Interplay between Phosphatases and the Anaphase-Promoting Complex/Cyclosome in Mitosis
Abstract
:1. Introduction
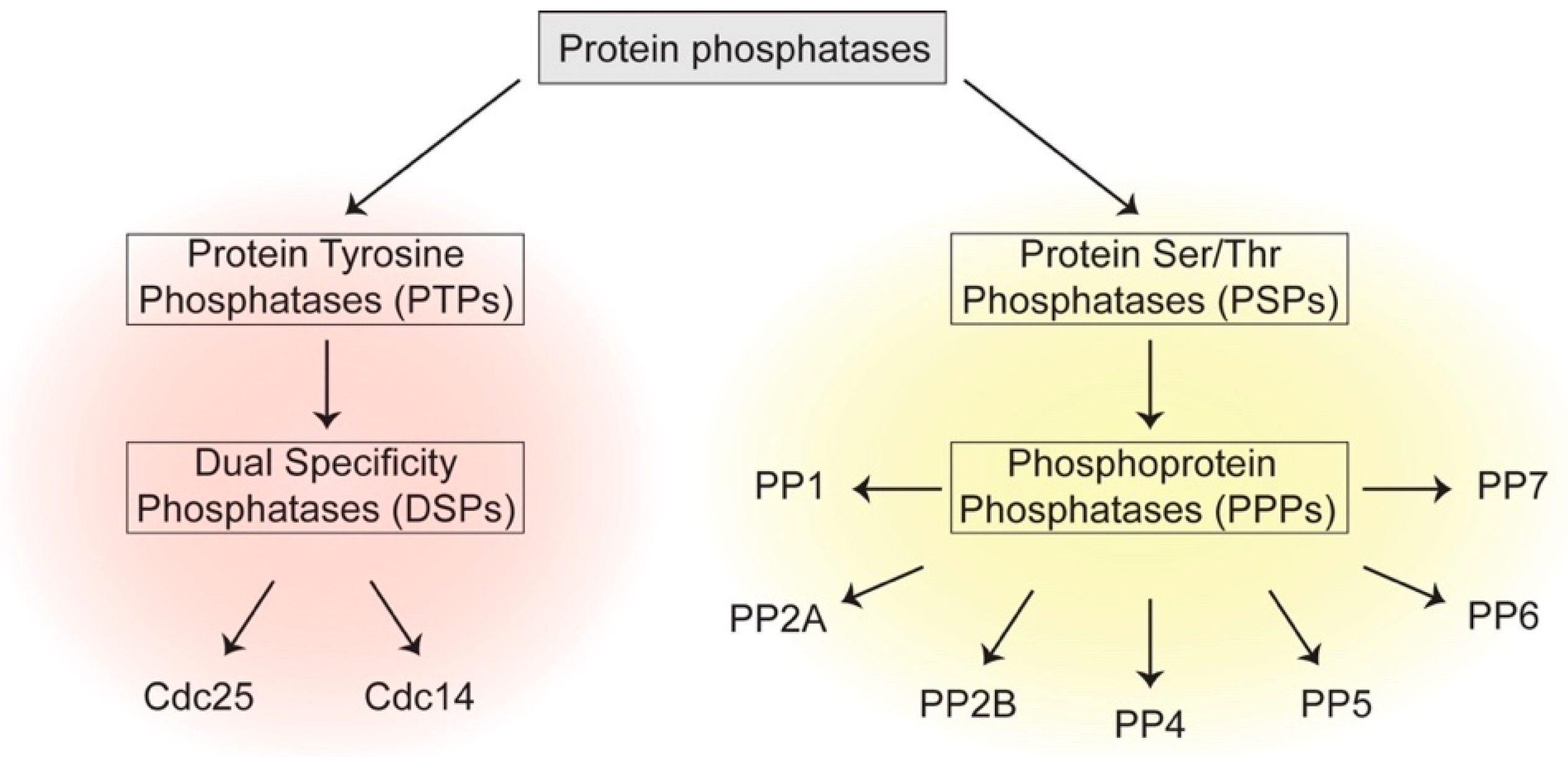
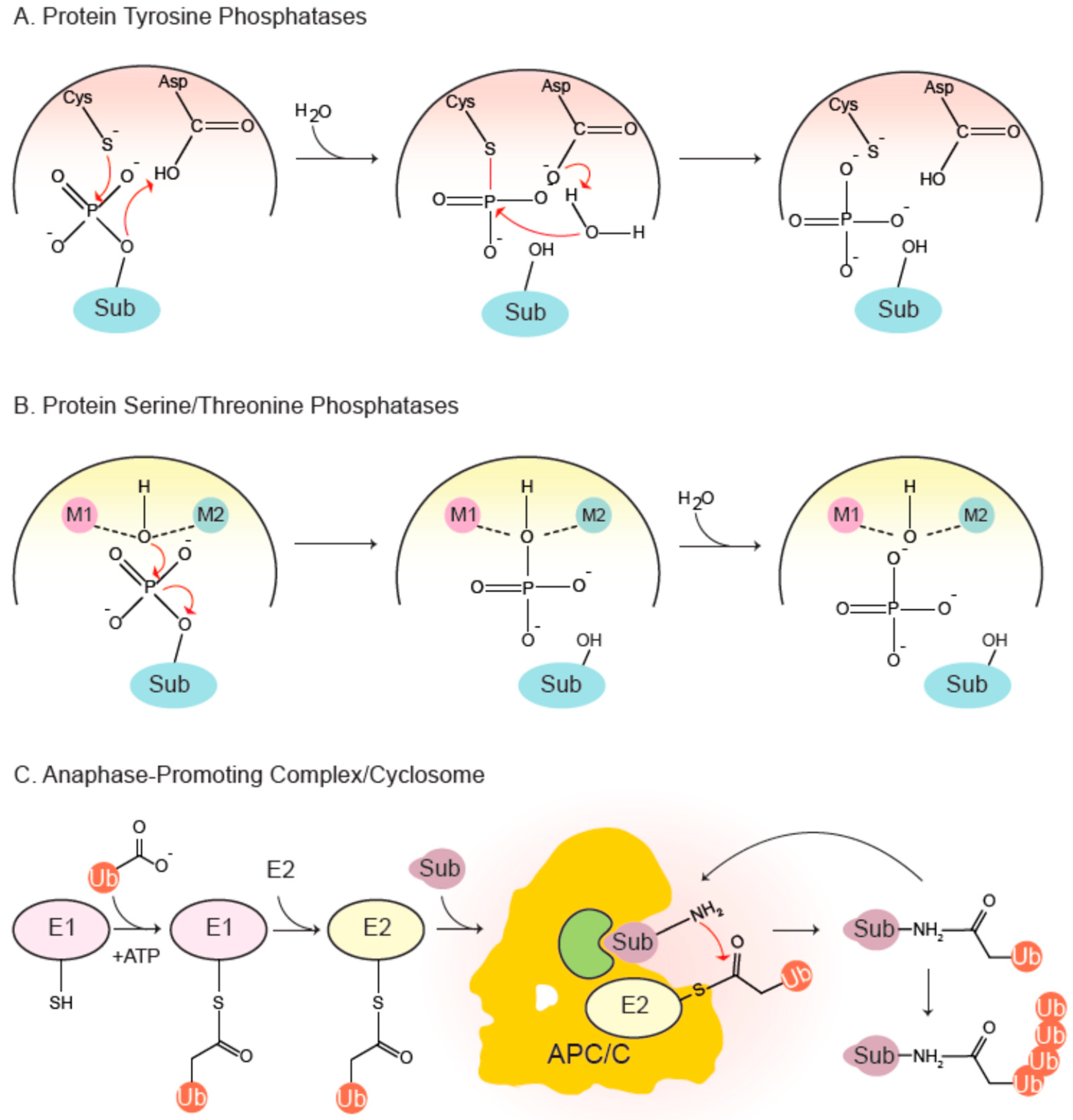
1.1. Protein Phosphatases—Ubiquitous Enzymes that Regulate Cell Physiology and Division
1.2. The Multiprotein Complex APC/C is a Master Regulator of the Cell Cycle
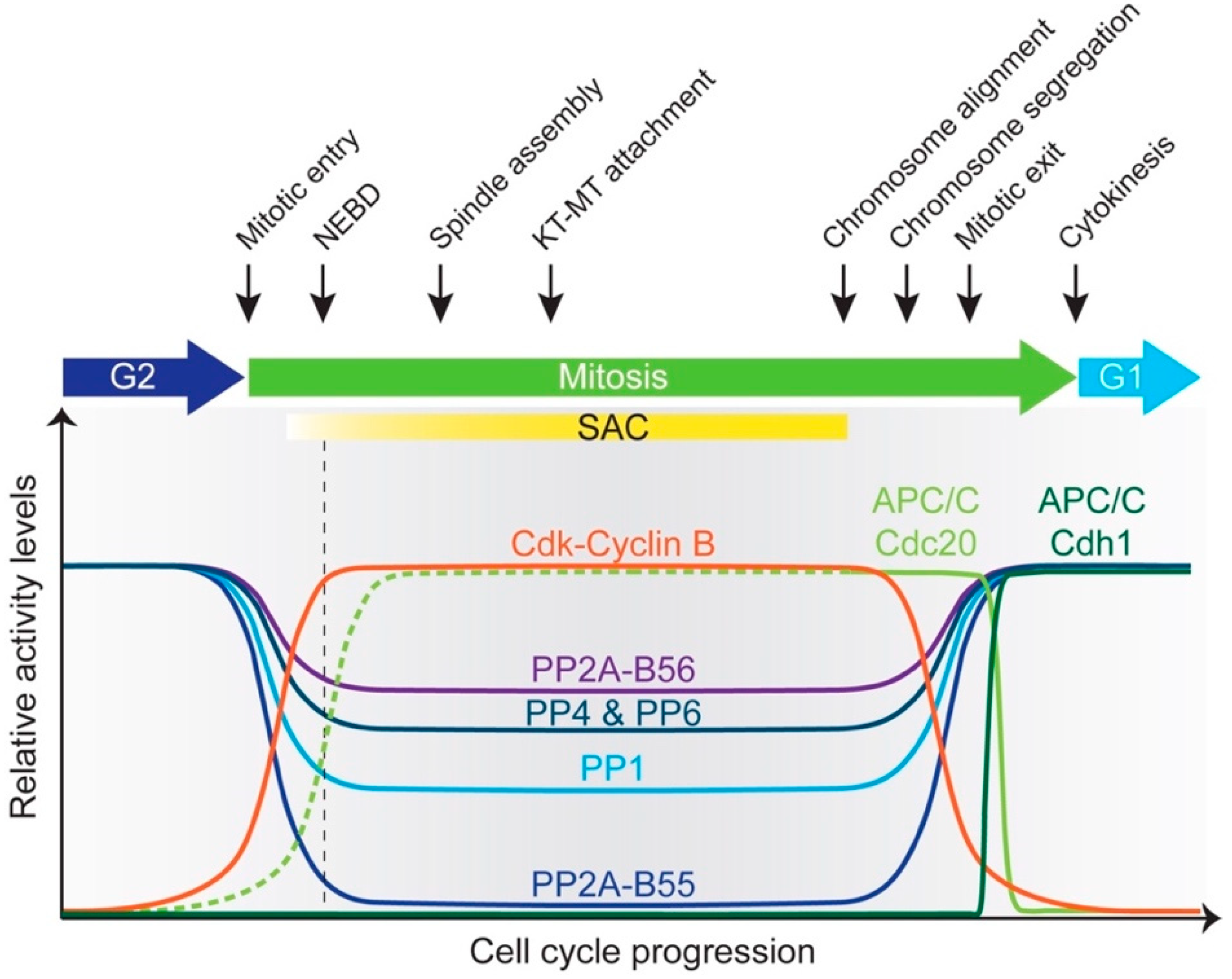
2. G2/M Transition
2.1. Cdc25 Promotes Cdk1-Cyclin B Activation and Mitotic Entry
2.2. Phosphatases are Downregulated at Mitotic Entry
3. Building a Metaphase Cell
3.1. PP1 and PP2A Phosphatases Regulate Kinetochore Capture by Microtubules
3.2. The Spindle Assembly Checkpoint Inhibits APC/C Activity in Metaphase and is Regulated by Phosphatases
3.3. Constructing and Segregating Mitotic Chromosomes
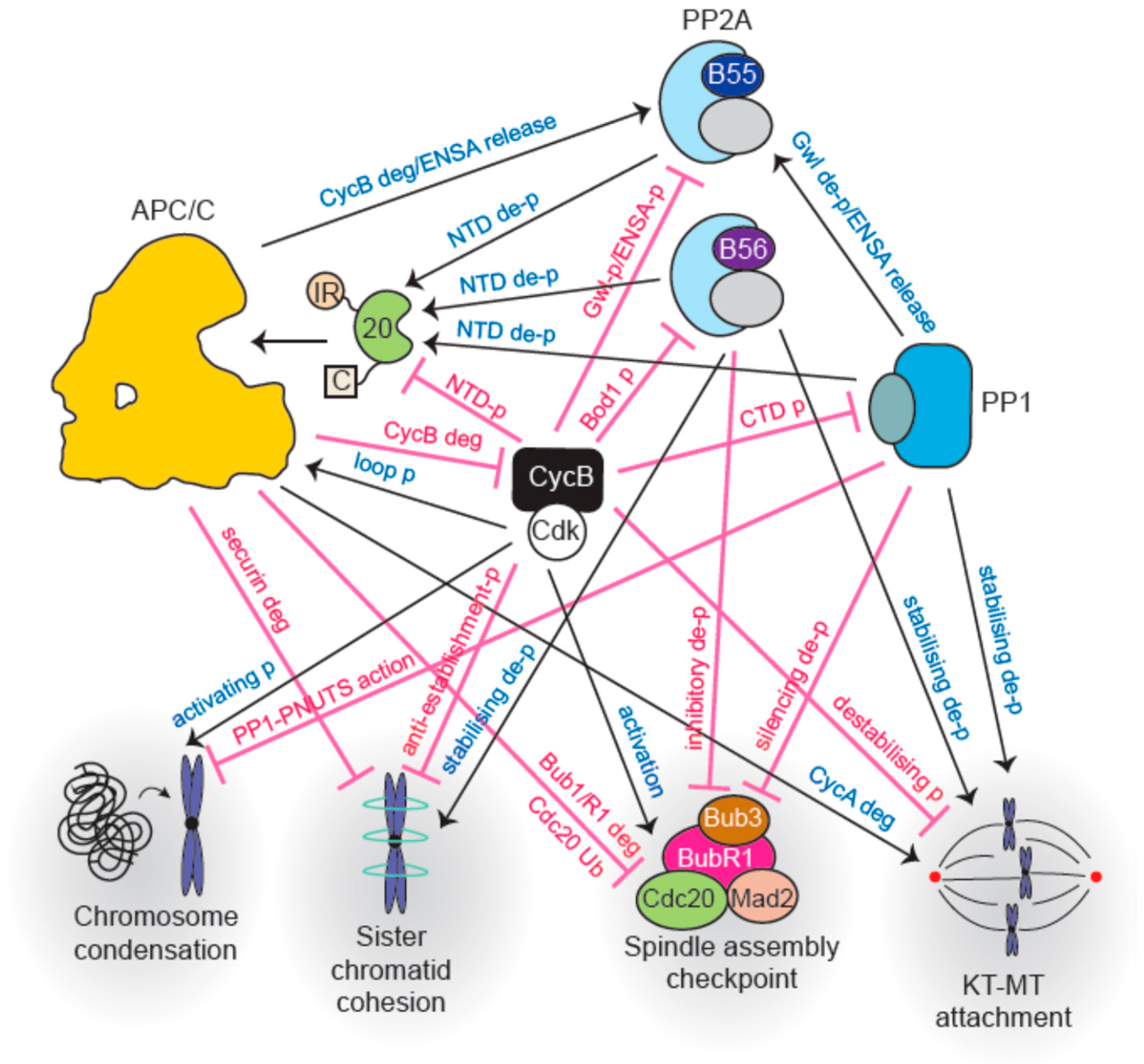
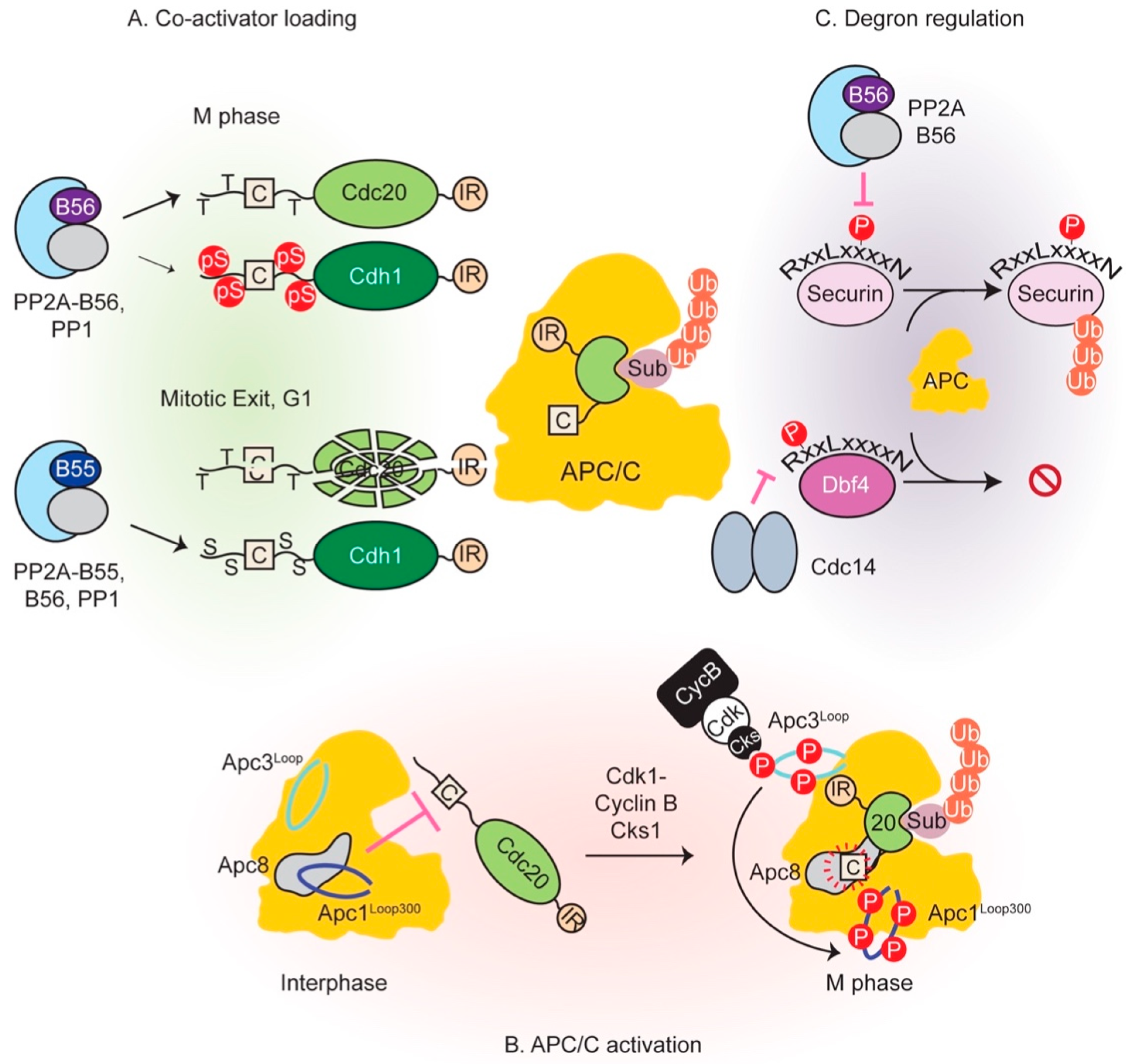
4. Intricate Interplay between Phosphatases and the APC/C Regulates Metaphase-Anaphase Transition and Mitotic Exit
4.1. Phosphorylation of APC/C Enables Cdc20 Binding
4.2. Co-Activator Dephosphorylation is Essential for APC/C Interaction
4.3. Multiple Mechanisms Regulate Degron-Mediated Substrate Degradation
5. Phosphatases Escort Cells through Exit from Mitosis
6. Roles in Diseases and Avenues for Therapies
7. Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Cori, G.T.; Green, A.A. Crystalline muscle phosphorylase: II. Prosthetic group. J. Biol. Chem. 1943, 151, 31–38. [Google Scholar]
- Fischer, E.H.; Krebs, E.G. Conversion of phosphorylase b to phosphorylase a in muscle extracts. J. Biol. Chem. 1955, 216, 121–132. [Google Scholar]
- Krebs, E.G.; Fischer, E.H. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim. Biophys. Acta 1956, 20, 150–157. [Google Scholar] [CrossRef]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833. [Google Scholar] [CrossRef]
- Shi, Y. Serine/Threonine Phosphatases: Mechanism through Structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10, eaag1796. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishito, Y.; Usui, H.; Shinzawa-Itoh, K.; Inoue, R.; Tanabe, O.; Nagase, T.; Murakami, T.; Takeda, M. Direct metal analyses of Mn2+-dependent and -independent protein phosphatase 2A from human erythrocytes detect zinc and iron only in the Mn2+-independent one. FEBS Lett. 1999, 447, 29–33. [Google Scholar] [CrossRef]
- Heroes, E.; Rip, J.; Beullens, M.; Van Meervelt, L.; De Gendt, S.; Bollen, M. Metals in the active site of native protein phosphatase-1. J. Inorg. Biochem. 2015, 149, 1–5. [Google Scholar] [CrossRef]
- Barford, D.; Das, A.K.; Egloff, M.-P. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 133–164. [Google Scholar] [CrossRef]
- Heroes, E.; Lesage, B.; Görnemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595. [Google Scholar] [CrossRef]
- Peti, W.; Nairn, A.C.; Page, R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013, 280, 596–611. [Google Scholar] [CrossRef]
- Weith, M.; Seiler, J.; van den Boom, J.; Kracht, M.; Hülsmann, J.; Primorac, I.; del Pino Garcia, J.; Kaschani, F.; Kaiser, M.; Musacchio, A.; et al. Ubiquitin-Independent Disassembly by a p97 AAA-ATPase Complex Drives PP1 Holoenzyme Formation. Mol. Cell 2018, 72, 766–777.e6. [Google Scholar] [CrossRef] [Green Version]
- Stone, E.M.; Yamano, H.; Kinoshita, N.; Yanagida, M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 1993, 3, 13–26. [Google Scholar] [CrossRef]
- Stern, B.; Nurse, P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996, 12, 345–350. [Google Scholar] [CrossRef]
- Coudreuse, D.; Nurse, P. Driving the cell cycle with a minimal CDK control network. Nature 2010, 468, 1074–1079. [Google Scholar] [CrossRef]
- Agostinis, P.; Goris, J.; Pinna, L.A.; Marchiori, F.; Perich, J.W.; Meyer, H.E.; Merlevede, W. Synthetic peptides as model substrates for the study of the specificity of the polycation-stimulated protein phosphatases. Eur. J. Biochem. 1990, 189, 235–241. [Google Scholar] [CrossRef]
- Godfrey, M.; Touati, S.A.; Kataria, M.; Jones, A.; Snijders, A.P.; Uhlmann, F. PP2ACdc55 Phosphatase Imposes Ordered Cell-Cycle Phosphorylation by Opposing Threonine Phosphorylation. Mol. Cell 2017, 65, 393–402.e3. [Google Scholar] [CrossRef]
- McCloy, R.A.; Parker, B.L.; Rogers, S.; Chaudhuri, R.; Gayevskiy, V.; Hoffman, N.J.; Ali, N.; Watkins, D.N.; Daly, R.J.; James, D.E.; et al. Global Phosphoproteomic Mapping of Early Mitotic Exit in Human Cells Identifies Novel Substrate Dephosphorylation Motifs. Mol. Cell. Proteom.: Mcp 2015, 14, 2194–2212. [Google Scholar] [CrossRef] [Green Version]
- Cundell, M.J.; Hutter, L.H.; Nunes Bastos, R.; Poser, E.; Holder, J.; Mohammed, S.; Novak, B.; Barr, F.A. A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J. Cell Biol. 2016, 214, 539–554. [Google Scholar] [CrossRef] [Green Version]
- Hein, J.B.; Hertz, E.P.T.; Garvanska, D.H.; Kruse, T.; Nilsson, J. Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat. Cell Biol. 2017. [Google Scholar] [CrossRef]
- Kataria, M.; Mouilleron, S.; Seo, M.-H.; Corbi-Verge, C.; Kim, P.M.; Uhlmann, F. A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase. Nat. Struct. Mol. Biol. 2018, 25, 1093–1102. [Google Scholar] [CrossRef]
- Pinna, L.A.; Donella, A.; Clari, G.; Moret, V. Preferential dephosphorylation of protein bound phosphorylthreonine and phosphorylserine residues by cytosol and mitochondrial “casein phosphatases”. Biochem. Biophys. Res. Commun. 1976, 70, 1308–1315. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Yu, T.; Yang, H.; Virshup, D.M.; Kops, G.J.; Lee, S.H.; Zhou, W.; Li, X.; Xu, W.; et al. Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein Cell 2016, 7, 516–526. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Bajaj, R.; Bollen, M.; Peti, W.; Page, R. Expanding the PP2A Interactome by Defining a B56-Specific SLiM. Structure 2016, 24, 2174–2181. [Google Scholar] [CrossRef] [Green Version]
- Kruse, T.; Zhang, G.; Larsen, M.S.; Lischetti, T.; Streicher, W.; Kragh Nielsen, T.; Bjorn, S.P.; Nilsson, J. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 2013, 126, 1086–1092. [Google Scholar] [CrossRef]
- Hertz, E.P.T.; Kruse, T.; Davey, N.E.; Lopez-Mendez, B.; Sigurethsson, J.O.; Montoya, G.; Olsen, J.V.; Nilsson, J. A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol. Cell 2016, 63, 686–695. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; D’Souza, R.C.; Tyanova, S.; Schaab, C.; Wisniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef]
- Touati, S.A.; Kataria, M.; Jones, A.W.; Snijders, A.P.; Uhlmann, F. Phosphoproteome dynamics during mitotic exit in budding yeast. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Ciehanover, A.; Hod, Y.; Hershko, A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978, 81, 1100–1105. [Google Scholar] [CrossRef]
- Ciechanover, A.; Finley, D.; Varshavsky, A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 1984, 37, 57–66. [Google Scholar] [CrossRef]
- Glotzer, M.; Murray, A.W.; Kirschner, M.W. Cyclin is degraded by the ubiquitin pathway. Nature 1991, 349, 132–138. [Google Scholar] [CrossRef]
- Holloway, S.L.; Glotzer, M.; King, R.W.; Murray, A.W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 1993, 73, 1393–1402. [Google Scholar] [CrossRef]
- Funabiki, H.; Yamano, H.; Kumada, K.; Nagao, K.; Hunt, T.; Yanagida, M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature 1996, 381, 438–441. [Google Scholar] [CrossRef]
- Cohen-Fix, O.; Peters, J.M.; Kirschner, M.W.; Koshland, D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996, 10, 3081–3093. [Google Scholar] [CrossRef]
- Uhlmann, F.; Lottspeich, F.; Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 1999, 400, 37–42. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Vlasschaert, C.; Cook, D.; Xia, X.; Gray, D.A. The evolution and functional diversification of the deubiquitinating enzyme superfamily. Genome Biol. Evol. 2017, 9, 558–573. [Google Scholar] [CrossRef]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Sudakin, V.; Ganoth, D.; Dahan, A.; Heller, H.; Hershko, J.; Luca, F.C.; Ruderman, J.V.; Hershko, A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 1995, 6, 185–197. [Google Scholar] [CrossRef]
- King, R.W.; Peters, J.-M.; Tugendreich, S.; Rolfe, M.; Hieter, P.; Kirschner, M.W. A 20s complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995, 81, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Irniger, S.; Piatti, S.; Michaelis, C.; Nasmyth, K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 1995, 81, 269–278. [Google Scholar] [CrossRef]
- Schwab, M.; Lutum, A.S.; Seufert, W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 1997, 90, 683–693. [Google Scholar] [CrossRef]
- Visintin, R.; Prinz, S.; Amon, A. CDC20 and CDH1: A Family of Substrate-Specific Activators of APC-Dependent Proteolysis. Science 1997, 278, 460–463. [Google Scholar] [CrossRef]
- Kraft, C.; Vodermaier, H.C.; Maurer-Stroh, S.; Eisenhaber, F.; Peters, J.M. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 2005, 18, 543–553. [Google Scholar] [CrossRef]
- Cooper, K.F.; Mallory, M.J.; Egeland, D.B.; Jarnik, M.; Strich, R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 14548–14553. [Google Scholar] [CrossRef] [Green Version]
- Blanco, M.A.; Pelloquin, L.; Moreno, S. Fission yeast mfr1 activates APC and coordinates meiotic nuclear division with sporulation. J. Cell Sci. 2001, 114, 2135–2143. [Google Scholar]
- Kimata, Y.; Kitamura, K.; Fenner, N.; Yamano, H. Mes1 controls the meiosis I to meiosis II transition by distinctly regulating the anaphase-promoting complex/cyclosome coactivators Fzr1/Mfr1 and Slp1 in fission yeast. Mol. Biol. Cell 2011, 22, 1486–1494. [Google Scholar] [CrossRef]
- Page, A.W.; Orr-Weaver, T.L. The Drosophila genes grauzone and cortex are necessary for proper female meiosis. J. Cell Sci. 1996, 109 Pt 7, 1707–1715. [Google Scholar]
- Van Voorhis, V.A.; Morgan, D.O. Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr. Biol. 2014, 24, 1556–1562. [Google Scholar] [CrossRef]
- Kimata, Y.; Baxter, J.E.; Fry, A.M.; Yamano, H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell 2008, 32, 576–583. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 1970, 66, 352–359. [Google Scholar] [CrossRef]
- Pfleger, C.M.; Kirschner, M.W. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000, 14, 655–665. [Google Scholar]
- Di Fiore, B.; Davey, N.E.; Hagting, A.; Izawa, D.; Mansfeld, J.; Gibson, T.J.; Pines, J. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell 2015, 32, 358–372. [Google Scholar] [CrossRef]
- Diaz-Martinez, L.A.; Tian, W.; Li, B.; Warrington, R.; Jia, L.; Brautigam, C.A.; Luo, X.; Yu, H. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 2015, 290, 2431–2443. [Google Scholar] [CrossRef]
- Davey, N.E.; Morgan, D.O. Building a Regulatory Network with Short Linear Sequence Motifs: Lessons from the Degrons of the Anaphase-Promoting Complex. Mol. Cell 2016, 64, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.F.; Zhang, Z.; Yang, J.; McLaughlin, S.H.; Barford, D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature 2014, 513, 388–393. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Zhang, Z.; Yang, J.; McLaughlin, S.H.; Barford, D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 2015, 522, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; VanderLinden, R.; Weissmann, F.; Qiao, R.; Dube, P.; Brown, N.G.; Haselbach, D.; Zhang, W.; Sidhu, S.S.; Peters, J.M.; et al. Cryo EM of Mitotic Checkpoint Complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 2016, 63, 593–607. [Google Scholar] [CrossRef]
- Frye, J.J.; Brown, N.G.; Petzold, G.; Watson, E.R.; Grace, C.R.; Nourse, A.; Jarvis, M.A.; Kriwacki, R.W.; Peters, J.M.; Stark, H.; et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat. Struct. Mol. Biol. 2013, 20, 827–835. [Google Scholar] [CrossRef]
- Brown, N.G.; VanderLinden, R.; Watson, E.R.; Weissmann, F.; Ordureau, A.; Wu, K.P.; Zhang, W.; Yu, S.; Mercredi, P.Y.; Harrison, J.S.; et al. Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C. Cell 2016, 165, 1440–1453. [Google Scholar] [CrossRef] [Green Version]
- Alfieri, C.; Chang, L.; Zhang, Z.; Yang, J.; Maslen, S.; Skehel, M.; Barford, D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 2016, 536, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Min, M.; Mayor, U.; Dittmar, G.; Lindon, C. Using in vivo biotinylated ubiquitin to describe a mitotic exit ubiquitome from human cells. Mol. Cell. Proteom.: Mcp 2014, 13, 2411–2425. [Google Scholar] [CrossRef]
- Hayes, M.J.; Kimata, Y.; Wattam, S.L.; Lindon, C.; Mao, G.; Yamano, H.; Fry, A.M. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat. Cell Biol. 2006, 8, 607–614. [Google Scholar] [CrossRef]
- Di Fiore, B.; Pines, J. How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 2010, 190, 501–509. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.; Morgan, D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007, 8, 894–903. [Google Scholar] [CrossRef]
- Peters, J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef]
- Primorac, I.; Musacchio, A. Panta rhei: The APC/C at steady state. J. Cell Biol. 2013, 201, 177–189. [Google Scholar] [CrossRef]
- Pines, J. Cubism and the cell cycle: The many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011, 12, 427–438. [Google Scholar] [CrossRef]
- López-Avilés, S.; Kapuy, O.; Novák, B.; Uhlmann, F. Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature 2009, 459, 592–595. [Google Scholar] [CrossRef] [Green Version]
- Potapova, T.A.; Sivakumar, S.; Flynn, J.N.; Li, R.; Gorbsky, G.J. Mitotic progression becomes irreversible in prometaphase and collapses when Wee1 and Cdc25 are inhibited. Mol. Biol. Cell 2011, 22, 1191–1206. [Google Scholar] [CrossRef]
- Saurin, A.T. Kinase and Phosphatase Cross-Talk at the Kinetochore. Front. Cell Dev. Biol. 2018, 6, 62. [Google Scholar] [CrossRef] [Green Version]
- Manic, G.; Corradi, F.; Sistigu, A.; Siteni, S.; Vitale, I. Molecular Regulation of the Spindle Assembly Checkpoint by Kinases and Phosphatases. Int. Rev. Cell Mol. Biol. 2017, 328, 105–161. [Google Scholar] [CrossRef]
- Joglekar, A.P.; Kukreja, A.A. How Kinetochore Architecture Shapes the Mechanisms of Its Function. Curr. Biol. 2017, 27, R816–R824. [Google Scholar] [CrossRef]
- Gelens, L.; Qian, J.; Bollen, M.; Saurin, A.T. The Importance of Kinase-Phosphatase Integration: Lessons from Mitosis. Trends Cell Biol. 2018, 28, 6–21. [Google Scholar] [CrossRef]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [Green Version]
- Atherton-Fessler, S.; Parker, L.L.; Geahlen, R.L.; Piwnica-Worms, H. Mechanisms of p34cdc2 regulation. Mol. Cell. Biol. 1993, 13, 1675–1685. [Google Scholar] [CrossRef]
- Solomon, M.J.; Glotzer, M.; Lee, T.H.; Philippe, M.; Kirschner, M.W. Cyclin activation of p34cdc2. Cell 1990, 63, 1013–1024. [Google Scholar] [CrossRef]
- Kinoshita, N.; Yamano, H.; Niwa, H.; Yoshida, T.; Yanagida, M. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev. 1993, 7, 1059–1071. [Google Scholar] [CrossRef]
- Felix, M.A.; Cohen, P.; Karsenti, E. Cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: Evidence from the effects of okadaic acid. EMBO J. 1990, 9, 675–683. [Google Scholar] [CrossRef]
- Clarke, P.R.; Hoffmann, I.; Draetta, G.; Karsenti, E. Dephosphorylation of cdc25-C by a type-2A protein phosphatase: Specific regulation during the cell cycle in Xenopus egg extracts. Mol. Biol. Cell 1993, 4, 397–411. [Google Scholar] [CrossRef]
- Mochida, S.; Rata, S.; Hino, H.; Nagai, T.; Novák, B. Two Bistable Switches Govern M Phase Entry. Curr. Biol. 2016, 26, 3361–3367. [Google Scholar] [CrossRef] [Green Version]
- Rata, S.; Suarez Peredo Rodriguez, M.F.; Joseph, S.; Peter, N.; Echegaray Iturra, F.; Yang, F.; Madzvamuse, A.; Ruppert, J.G.; Samejima, K.; Platani, M.; et al. Two Interlinked Bistable Switches Govern Mitotic Control in Mammalian Cells. Curr. Biol. 2018. [Google Scholar] [CrossRef]
- Vigneron, S.; Sundermann, L.; Labbe, J.C.; Pintard, L.; Radulescu, O.; Castro, A.; Lorca, T. Cyclin A-cdk1-Dependent Phosphorylation of Bora Is the Triggering Factor Promoting Mitotic Entry. Dev. Cell 2018, 45, 637–650.e7. [Google Scholar] [CrossRef]
- Gavet, O.; Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 2010, 18, 533–543. [Google Scholar] [CrossRef]
- Santos, S.D.M.; Wollman, R.; Meyer, T.; Ferrell, J.E., Jr. Spatial positive feedback at the onset of mitosis. Cell 2012, 149, 1500–1513. [Google Scholar] [CrossRef]
- Hagting, A.; Jackman, M.; Simpson, K.; Pines, J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 1999, 9, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Meyer, A.N.; Donoghue, D.J. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 1997, 94, 502–507. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.Y.; Graves, P.R.; Thoma, R.S.; Wu, Z.; Shaw, A.S.; Piwnica-Worms, H. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997, 277, 1501–1505. [Google Scholar] [CrossRef]
- Kumagai, A.; Yakowec, P.S.; Dunphy, W.G. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol. Biol. Cell 1998, 9, 345–354. [Google Scholar] [CrossRef]
- Margolis, S.S.; Walsh, S.; Weiser, D.C.; Yoshida, M.; Shenolikar, S.; Kornbluth, S. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. EMBO J. 2003, 22, 5734–5745. [Google Scholar] [CrossRef] [Green Version]
- Margolis, S.S.; Perry, J.A.; Weitzel, D.H.; Freel, C.D.; Yoshida, M.; Haystead, T.A.; Kornbluth, S. A role for PP1 in the Cdc2/Cyclin B-mediated positive feedback activation of Cdc25. Mol. Biol. Cell 2006, 17, 1779–1789. [Google Scholar] [CrossRef]
- Donzelli, M.; Squatrito, M.; Ganoth, D.; Hershko, A.; Pagano, M.; Draetta, G.F. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002, 21, 4875–4884. [Google Scholar] [CrossRef] [Green Version]
- Vigneron, S.; Gharbi-Ayachi, A.; Raymond, A.-A.; Burgess, A.; Labbé, J.-C.; Labesse, G.; Monsarrat, B.; Lorca, T.; Castro, A. Characterization of the Mechanisms Controlling Greatwall Activity. Mol. Cell. Biol. 2011, 31, 2262. [Google Scholar] [CrossRef]
- Blake-Hodek, K.A.; Williams, B.C.; Zhao, Y.; Castilho, P.V.; Chen, W.; Mao, Y.; Yamamoto, T.M.; Goldberg, M.L. Determinants for Activation of the Atypical AGC Kinase Greatwall during M Phase Entry. Mol. Cell. Biol. 2012, 32, 1337–1353. [Google Scholar] [CrossRef] [Green Version]
- Mochida, S.; Maslen, S.L.; Skehel, M.; Hunt, T. Greatwall Phosphorylates an Inhibitor of Protein Phosphatase 2Α That Is Essential for Mitosis. Science 2010, 330, 1670. [Google Scholar] [CrossRef]
- Gharbi-Ayachi, A.; Labbé, J.-C.; Burgess, A.; Vigneron, S.; Strub, J.-M.; Brioudes, E.; Van-Dorsselaer, A.; Castro, A.; Lorca, T. The Substrate of Greatwall Kinase, Arpp19, Controls Mitosis by Inhibiting Protein Phosphatase 2A. Science 2010, 330, 1673. [Google Scholar] [CrossRef]
- Mochida, S. Regulation of α–endosulfine, an inhibitor of protein phosphatase 2A, by multisite phosphorylation. FEBS J. 2014, 281, 1159–1169. [Google Scholar] [CrossRef]
- Williams, B.C.; Filter, J.J.; Blake-Hodek, K.A.; Wadzinski, B.E.; Fuda, N.J.; Shalloway, D.; Goldberg, M.L. Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A-B55 heterotrimers. eLife 2014, 3, e01695. [Google Scholar] [CrossRef]
- Schleicher, K.; Porter, M.; Ten Have, S.; Sundaramoorthy, R.; Porter, I.M.; Swedlow, J.R. The Ndc80 complex targets Bod1 to human mitotic kinetochores. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Porter, I.M.; Schleicher, K.; Porter, M.; Swedlow, J.R. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat. Commun. 2013, 4, 2677. [Google Scholar] [CrossRef] [Green Version]
- Longin, S.; Zwaenepoel, K.; Louis, J.V.; Dilworth, S.; Goris, J.; Janssens, V. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J. Biol. Chem. 2007, 282, 26971–26980. [Google Scholar] [CrossRef]
- Jativa, S.; Calabria, I.; Moyano-Rodriguez, Y.; Garcia, P.; Queralt, E. Cdc14 activation requires coordinated Cdk1-dependent phosphorylation of Net1 and PP2A-Cdc55 at anaphase onset. Cell Mol. Life Sci. 2019. [Google Scholar] [CrossRef]
- Schmitz, M.H.A.; Held, M.; Janssens, V.; Hutchins, J.R.A.; Hudecz, O.; Ivanova, E.; Goris, J.; Trinkle-Mulcahy, L.; Lamond, A.I.; Poser, I.; et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 2010, 12, 886–893. [Google Scholar] [CrossRef]
- Kwon, Y.G.; Lee, S.Y.; Choi, Y.; Greengard, P.; Nairn, A.C. Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 2168–2173. [Google Scholar] [CrossRef] [Green Version]
- Yamano, H.; Ishii, K.; Yanagida, M. Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J. 1994, 13, 5310–5318. [Google Scholar] [CrossRef]
- Dohadwala, M.; da Cruz e Silva, E.F.; Hall, F.L.; Williams, R.T.; Carbonaro-Hall, D.A.; Nairn, A.C.; Greengard, P.; Berndt, N. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA 1994, 91, 6408–6412. [Google Scholar] [CrossRef]
- Wu, J.Q.; Guo, J.Y.; Tang, W.; Yang, C.-S.; Freel, C.D.; Chen, C.; Nairn, A.C.; Kornbluth, S. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 2009, 11, 644–651. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P. The discovery of protein phosphatases: From chaos and confusion to an understanding of their role in cell regulation and human disease. Bioessays 1994, 16, 583–588. [Google Scholar] [CrossRef]
- Miller, M.L.; Jensen, L.J.; Diella, F.; Jørgensen, C.; Tinti, M.; Li, L.; Hsiung, M.; Parker, S.A.; Bordeaux, J.; Sicheritz-Ponten, T.; et al. Linear Motif Atlas for Phosphorylation-Dependent Signaling. Sci. Signal. 2008, 1, ra2. [Google Scholar] [CrossRef]
- Nasa, I.; Rusin, S.F.; Kettenbach, A.N.; Moorhead, G.B. Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Zeng, K.; Bastos, R.N.; Barr, F.A.; Gruneberg, U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 2010, 191, 1315–1332. [Google Scholar] [CrossRef] [Green Version]
- Kettenbach, A.N.; Schlosser, K.A.; Lyons, S.P.; Nasa, I.; Gui, J.; Adamo, M.E.; Gerber, S.A. Global assessment of its network dynamics reveals that the kinase Plk1 inhibits the phosphatase PP6 to promote Aurora A activity. Sci. Signal. 2018, 11, eaaq1441. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P.T.; Philp, A.; Vazquez-Martin, C. Protein phosphatase 4--from obscurity to vital functions. FEBS Lett. 2005, 579, 3278–3286. [Google Scholar] [CrossRef]
- Lee, D.-H.; Acharya, S.S.; Kwon, M.; Drane, P.; Guan, Y.; Adelmant, G.; Kalev, P.; Shah, J.; Pellman, D.; Marto, J.A.; et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol. Cell 2014, 54, 512–525. [Google Scholar] [CrossRef]
- Lipinszki, Z.; Lefevre, S.; Savoian, M.S.; Singleton, M.R.; Glover, D.M.; Przewloka, M.R. Centromeric binding and activity of Protein Phosphatase 4. Nat. Commun. 2015, 6, 5894. [Google Scholar] [CrossRef]
- Helps, N.R.; Brewis, N.D.; Lineruth, K.; Davis, T.; Kaiser, K.; Cohen, P.T. Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J. Cell Sci. 1998, 111 Pt 10, 1331–1340. [Google Scholar]
- Sumiyoshi, E.; Sugimoto, A.; Yamamoto, M. Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J. Cell Sci. 2002, 115, 1403–1410. [Google Scholar]
- Voss, M.; Campbell, K.; Saranzewa, N.; Campbell, D.G.; Hastie, C.J.; Peggie, M.W.; Martin-Granados, C.; Prescott, A.R.; Cohen, P.T. Protein phosphatase 4 is phosphorylated and inactivated by Cdk in response to spindle toxins and interacts with gamma-tubulin. Cell Cycle 2013, 12, 2876–2887. [Google Scholar] [CrossRef]
- Kim, T.; Lara-Gonzalez, P.; Prevo, B.; Meitinger, F.; Cheerambathur, D.K.; Oegema, K.; Desai, A. Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev. 2017, 31, 1089–1094. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Rodriguez-Bravo, V.; Kim, H.; Datta, S.; Foley, E.A. The PP2AB56 phosphatase promotes the association of Cdc20 with APC/C in mitosis. J. Cell Sci. 2017, 130, 1760. [Google Scholar] [CrossRef]
- Labit, H.; Fujimitsu, K.; Bayin, N.S.; Takaki, T.; Gannon, J.; Yamano, H. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 2012, 31, 3351–3362. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Craney, A.; Rape, M. Microtubule-dependent regulation of mitotic protein degradation. Mol. Cell 2014, 53, 179–192. [Google Scholar] [CrossRef]
- Magidson, V.; O’Connell, C.B.; Lončarek, J.; Paul, R.; Mogilner, A.; Khodjakov, A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 2011, 146, 555–567. [Google Scholar] [CrossRef]
- Tanaka, K.; Mukae, N.; Dewar, H.; van Breugel, M.; James, E.K.; Prescott, A.R.; Antony, C.; Tanaka, T.U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 2005, 434, 987–994. [Google Scholar] [CrossRef]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef]
- Kawashima, S.A.; Yamagishi, Y.; Honda, T.; Ishiguro, K.-i.; Watanabe, Y. Phosphorylation of H2A by Bub1 Prevents Chromosomal Instability Through Localizing Shugoshin. Science 2010, 327, 172. [Google Scholar] [CrossRef]
- Hindriksen, S.; Lens, S.M.A.; Hadders, M.A. The Ins and Outs of Aurora B Inner Centromere Localization. Front. Cell Dev. Biol. 2017, 5, 112. [Google Scholar] [CrossRef] [Green Version]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006, 127, 969–982. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Arthanari, H.; Boeszoermenyi, A.; Dashkevich, N.M.; Wilson-Kubalek, E.M.; Monnier, N.; Markus, M.; Oberer, M.; Milligan, R.A.; Bathe, M.; et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 2012, 23, 968–980. [Google Scholar] [CrossRef]
- Chan, Y.W.; Jeyaprakash, A.A.; Nigg, E.A.; Santamaria, A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J. Cell Biol. 2012, 196, 563–571. [Google Scholar] [CrossRef]
- Foley, E.A.; Maldonado, M.; Kapoor, T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011, 13, 1265–1271. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Vleugel, M.; Backer, C.B.; Hori, T.; Fukagawa, T.; Cheeseman, I.M.; Lampson, M.A. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010, 188, 809–820. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Holland, A.J.; Lan, W.; Cleveland, D.W. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 2010, 142, 444–455. [Google Scholar] [CrossRef]
- Sivakumar, S.; Janczyk, P.L.; Qu, Q.; Brautigam, C.A.; Stukenberg, P.T.; Yu, H.; Gorbsky, G.J. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife 2016, 5. [Google Scholar] [CrossRef]
- Kabeche, L.; Compton, D.A. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 2013, 502, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Wolthuis, R.; Clay-Farrace, L.; van Zon, W.; Yekezare, M.; Koop, L.; Ogink, J.; Medema, R.; Pines, J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell 2008, 30, 290–302. [Google Scholar] [CrossRef]
- Wan, X.; O’Quinn, R.P.; Pierce, H.L.; Joglekar, A.P.; Gall, W.E.; DeLuca, J.G.; Carroll, C.W.; Liu, S.-T.; Yen, T.J.; McEwen, B.F.; et al. Protein Architecture of the Human Kinetochore Microtubule Attachment Site. Cell 2009, 137, 672–684. [Google Scholar] [CrossRef] [Green Version]
- Musacchio, A.; Desai, A. A Molecular View of Kinetochore Assembly and Function. Biol. (Basel) 2017, 6, 5. [Google Scholar] [CrossRef]
- Choi, E.; Choe, H.; Min, J.; Choi, J.Y.; Kim, J.; Lee, H. BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 2009, 28, 2077–2089. [Google Scholar] [CrossRef] [Green Version]
- King, E.M.; van der Sar, S.J.; Hardwick, K.G. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE 2007, 2, e342. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Ribeiro, S.; Sennels, L.; Sanchez-Pulido, L.; de Lima Alves, F.; Verheyen, T.; Kelly, D.A.; Ponting, C.P.; Rappsilber, J.; Earnshaw, W.C. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 2011, 21, 328–342. [Google Scholar] [CrossRef]
- Qian, J.; Lesage, B.; Beullens, M.; Van Eynde, A.; Bollen, M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 2011, 21, 766–773. [Google Scholar] [CrossRef]
- Mirchenko, L.; Uhlmann, F. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 2010, 20, 1396–1401. [Google Scholar] [CrossRef]
- Luo, X.; Tang, Z.; Rizo, J.; Yu, H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 2002, 9, 59–71. [Google Scholar] [CrossRef]
- Faesen, A.C.; Thanasoula, M.; Maffini, S.; Breit, C.; Müller, F.; van Gerwen, S.; Bange, T.; Musacchio, A. Basis of catalytic assembly of the mitotic checkpoint complex. Nature 2017, 542, 498–502. [Google Scholar] [CrossRef] [Green Version]
- De Antoni, A.; Pearson, C.G.; Cimini, D.; Canman, J.C.; Sala, V.; Nezi, L.; Mapelli, M.; Sironi, L.; Faretta, M.; Salmon, E.D.; et al. The Mad1/Mad2 Complex as a Template for Mad2 Activation in the Spindle Assembly Checkpoint. Curr. Biol. 2005, 15, 214–225. [Google Scholar] [CrossRef] [Green Version]
- Izawa, D.; Pines, J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 2015, 517, 631–634. [Google Scholar] [CrossRef]
- Morin, V.; Prieto, S.; Melines, S.; Hem, S.; Rossignol, M.; Lorca, T.; Espeut, J.; Morin, N.; Abrieu, A. CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr. Biol. 2012, 22, 289–295. [Google Scholar] [CrossRef]
- von Schubert, C.; Cubizolles, F.; Bracher, J.M.; Sliedrecht, T.; Kops, G.; Nigg, E.A. Plk1 and Mps1 Cooperatively Regulate the Spindle Assembly Checkpoint in Human Cells. Cell Rep. 2015, 12, 66–78. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, J.; Yekezare, M.; Minshull, J.; Pines, J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 2008, 10, 1411–1420. [Google Scholar] [CrossRef]
- Reddy, S.K.; Rape, M.; Margansky, W.A.; Kirschner, M.W. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 2007, 446, 921–925. [Google Scholar] [CrossRef]
- Stegmeier, F.; Rape, M.; Draviam, V.M.; Nalepa, G.; Sowa, M.E.; Ang, X.L.; McDonald, E.R., 3rd; Li, M.Z.; Hannon, G.J.; Sorger, P.K.; et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 2007, 446, 876–881. [Google Scholar] [CrossRef]
- Yang, M.; Li, B.; Tomchick, D.R.; Machius, M.; Rizo, J.; Yu, H.; Luo, X. p31comet blocks Mad2 activation through structural mimicry. Cell 2007, 131, 744–755. [Google Scholar] [CrossRef]
- Habu, T.; Kim, S.H.; Weinstein, J.; Matsumoto, T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002, 21, 6419–6428. [Google Scholar] [CrossRef] [Green Version]
- Alfieri, C.; Chang, L.; Barford, D. Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 2018, 559, 274–278. [Google Scholar] [CrossRef]
- Mo, M.; Arnaoutov, A.; Dasso, M. Phosphorylation of Xenopus p31comet potentiates mitotic checkpoint exit. Cell Cycle 2015, 14, 3978–3985. [Google Scholar] [CrossRef]
- Date, D.A.; Burrows, A.C.; Summers, M.K. Phosphorylation regulates the p31Comet-mitotic arrest-deficient 2 (Mad2) interaction to promote spindle assembly checkpoint (SAC) activity. J. Biol. Chem. 2014, 289, 11367–11373. [Google Scholar] [CrossRef]
- Kaisari, S.; Shomer, P.; Ziv, T.; Sitry-Shevah, D.; Miniowitz-Shemtov, S.; Teichner, A.; Hershko, A. Role of Polo-like kinase 1 in the regulation of the action of p31(comet) in the disassembly of mitotic checkpoint complexes. Proc. Natl. Acad. Sci. USA 2019, 116, 11725–11730. [Google Scholar] [CrossRef]
- Hiruma, Y.; Sacristan, C.; Pachis, S.T.; Adamopoulos, A.; Kuijt, T.; Ubbink, M.; von Castelmur, E.; Perrakis, A.; Kops, G.J. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 2015, 348, 1264–1267. [Google Scholar] [CrossRef]
- Qian, J.; Garcia-Gimeno, M.A.; Beullens, M.; Manzione, M.G.; Van der Hoeven, G.; Igual, J.C.; Heredia, M.; Sanz, P.; Gelens, L.; Bollen, M. An Attachment-Independent Biochemical Timer of the Spindle Assembly Checkpoint. Mol. Cell 2017, 68, 715–730.e5. [Google Scholar] [CrossRef]
- Espert, A.; Uluocak, P.; Bastos, R.N.; Mangat, D.; Graab, P.; Gruneberg, U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 2014, 206, 833–842. [Google Scholar] [CrossRef]
- Nijenhuis, W.; Vallardi, G.; Teixeira, A.; Kops, G.J.; Saurin, A.T. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 2014, 16, 1257–1264. [Google Scholar] [CrossRef] [Green Version]
- Clute, P.; Pines, J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999, 1, 82–87. [Google Scholar] [CrossRef]
- Uhlmann, F. SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef]
- Hirano, T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell 2016, 164, 847–857. [Google Scholar] [CrossRef] [Green Version]
- Shintomi, K.; Takahashi, T.S.; Hirano, T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol. 2015, 17, 1014–1023. [Google Scholar] [CrossRef]
- St-Pierre, J.; Douziech, M.; Bazile, F.; Pascariu, M.; Bonneil, E.; Sauve, V.; Ratsima, H.; D’Amours, D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol. Cell 2009, 34, 416–426. [Google Scholar] [CrossRef]
- Mora-Bermudez, F.; Gerlich, D.; Ellenberg, J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 2007, 9, 822–831. [Google Scholar] [CrossRef]
- Lipp, J.J.; Hirota, T.; Poser, I.; Peters, J.-M. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J. Cell Sci. 2007, 120, 1245. [Google Scholar] [CrossRef]
- Abe, S.; Nagasaka, K.; Hirayama, Y.; Kozuka-Hata, H.; Oyama, M.; Aoyagi, Y.; Obuse, C.; Hirota, T. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 2011, 25, 863–874. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Hirano, M.; Kobayashi, R.; Hirano, T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 1998, 282, 487–490. [Google Scholar] [CrossRef]
- Sutani, T.; Yuasa, T.; Tomonaga, T.; Dohmae, N.; Takio, K.; Yanagida, M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999, 13, 2271–2283. [Google Scholar] [CrossRef]
- Kao, L.; Wang, Y.-T.; Chen, Y.-C.; Tseng, S.-F.; Jhang, J.-C.; Chen, Y.-J.; Teng, S.-C. Global analysis of cdc14 dephosphorylation sites reveals essential regulatory role in mitosis and cytokinesis. Mol. Cell. Proteom.: Mcp 2014, 13, 594–605. [Google Scholar] [CrossRef]
- Xing, H.; Vanderford, N.L.; Sarge, K.D. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 2008, 10, 1318–1323. [Google Scholar] [CrossRef]
- Takemoto, A.; Kimura, K.; Yanagisawa, J.; Yokoyama, S.; Hanaoka, F. Negative regulation of condensin I by CK2-mediated phosphorylation. EMBO J. 2006, 25, 5339–5348. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, T.; Sykora, M.M.; Huis in ‘t Veld, P.J.; Mechtler, K.; Peters, J.-M. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc. Natl. Acad. Sci. USA 2013, 110, 13404–13409. [Google Scholar] [CrossRef] [Green Version]
- Sumara, I.; Vorlaufer, E.; Stukenberg, P.T.; Kelm, O.; Redemann, N.; Nigg, E.A.; Peters, J.M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell 2002, 9, 515–525. [Google Scholar] [CrossRef]
- Liu, H.; Rankin, S.; Yu, H. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat. Cell Biol. 2013, 15, 40–49. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Kawashima, S.A.; Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004, 427, 510–517. [Google Scholar] [CrossRef]
- Xu, Z.; Cetin, B.; Anger, M.; Cho, U.S.; Helmhart, W.; Nasmyth, K.; Xu, W. Structure and function of the PP2A-shugoshin interaction. Mol. Cell 2009, 35, 426–441. [Google Scholar] [CrossRef]
- Qi, W.; Yu, H. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007, 282, 3672–3679. [Google Scholar] [CrossRef]
- Hellmuth, S.; Böttger, F.; Pan, C.; Mann, M.; Stemmann, O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014, 33, 1134–1147. [Google Scholar] [CrossRef]
- Holland, A.J.; Böttger, F.; Stemmann, O.; Taylor, S.S. Protein Phosphatase 2A and Separase Form a Complex Regulated by Separase Autocleavage. J. Biol. Chem. 2007, 282, 24623–24632. [Google Scholar] [CrossRef] [Green Version]
- Shindo, N.; Kumada, K.; Hirota, T. Separase sensor reveals dual roles for separase coordinating cohesin cleavage and cdk1 inhibition. Dev. Cell 2012, 23, 112–123. [Google Scholar] [CrossRef]
- Holt, L.J.; Krutchinsky, A.N.; Morgan, D.O. Positive feedback sharpens the anaphase switch. Nature 2008, 454, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Schwab, M.; Neutzner, M.; Mocker, D.; Seufert, W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001, 20, 5165–5175. [Google Scholar] [CrossRef] [Green Version]
- Vodermaier, H.C.; Gieffers, C.; Maurer-Stroh, S.; Eisenhaber, F.; Peters, J.M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 2003, 13, 1459–1468. [Google Scholar] [CrossRef]
- Rudner, A.D.; Murray, A.W. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 2000, 149, 1377–1390. [Google Scholar] [CrossRef]
- Kramer, E.R.; Scheuringer, N.; Podtelejnikov, A.V.; Mann, M.; Peters, J.-M. Mitotic Regulation of the APC Activator Proteins CDC20 and CDH1. Mol. Biol. Cell 2000, 11, 1555–1569. [Google Scholar] [CrossRef] [Green Version]
- Shteinberg, M.; Protopopov, Y.; Listovsky, T.; Brandeis, M.; Hershko, A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 1999, 260, 193–198. [Google Scholar] [CrossRef]
- Kraft, C.; Herzog, F.; Gieffers, C.; Mechtler, K.; Hagting, A.; Pines, J.; Peters, J.-M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003, 22, 6598–6609. [Google Scholar] [CrossRef] [Green Version]
- Golan, A.; Yudkovsky, Y.; Hershko, A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 2002, 277, 15552–15557. [Google Scholar] [CrossRef]
- Fujimitsu, K.; Grimaldi, M.; Yamano, H. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science 2016, 352, 1121–1124. [Google Scholar] [CrossRef]
- Zhang, S.; Chang, L.; Alfieri, C.; Zhang, Z.; Yang, J.; Maslen, S.; Skehel, M.; Barford, D. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 2016, 533, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Qiao, R.; Weissmann, F.; Yamaguchi, M.; Brown, N.G.; VanderLinden, R.; Imre, R.; Jarvis, M.A.; Brunner, M.R.; Davidson, I.F.; Litos, G.; et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2016, 113, E2570–E2578. [Google Scholar] [CrossRef]
- Shteinberg, M.; Hershko, A. Role of Suc1 in the activation of the cyclosome by protein kinase Cdk1/cyclin B. Biochem. Biophys. Res. Commun. 1999, 257, 12–18. [Google Scholar] [CrossRef]
- Patra, D.; Dunphy, W.G. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev. 1998, 12, 2549–2559. [Google Scholar] [CrossRef]
- McGrath, D.A.; Balog, E.R.M.; Kõivomägi, M.; Lucena, R.; Mai, M.V.; Hirschi, A.; Kellogg, D.R.; Loog, M.; Rubin, S.M. Cks confers specificity to phosphorylation-dependent CDK signaling pathways. Nat. Struct. Mol. Biol. 2013, 20, 1407–1414. [Google Scholar] [CrossRef] [Green Version]
- Kõivomägi, M.; Ord, M.; Iofik, A.; Valk, E.; Venta, R.; Faustova, I.; Kivi, R.; Balog, E.R.M.; Rubin, S.M.; Loog, M. Multisite phosphorylation networks as signal processors for Cdk1. Nat. Struct. Mol. Biol. 2013, 20, 1415–1424. [Google Scholar] [CrossRef] [Green Version]
- Alfieri, C.; Zhang, S.; Barford, D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Passmore, L.A.; Booth, C.R.; Venien-Bryan, C.; Ludtke, S.J.; Fioretto, C.; Johnson, L.N.; Chiu, W.; Barford, D. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol. Cell 2005, 20, 855–866. [Google Scholar] [CrossRef]
- Ohi, M.D.; Feoktistova, A.; Ren, L.; Yip, C.; Cheng, Y.; Chen, J.S.; Yoon, H.J.; Wall, J.S.; Huang, Z.; Penczek, P.A.; et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell 2007, 28, 871–885. [Google Scholar] [CrossRef]
- Schreiber, A.; Stengel, F.; Zhang, Z.; Enchev, R.I.; Kong, E.H.; Morris, E.P.; Robinson, C.V.; da Fonseca, P.C.; Barford, D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 2011, 470, 227–232. [Google Scholar] [CrossRef]
- Yamada, H.; Kumada, K.; Yanagida, M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J. Cell Sci. 1997, 110 Pt 15, 1793–1804. [Google Scholar]
- Sivakumar, S.; Daum, J.R.; Tipton, A.R.; Rankin, S.; Gorbsky, G.J. The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol. Biol. Cell 2014, 25, 594–605. [Google Scholar] [CrossRef]
- Lu, D.; Hsiao, J.Y.; Davey, N.E.; Van Voorhis, V.A.; Foster, S.A.; Tang, C.; Morgan, D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014, 207, 23–39. [Google Scholar] [CrossRef] [Green Version]
- Floyd, S.; Pines, J.; Lindon, C. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr. Biol. 2008, 18, 1649–1658. [Google Scholar] [CrossRef]
- Zachariae, W.; Schwab, M.; Nasmyth, K.; Seufert, W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 1998, 282, 1721–1724. [Google Scholar] [CrossRef]
- Visintin, R.; Craig, K.; Hwang, E.S.; Prinz, S.; Tyers, M.; Amon, A. The Phosphatase Cdc14 Triggers Mitotic Exit by Reversal of Cdk-Dependent Phosphorylation. Mol. Cell 1998, 2, 709–718. [Google Scholar] [CrossRef]
- Yudkovsky, Y.; Shteinberg, M.; Listovsky, T.; Brandeis, M.; Hershko, A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 2000, 271, 299–304. [Google Scholar] [CrossRef]
- Rogers, S.; McCloy, R.; Watkins, D.N.; Burgess, A. Mechanisms regulating phosphatase specificity and the removal of individual phosphorylation sites during mitotic exit. BioEssays 2016, 38, S24–S32. [Google Scholar] [CrossRef]
- Nilsson, J. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 2019, 218, 395. [Google Scholar] [CrossRef]
- Craney, A.; Kelly, A.; Jia, L.; Fedrigo, I.; Yu, H.; Rape, M. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc. Natl. Acad. Sci. 2016, 113, 1540. [Google Scholar] [CrossRef]
- Lu, D.; Girard, J.R.; Li, W.; Mizrak, A.; Morgan, D.O. Quantitative framework for ordered degradation of APC/C substrates. BMC Biol. 2015, 13, 96. [Google Scholar] [CrossRef]
- He, J.; Chao, W.C.H.; Zhang, Z.; Yang, J.; Cronin, N.; Barford, D. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol. Cell 2013, 50, 649–660. [Google Scholar] [CrossRef]
- Tian, W.; Li, B.; Warrington, R.; Tomchick, D.R.; Yu, H.; Luo, X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc. Natl. Acad. Sci. USA 2012, 109, 18419–18424. [Google Scholar] [CrossRef] [Green Version]
- Chao, W.C.; Kulkarni, K.; Zhang, Z.; Kong, E.H.; Barford, D. Structure of the mitotic checkpoint complex. Nature 2012, 484, 208–213. [Google Scholar] [CrossRef]
- Sedgwick, G.G.; Hayward, D.G.; Di Fiore, B.; Pardo, M.; Yu, L.; Pines, J.; Nilsson, J. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 2013, 32, 303–314. [Google Scholar] [CrossRef]
- Mailand, N.; Diffley, J.F. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005, 122, 915–926. [Google Scholar] [CrossRef]
- Ma, S.; Vigneron, S.; Robert, P.; Strub, J.M.; Cianferani, S.; Castro, A.; Lorca, T. Greatwall dephosphorylation and inactivation upon mitotic exit is triggered by PP1. J. Cell Sci. 2016, 129, 1329–1339. [Google Scholar] [CrossRef]
- Heim, A.; Konietzny, A.; Mayer, T.U. Protein phosphatase 1 is essential for Greatwall inactivation at mitotic exit. EMBO Rep. 2015, 16, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Rogers, S.; Fey, D.; McCloy, R.A.; Parker, B.L.; Mitchell, N.J.; Payne, R.J.; Daly, R.J.; James, D.E.; Caldon, C.E.; Watkins, D.N.; et al. PP1 initiates the dephosphorylation of MASTL, triggering mitotic exit and bistability in human cells. J. Cell Sci. 2016, 129, 1340–1354. [Google Scholar] [CrossRef] [Green Version]
- Grallert, A.; Boke, E.; Hagting, A.; Hodgson, B.; Connolly, Y.; Griffiths, J.R.; Smith, D.L.; Pines, J.; Hagan, I.M. A PP1-PP2A phosphatase relay controls mitotic progression. Nature 2015, 517, 94–98. [Google Scholar] [CrossRef]
- De Wulf, P.; Montani, F.; Visintin, R. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 2009, 21, 806–815. [Google Scholar] [CrossRef]
- Queralt, E.; Uhlmann, F. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 2008, 20, 661–668. [Google Scholar] [CrossRef] [Green Version]
- Skoufias, D.A.; Indorato, R.-L.; Lacroix, F.; Panopoulos, A.; Margolis, R.L. Mitosis persists in the absence of Cdk1 activity when proteolysis or protein phosphatase activity is suppressed. J. Cell Biol. 2007, 179, 671–685. [Google Scholar] [CrossRef] [Green Version]
- Mochida, S.; Ikeo, S.; Gannon, J.; Hunt, T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009, 28, 2777–2785. [Google Scholar] [CrossRef]
- Cundell, M.J.; Bastos, R.N.; Zhang, T.; Holder, J.; Gruneberg, U.; Novak, B.; Barr, F.A. The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol. Cell 2013, 52, 393–405. [Google Scholar] [CrossRef]
- Bastos, R.N.; Cundell, M.J.; Barr, F.A. KIF4A and PP2A–B56 form a spatially restricted feedback loop opposing Aurora B at the anaphase central spindle. J. Cell Biol. 2014, 207, 683. [Google Scholar] [CrossRef]
- Gil, R.S.; Vagnarelli, P. Protein phosphatases in chromatin structure and function. Biochim. Et Biophys. Acta. Mol. Cell Res. 2019, 1866, 90–101. [Google Scholar] [CrossRef]
- Bhowmick, R.; Thakur, R.S.; Venegas, A.B.; Liu, Y.; Nilsson, J.; Barisic, M.; Hickson, I.D. The RIF1-PP1 Axis Controls Abscission Timing in Human Cells. Curr. Biol. 2019, 29, 1232–1242.e5. [Google Scholar] [CrossRef] [Green Version]
- Qian, J.; Beullens, M.; Huang, J.; De Munter, S.; Lesage, B.; Bollen, M. Cdk1 orders mitotic events through coordination of a chromosome-associated phosphatase switch. Nat. Commun. 2015, 6, 10215. [Google Scholar] [CrossRef]
- Landsverk, H.B.; Kirkhus, M.; Bollen, M.; Küntziger, T.; Collas, P. PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochem. J. 2005, 390, 709–717. [Google Scholar] [CrossRef]
- Wolf, F.; Wandke, C.; Isenberg, N.; Geley, S. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 2006, 25, 2802–2813. [Google Scholar] [CrossRef]
- Stemmann, O.; Zou, H.; Gerber, S.A.; Gygi, S.P.; Kirschner, M.W. Dual inhibition of sister chromatid separation at metaphase. Cell 2001, 107, 715–726. [Google Scholar] [CrossRef]
- Bouchoux, C.; Uhlmann, F. A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell 2011, 147, 803–814. [Google Scholar] [CrossRef]
- Nakajima, H.; Toyoshima-Morimoto, F.; Taniguchi, E.; Nishida, E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003, 278, 25277–25280. [Google Scholar] [CrossRef]
- Rusin, S.F.; Adamo, M.E.; Kettenbach, A.N. Identification of Candidate Casein Kinase 2 Substrates in Mitosis by Quantitative Phosphoproteomics. Front. Cell Dev. Biol. 2017, 5, 97. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Bremmer, S.C.; Hall, H.; Martinez, J.S.; Eissler, C.L.; Hinrichsen, T.H.; Rossie, S.; Parker, L.L.; Hall, M.C.; Charbonneau, H. Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J. Biol. Chem. 2012, 287, 1662–1669. [Google Scholar] [CrossRef]
- Eissler, C.L.; Mazon, G.; Powers, B.L.; Savinov, S.N.; Symington, L.S.; Hall, M.C. The Cdk/Cdc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Mol. Cell 2014, 54, 80–93. [Google Scholar] [CrossRef]
- Bassermann, F.; Frescas, D.; Guardavaccaro, D.; Busino, L.; Peschiaroli, A.; Pagano, M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 2008, 134, 256–267. [Google Scholar] [CrossRef]
- Vallardi, G.; Allan, L.A.; Crozier, L.; Saurin, A.T. Division of labour between PP2A-B56 isoforms at the centromere and kinetochore. eLife 2019, 8. [Google Scholar] [CrossRef]
- Kumar, G.S.; Gokhan, E.; De Munter, S.; Bollen, M.; Vagnarelli, P.; Peti, W.; Page, R. The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. eLife 2016, 5. [Google Scholar] [CrossRef]
- Wang, Q.; Moyret-Lalle, C.; Couzon, F.; Surbiguet-Clippe, C.; Saurin, J.-C.; Lorca, T.; Navarro, C.; Puisieux, A. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene 2003, 22, 1486–1490. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Daigo, Y.; Aragaki, M.; Ishikawa, K.; Sato, M.; Kaji, M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J. Surg. Oncol. 2012, 106, 423–430. [Google Scholar] [CrossRef]
- Wu, W.J.; Hu, K.S.; Wang, D.S.; Zeng, Z.L.; Zhang, D.S.; Chen, D.L.; Bai, L.; Xu, R.H. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J. Transl. Med. 2013, 11, 142. [Google Scholar] [CrossRef]
- Lub, S.; Maes, A.; Maes, K.; De Veirman, K.; De Bruyne, E.; Menu, E.; Fostier, K.; Kassambara, A.; Moreaux, J.; Hose, D.; et al. Inhibiting the anaphase promoting complex/cyclosome induces a metaphase arrest and cell death in multiple myeloma cells. Oncotarget 2016, 7, 4062–4076. [Google Scholar] [CrossRef]
- Manchado, E.; Guillamot, M.; de Carcer, G.; Eguren, M.; Trickey, M.; Garcia-Higuera, I.; Moreno, S.; Yamano, H.; Canamero, M.; Malumbres, M. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell 2010, 18, 641–654. [Google Scholar] [CrossRef]
- Fujita, T.; Liu, W.; Doihara, H.; Date, H.; Wan, Y. Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin. Cancer Res. 2008, 14, 1966–1975. [Google Scholar] [CrossRef]
- Fujita, T.; Liu, W.; Doihara, H.; Wan, Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am. J. Pathol. 2008, 173, 217–228. [Google Scholar] [CrossRef]
- Engelbert, D.; Schnerch, D.; Baumgarten, A.; Wasch, R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene 2008, 27, 907–917. [Google Scholar] [CrossRef]
- De, K.; Grubb, T.M.; Zalenski, A.A.; Pfaff, K.E.; Pal, D.; Majumder, S.; Summers, M.K.; Venere, M. Hyperphosphorylation of CDH1 in Glioblastoma Cancer Stem Cells Attenuates APC/C(CDH1) Activity and Pharmacologic Inhibition of APC/C(CDH1/CDC20) Compromises Viability. Mol. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Garcia-Higuera, I.; Manchado, E.; Dubus, P.; Canamero, M.; Mendez, J.; Moreno, S.; Malumbres, M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008, 10, 802–811. [Google Scholar] [CrossRef]
- Fuchsberger, T.; Lloret, A.; Vina, J. New Functions of APC/C Ubiquitin Ligase in the Nervous System and Its Role in Alzheimer’s Disease. Int. J. Mol. Sci. 2017, 18, 1057. [Google Scholar] [CrossRef]
- Silva, P.N.; Gigek, C.O.; Leal, M.F.; Bertolucci, P.H.; de Labio, R.W.; Payao, S.L.; Smith Mde, A. Promoter methylation analysis of SIRT3, SMARCA5, HTERT and CDH1 genes in aging and Alzheimer’s disease. J. Alzheimers Dis. 2008, 13, 173–176. [Google Scholar] [CrossRef]
- Vina, J.; Fuchsberger, T.; Giraldo, E.; Lloret, A. APC/Cdh E3 ubiquitin ligase in the pathophysiology of Alzheimers disease. Free Radic. Biol. Med. 2014, 75 (Suppl. 1), S4. [Google Scholar] [CrossRef]
- Zeng, X.; Sigoillot, F.; Gaur, S.; Choi, S.; Pfaff, K.L.; Oh, D.C.; Hathaway, N.; Dimova, N.; Cuny, G.D.; King, R.W. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 2010, 18, 382–395. [Google Scholar] [CrossRef]
- Verma, R.; Peters, N.R.; D’Onofrio, M.; Tochtrop, G.P.; Sakamoto, K.M.; Varadan, R.; Zhang, M.; Coffino, P.; Fushman, D.; Deshaies, R.J.; et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 2004, 306, 117–120. [Google Scholar] [CrossRef]
- Sackton, K.L.; Dimova, N.; Zeng, X.; Tian, W.; Zhang, M.; Sackton, T.B.; Meaders, J.; Pfaff, K.L.; Sigoillot, F.; Yu, H.; et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 2014, 514, 646–649. [Google Scholar] [CrossRef] [Green Version]
- Brito, D.A.; Rieder, C.L. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 2006, 16, 1194–1200. [Google Scholar] [CrossRef]
- Sangodkar, J.; Farrington, C.C.; McClinch, K.; Galsky, M.D.; Kastrinsky, D.B.; Narla, G. All roads lead to PP2A: Exploiting the therapeutic potential of this phosphatase. FEBS J. 2016, 283, 1004–1024. [Google Scholar] [CrossRef]
- Baskaran, R.; Velmurugan, B.K. Protein phosphatase 2A as therapeutic targets in various disease models. Life Sci. 2018, 210, 40–46. [Google Scholar] [CrossRef]
- Chen, W.; Arroyo, J.D.; Timmons, J.C.; Possemato, R.; Hahn, W.C. Cancer-associated PP2A Aalpha subunits induce functional haploinsufficiency and tumorigenicity. Cancer Res. 2005, 65, 8183–8192. [Google Scholar] [CrossRef]
- Ruediger, R.; Ruiz, J.; Walter, G. Human cancer-associated mutations in the Aα subunit of protein phosphatase 2A increase lung cancer incidence in Aα knock-in and knockout mice. Mol. Cell. Biol. 2011, 31, 3832–3844. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Singh, S.; Srivastava, S.K.; Honkanen, R.E.; Reed, E.; Singh, A.P. Modulation of protein phosphatase 2A activity alters androgen-independent growth of prostate cancer cells: Therapeutic implications. Mol. Cancer 2011, 10, 720–731. [Google Scholar] [CrossRef]
- Nobumori, Y.; Shouse, G.P.; Wu, Y.; Lee, K.J.; Shen, B.; Liu, X. B56γ tumor-associated mutations provide new mechanisms for B56γ-PP2A tumor suppressor activity. Mol. Cancer Res. Mcr. 2013, 11, 995–1003. [Google Scholar] [CrossRef]
- Li, M.; Makkinje, A.; Damuni, Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 1996, 271, 11059–11062. [Google Scholar] [CrossRef]
- Liu, H.; Gu, Y.; Wang, H.; Yin, J.; Zheng, G.; Zhang, Z.; Lu, M.; Wang, C.; He, Z. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget 2015, 6, 14913–14925. [Google Scholar] [CrossRef]
- Szymiczek, A.; Pastorino, S.; Larson, D.; Tanji, M.; Pellegrini, L.; Xue, J.; Li, S.; Giorgi, C.; Pinton, P.; Takinishi, Y.; et al. FTY720 inhibits mesothelioma growth in vitro and in a syngeneic mouse model. J. Transl. Med. 2017, 15, 58. [Google Scholar] [CrossRef]
- Kauko, O.; O’Connor, C.M.; Kulesskiy, E.; Sangodkar, J.; Aakula, A.; Izadmehr, S.; Yetukuri, L.; Yadav, B.; Padzik, A.; Laajala, T.D.; et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Wei, D.; Parsels, L.A.; Karnak, D.; Davis, M.A.; Parsels, J.D.; Marsh, A.C.; Zhao, L.; Maybaum, J.; Lawrence, T.S.; Sun, Y.; et al. Inhibition of protein phosphatase 2A radiosensitizes pancreatic cancers by modulating CDC25C/CDK1 and homologous recombination repair. Clin. Cancer Res. 2013, 19, 4422–4432. [Google Scholar] [CrossRef]
- Chung, V.; Mansfield, A.S.; Braiteh, F.; Richards, D.; Durivage, H.; Ungerleider, R.S.; Johnson, F.; Kovach, J.S. Safety, Tolerability, and Preliminary Activity of LB-100, an Inhibitor of Protein Phosphatase 2A, in Patients with Relapsed Solid Tumors: An Open-Label, Dose Escalation, First-in-Human, Phase I Trial. Clin. Cancer Res. 2017, 23, 3277–3284. [Google Scholar] [CrossRef]
- D’Arcy, B.M.; Swingle, M.R.; Papke, C.M.; Abney, K.A.; Bouska, E.S.; Prakash, A.; Honkanen, R.E. The Antitumor Drug LB-100 Is a Catalytic Inhibitor of Protein Phosphatase 2A (PPP2CA) and 5 (PPP5C) Coordinating with the Active-Site Catalytic Metals in PPP5C. Mol. Cancer 2019, 18, 556–566. [Google Scholar] [CrossRef] [Green Version]
- Winkler, C.; De Munter, S.; Van Dessel, N.; Lesage, B.; Heroes, E.; Boens, S.; Beullens, M.; Van Eynde, A.; Bollen, M. The selective inhibition of protein phosphatase-1 results in mitotic catastrophe and impaired tumor growth. J. Cell Sci. 2015, 128, 4526–4537. [Google Scholar] [CrossRef] [Green Version]
- Krzyzosiak, A.; Sigurdardottir, A.; Luh, L.; Carrara, M.; Das, I.; Schneider, K.; Bertolotti, A. Target-Based Discovery of an Inhibitor of the Regulatory Phosphatase PPP1R15B. Cell 2018, 174, 1216–1228.e19. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kataria, M.; Yamano, H. Interplay between Phosphatases and the Anaphase-Promoting Complex/Cyclosome in Mitosis. Cells 2019, 8, 814. https://doi.org/10.3390/cells8080814
Kataria M, Yamano H. Interplay between Phosphatases and the Anaphase-Promoting Complex/Cyclosome in Mitosis. Cells. 2019; 8(8):814. https://doi.org/10.3390/cells8080814
Chicago/Turabian StyleKataria, Meghna, and Hiroyuki Yamano. 2019. "Interplay between Phosphatases and the Anaphase-Promoting Complex/Cyclosome in Mitosis" Cells 8, no. 8: 814. https://doi.org/10.3390/cells8080814




