin. Chem. 2017, 63, 1633–1641. [Google Scholar] [CrossRef] [Green Version]
- Halim, A.T.A.; Ariffin, N.A.F.M.; Azlan, M. Review: The Multiple Roles of Monocytic Microparticles. Inflammation 2016, 39, 1277–1284. [Google Scholar] [CrossRef]
- Tissot, J.D.; Canellini, G.; Rubin, O.; Angelillo-Scherrer, A.; Delobel, J.; Prudent, M.; Lion, N. Blood microvesicles: From proteomics to physiology. Transl. Proteom. 2013, 1, 38–52. [Google Scholar] [CrossRef] [Green Version]
- Burnier, L.; Fontana, P.; Kwak, B.R.; Angelillo-Scherrer, A. Cell-derived microparticles in haemostasis and vascular medicine. Thromb. Haemost. 2009, 101, 439–451. [Google Scholar] [CrossRef]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodebski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Ullal, A.J.; Pisetsky, D.S.; Reich, C.F., III. Use of SYTO 13, a fluorescent dye binding nucleic acids, for the detection of microparticles in in vitro systems. Cytom. Part A J. Int. Soc. Anal. Cytol. 2010, 7, 294–301. [Google Scholar] [CrossRef] [PubMed]
- McCracken, J.M.; Allen, L.A. Regulation of human neutrophil apoptosis and lifespan in health and disease. J. Cell Death 2014, 7, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef] [PubMed]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Cameron, M.D.; Schmidt, E.E.; Kerkvliet, N.; Nadkarni, K.V.; Morris, V.L.; Groom, A.C.; Chambers, A.F.; MacDonald, I.C. Temporal progression of metastasis in lung: Cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000, 60, 2541–2546. [Google Scholar] [PubMed]
- Li, Q.S.; Lee, G.Y.H.; Ong, C.N.; Lim, C.T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef]
- Zhang, W.; Kai, K.; Choi, D.S.; Iwamoto, T.; Nguyen, Y.H.; Wong, H.; Landis, M.D.; Ueno, N.T.; Chang, J.; Qin, L. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2012, 109, 18707–18712. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef]
- Gossett, D.R.; Tse, H.T.K.; Lee, S.A.; Ying, Y.; Lindgren, A.G.; Yang, O.O.; Rao, J.; Clark, A.T.; Di Carlo, D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. USA 2012, 109, 7630–7635. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Luo, T.; Ren, Y.; Florey, O.; Shirasawa, S.; Sasazuki, T.; Robinson, D.N.; Overholtzer, M. Competition between human cells by entosis. Cell Res. 2014, 24, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Choi, W.; Shah, J.B.; Lee, E.K.; Willis, D.L.; Kamat, A.M. Blebbishields, the emergency program for cancer stem cells: Sphere formation and tumorigenesis after apoptosis. Cell Death Differ. 2013, 20, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Kim, J.; Hager, M.H.; Morello, M.; Yang, W.; Lafargue, C.J.; True, L.D.; Rubin, M.A.; Adam, R.M.; Beroukhim, R.; et al. Oncosome formation in prostate cancer: Association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009, 69, 5601–5609. [Google Scholar] [CrossRef] [PubMed]
- Reis-Sobreiro, M.; Chen, J.F.; Novitskaya, T.; You, S.; Morley, S.; Steadman, K.; Gill, N.K.; Eskaros, A.; Rotinen, M.; Chu, C.Y.; et al. Emerin Deregulation Links Nuclear Shape Instability to Metastatic Potential. Cancer Res. 2018, 78, 6086–6097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| A. EpCAM Enrichment | B. No Enrichment | ||||
|---|---|---|---|---|---|
| Leukocytes | DAPI/Hoechst a | Mean Intensity | >30 | Mean Intensity | >30 |
| Max Intensity | >50 | Max Intensity | >50 | ||
| Size | >16 μm2 | Size | >16 μm2 | ||
| CD45 | Mean Intensity | >30 | Mean Intensity | >30 | |
| Max Intensity | >50 | Max Intensity | >50 | ||
| Size | ≤400 μm2 | ||||
| CK | Standard Deviation | ≤5 | n/a b | ||
| CD61 | n/a b | Standard Deviation | ≤5 | ||
| CD235a | n/a b | Standard Deviation | ≤5 | ||
| Extra channel | Standard Deviation | ≤5 | Standard Deviation | ≤5 | |
| ldEVs | DAPI/Hoechst a | Standard Deviation | ≤5 | Standard Deviation | ≤5 |
| CD45 | Mean Intensity | >30 | Mean Intensity | >30 | |
| Max Intensity | >50 | Max Intensity | >50 | ||
| Perimeter | >5 pixels | Perimeter | >5 pixels | ||
| Size | ≤150 | Size | ≤150 μm2 | ||
| Eccentricity | ≤0.85 | Eccentricity | ≤0.85 | ||
| CK | Standard Deviation | ≤5 | n/a b | ||
| CD61 | n/a b | Standard Deviation | ≤5 | ||
| CD235a | n/a b | Standard Deviation | ≤5 | ||
| Extra channel | Standard Deviation | ≤5 | Standard Deviation | ≤5 | |
| Platelets | CD45 | n/a b | Standard Deviation | ≤5 | |
| CD61 | Mean Intensity | >30 | |||
| Max Intensity | >50 | ||||
| Perimeter | >5 pixels | ||||
| Size | ≤150 μm2 | ||||
| Eccentricity | ≤0.85 | ||||
| CD235a | Standard Deviation | ≤5 | |||
| Standard Deviation | ≤5 | ||||
| Extra Channel | Standard Deviation | ≤5 | |||
| Red blood cells | Hoechst | n/a b | Standard Deviation | ≤5 | |
| CD45 | Standard Deviation | ≤5 | |||
| CD61 | Standard Deviation | ≤5 | |||
| CD235a | Mean Intensity | >30 | |||
| Max Intensity | >50 | ||||
| Perimeter | >5 pixels | ||||
| Extra Channel | Standard Deviation | ≤5 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanou, A.; Zeune, L.L.; Terstappen, L.W.M.M. Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment. Cells 2019, 8, 937. https://doi.org/10.3390/cells8080937
Nanou A, Zeune LL, Terstappen LWMM. Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment. Cells. 2019; 8(8):937. https://doi.org/10.3390/cells8080937
Chicago/Turabian StyleNanou, Afroditi, Leonie L. Zeune, and Leon W.M.M. Terstappen. 2019. "Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment" Cells 8, no. 8: 937. https://doi.org/10.3390/cells8080937
APA StyleNanou, A., Zeune, L. L., & Terstappen, L. W. M. M. (2019). Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment. Cells, 8(8), 937. https://doi.org/10.3390/cells8080937






