Dynamin-Like Protein B of Dictyostelium Contributes to Cytokinesis Cooperatively with Other Dynamins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Construction and Transformation
2.3. Antibodies
2.4. Cells Preparations for Microscopy
2.5. Microscopy
2.6. Fluorescence Recovery after Photobleaching (FRAP) Analysis
2.7. Immunoblotting and Co-Immunoprecipitation
2.8. Statistical Analysis
3. Results
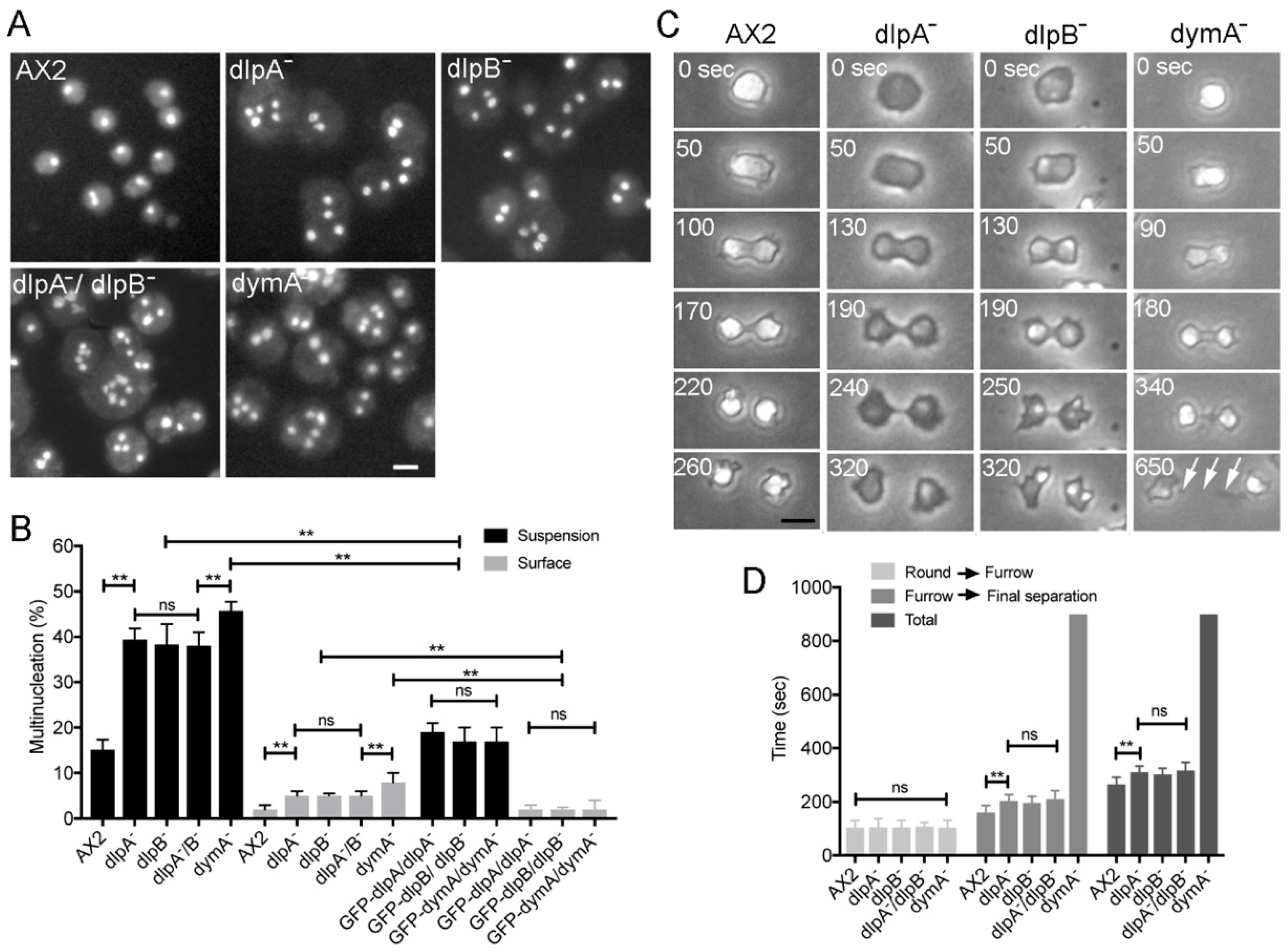
3.1. DlpA, DlpB, and DymA Contribute to Cytokinesis in Different Manners
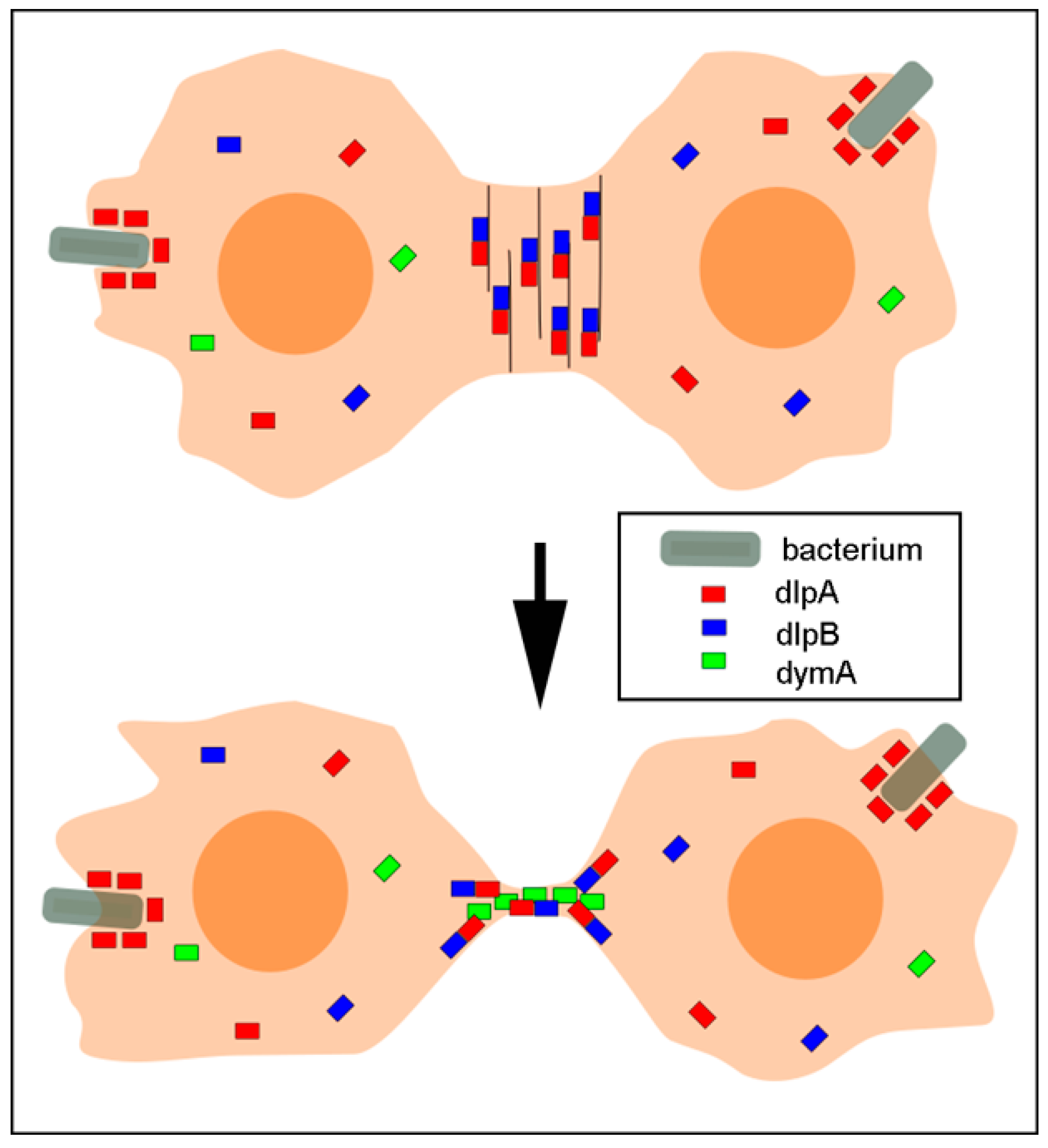
3.2. Three Dynamins Localize at the Cleavage Furrow
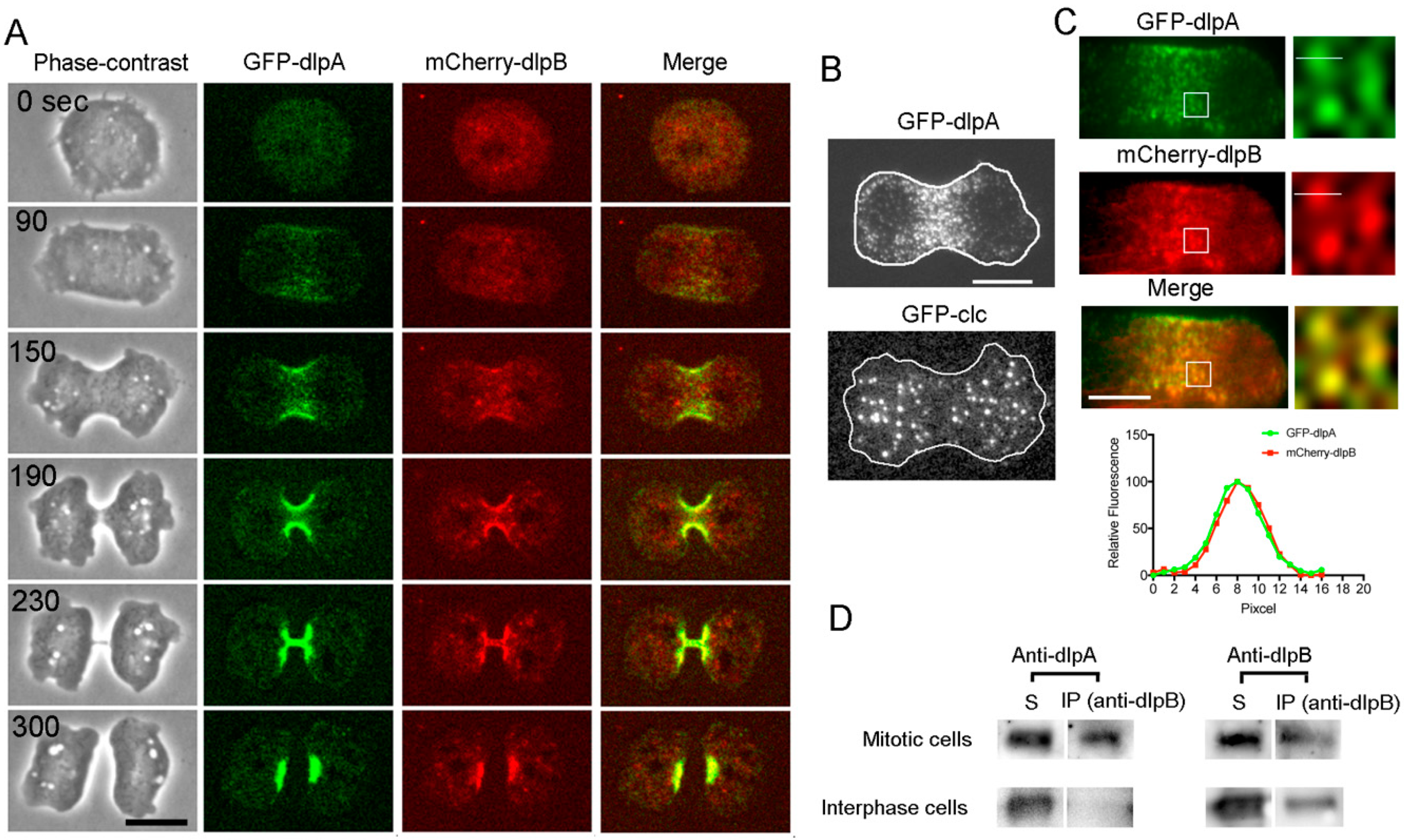
3.3. DlpA and DlpB Colocalize at the Cleavage Furrow
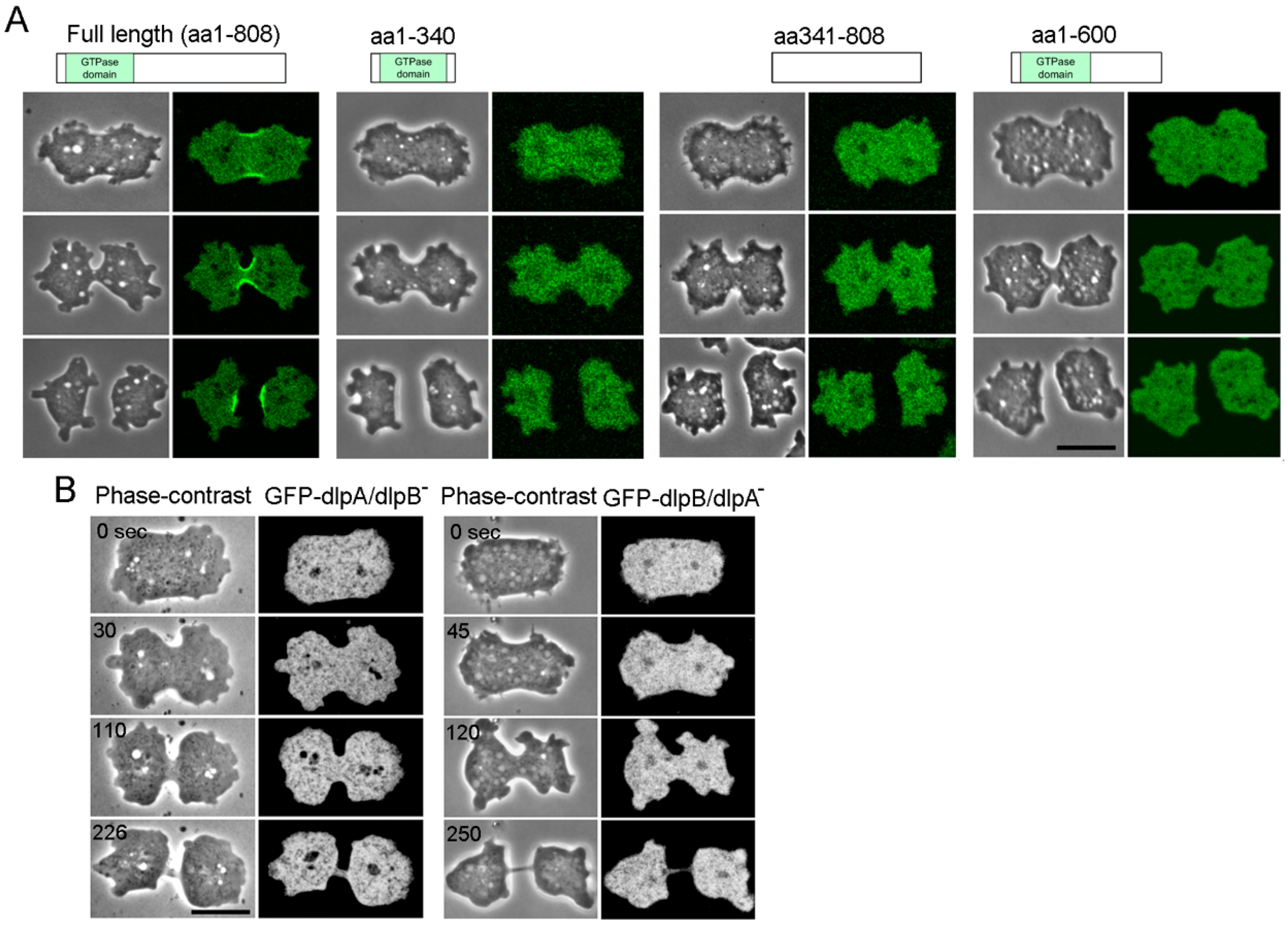
3.4. Both DlpA and DlpB are Required for their Localization to the Furrow
3.5. DlpA and DlpB Associate with Actin Filaments at the Cleavage Furrow
3.6. DlpA and dlpB Stabilize the Actin Filaments at the Cleavage Furrow
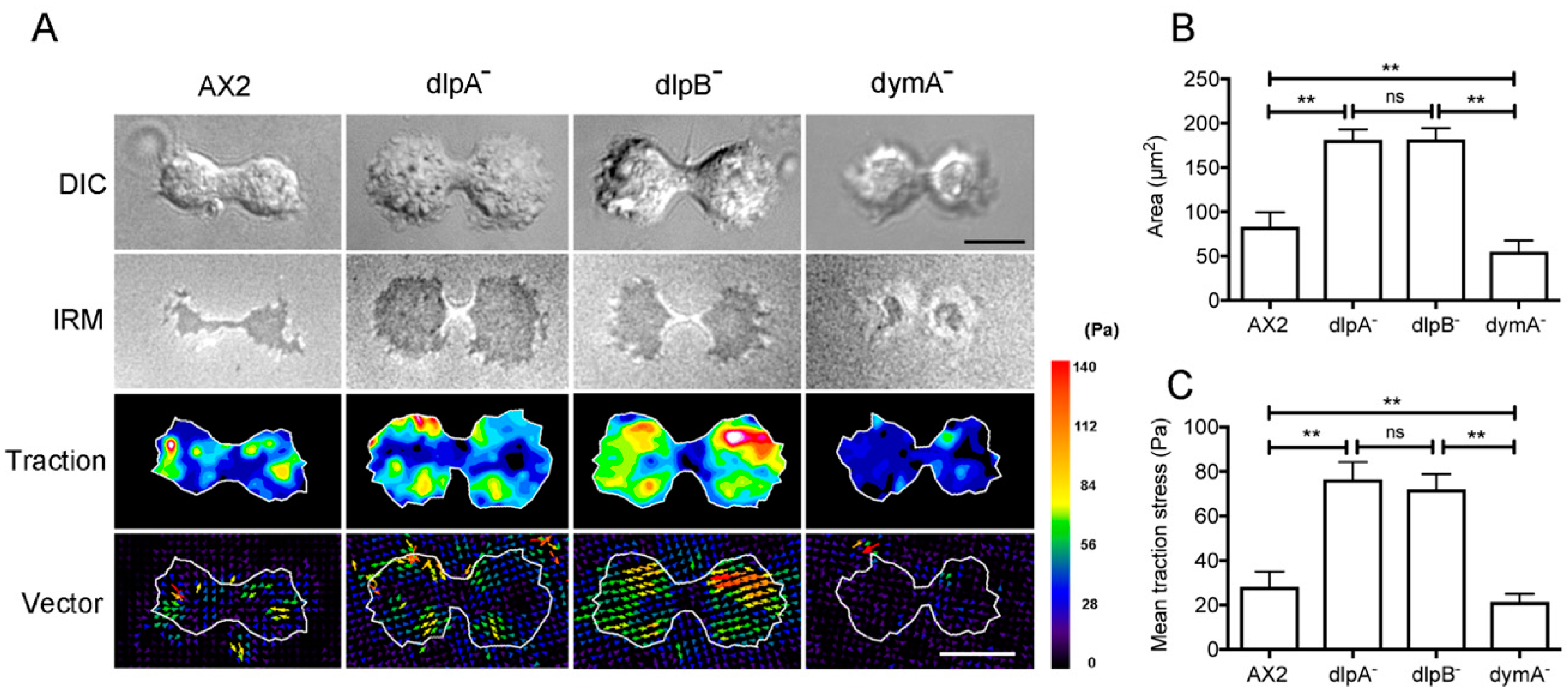
3.7. Dynamins Contribute to Cell-Substratum Adhesion and Traction Force
3.8. DlpA Localizes at the Phagocytic Cup Independently of DlpB
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heymann, J.A.; Hinshaw, J.E. Dynamins at a glance. J. Cell Sci. 2009, 122, 3427–3431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimah, J.R.; Hinshaw, J.E. Structural Insights into the Mechanism of Dynamin Superfamily Proteins. Trends Cell Biol. 2019, 29, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Chircop, M.; Sarcevic, B.; Larsen, M.R.; Malladi, C.S.; Chau, N.; Zavortink, M.; Smith, C.M.; Quan, A.; Anggono, V.; Hains, P.G.; et al. Phosphorylation of dynamin II at serine-764 is associated with cytokinesis. Biochim. Biophys. Acta. 2011, 1813, 1689–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, N.; Nakamura, Y.; Tanabe, K.; Li, S.A.; Takei, K. Dynamin 2 associates with microtubules at mitosis and regulates cell cycle progression. Cell Struct. Funct. 2011, 36, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Schwarz, H.; Jesuthasan, S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp. Cell Res. 2002, 279, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.M.; Skop, A.R.; Euteneuer, U.; Meyer, B.J.; McNiven, M.A. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr. Biol. 2002, 12, 2111–2117. [Google Scholar] [CrossRef]
- Pelissier, A.; Chauvin, J.P.; Lecuit, T. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr. Biol. 2003, 13, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Verma, D.P. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. Embo J. 1996, 15, 695–704. [Google Scholar] [CrossRef]
- Ahn, G.; Kim, H.; Kim, D.H.; Hanh, H.; Yoon, Y.; Singaram, I.; Wijesinghe, K.J.; Johnson, K.A.; Zhuang, X.; Liang, Z.; et al. SH3 Domain-Containing Protein 2 Plays a Crucial Role at the Step of Membrane Tubulation during Cell Plate Formation. Plant Cell 2017, 29, 1388–1405. [Google Scholar] [Green Version]
- Kang, B.H.; Busse, J.S.; Bednarek, S.Y. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 2003, 15, 899–913. [Google Scholar] [CrossRef]
- Miyagishima, S.Y.; Kuwayama, H.; Urushihara, H.; Nakanishi, H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 15202–15207. [Google Scholar] [CrossRef] [Green Version]
- Arakaki, Y.; Fujiwara, T.; Kawai-Toyooka, H.; Kawafune, K.; Featherston, J.; Durand, P.M.; Miyagishima, S.Y.; Nozaki, H. Evolution of cytokinesis-related protein localization during the emergence of multicellularity in volvocine green algae. BMC Evol. Biol. 2017, 17, 243. [Google Scholar] [CrossRef]
- Schlimpert, S.; Wasserstrom, S.; Chandra, G.; Bibb, M.J.; Findlay, K.C.; Flärdh, K.; Buttner, M.J. Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl. Acad. Sci. USA 2017, 114, E6176–E6183. [Google Scholar] [CrossRef]
- Rai, A.; Nothe, H.; Tzvetkov, N.; Korenbaum, E.; Manstein, D.J. Dictyostelium dynamin B modulates cytoskeletal structures and membranous organelles. Cell Mol. Life Sci. 2011, 68, 2751–2767. [Google Scholar] [CrossRef]
- Schimmel, B.G.; Berbusse, G.W.; Naylor, K. Mitochondrial fission and fusion in Dictyostelium discoideum: A search for proteins involved in membrane dynamics. BMC Res. Notes 2012, 5, 505. [Google Scholar] [CrossRef]
- Wienke, D.C.; Knetsch, M.L.; Neuhaus, E.M.; Reedy, M.C.; Manstein, D.J. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 1999, 10, 225–243. [Google Scholar] [CrossRef]
- Masud, A.Y.K.M.; Tsujioka, M.; Miyagishima, S.; Ueda, M.; Yumura, S. Dynamin contributes to cytokinesis by stabilizing actin filaments in the contractile ring. Genes Cells 2013, 18, 621–635. [Google Scholar] [CrossRef]
- Neujahr, R.; Heizer, C.; Gerisch, G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: Redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 1997, 110, 123–137. [Google Scholar]
- Taira, R.; Yumura, S. A novel mode of cytokinesis without cell-substratum adhesion. Sci. Rep. 2017, 7, 17694. [Google Scholar] [CrossRef]
- Tanaka, Y.; Jahan, M.G.S.; Kondo, T.; Nakano, M.; Yumura, S. Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis. Cells 2019, 8, 473. [Google Scholar] [CrossRef]
- Uyeda, T.Q.; Nagasaki, A. Variations on a theme: The many modes of cytokinesis. Curr. Opin. Cell Biol. 2004, 16, 55–60. [Google Scholar] [CrossRef]
- Jahan, M.G.S.; Yumura, S. Traction force and its regulation during cytokinesis in Dictyostelium cells. Eur. J. Cell Biol. 2017, 96, 515–528. [Google Scholar] [CrossRef]
- Pollard, T.D. Nine unanswered questions about cytokinesis. J. Cell Biol 2017, 216, 3007–3016. [Google Scholar] [CrossRef] [Green Version]
- Yumura, S.; Matsuzaki, R.; Kitanishi-Yumura, T. Introduction of macromolecules into living Dictyostelium cells by electroporation. Cell Struct. Funct. 1995, 20, 185–190. [Google Scholar] [CrossRef]
- Yumura, S. A novel low-power laser-mediated transfer of foreign molecules into cells. Sci. Rep. 2016, 6, 22055. [Google Scholar] [CrossRef]
- Linkner, J.; Nordholz, B.; Junemann, A.; Winterhoff, M.; Faix, J. Highly effective removal of floxed Blasticidin S resistance cassettes from Dictyostelium discoideum mutants by extrachromosomal expression of Cre. Eur. J. Cell Biol. 2012, 91, 156–160. [Google Scholar] [CrossRef]
- Manstein, D.J.; Titus, M.A.; De Lozanne, A.; Spudich, J.A. Gene replacement in Dictyostelium: Generation of myosin null mutants. Embo J. 1989, 8, 923–932. [Google Scholar] [CrossRef]
- Okazaki, K.; Yumura, S. Differential association of three actin-bundling proteins with microfilaments in Dictyostelium amoebae. Eur. J. Cell Biol. 1995, 66, 75–81. [Google Scholar]
- Yumura, S.; Yoshida, M.; Betapudi, V.; Licate, L.S.; Iwadate, Y.; Nagasaki, A.; Uyeda, T.Q.; Egelhoff, T.T. Multiple myosin II heavy chain kinases: Roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol. Biol. Cell 2005, 16, 4256–4266. [Google Scholar] [CrossRef]
- Yumura, S.; Mori, H.; Fukui, Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 1984, 99, 894–899. [Google Scholar] [CrossRef]
- Yumura, S.; Ueda, M.; Sako, Y.; Kitanishi-Yumura, T.; Yanagida, T. Multiple mechanisms for accumulation of myosin II filaments at the equator during cytokinesis. Traffic 2008, 9, 2089–2099. [Google Scholar] [CrossRef]
- Yumura, S. Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J. Cell Biol. 2001, 154, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Sochacki, K.A.; Wang, H.; Fang, S.; Canagarajah, B.; Kehr, A.D.; Rice, W.J.; Strub, M.-P.; Taraska, J.W.; Hinshaw, J.E. Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 2018, 560, 258–262. [Google Scholar] [CrossRef]
- Podolski, J.L.; Steck, T.L. Association of deoxyribonuclease I with the pointed ends of actin filaments in human red blood cell membrane skeletons. J. Biol. Chem. 1988, 263, 638–645. [Google Scholar]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin:DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Yumura, S.; Itoh, G.; Kikuta, Y.; Kikuchi, T.; Kitanishi-Yumura, T.; Tsujioka, M. Cell-scale dynamic recycling and cortical flow of the actin-myosin cytoskeleton for rapid cell migration. Biol. Open 2013, 2, 200–209. [Google Scholar] [CrossRef]
- Courtemanche, N.; Pollard, T.D.; Chen, Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat. Cell Biol. 2016, 18, 676–683. [Google Scholar] [CrossRef] [Green Version]
- Uyeda, T.Q.; Kitayama, C.; Yumura, S. Myosin II-independent cytokinesis in Dictyostelium: Its mechanism and implications. Cell Struct. Funct. 2000, 25, 1–10. [Google Scholar] [CrossRef]
- Zang, J.H.; Cavet, G.; Sabry, J.H.; Wagner, P.; Moores, S.L.; Spudich, J.A. On the role of myosin-II in cytokinesis: Division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell 1997, 8, 2617–2629. [Google Scholar] [CrossRef]
- Yumura, S.; Uyeda, T.Q. Myosins and cell dynamics in cellular slime molds. Int. Rev. Cytol. 2003, 224, 173–225. [Google Scholar]
- Makiuchi, T.; Santos, H.J.; Tachibana, H.; Nozaki, T. Hetero-oligomer of dynamin-related proteins participates in the fission of highly divergent mitochondria from Entamoeba histolytica. Sci. Rep. 2017, 7, 13439. [Google Scholar] [CrossRef] [Green Version]
- Gerald, N.J.; Damer, C.K.; O’Halloran, T.J.; De Lozanne, A. Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cell Motil. Cytoskelet. 2001, 48, 213–223. [Google Scholar] [CrossRef]
- Kwak, E.; Gerald, N.; Larochelle, D.A.; Vithalani, K.K.; Niswonger, M.L.; Maready, M.; De Lozanne, A. LvsA, a protein related to the mouse beige protein, is required for cytokinesis in Dictyostelium. Mol. Biol. Cell 1999, 10, 4429–4439. [Google Scholar] [CrossRef]
- Konopka, C.A.; Schleede, J.B.; Skop, A.R.; Bednarek, S.Y. Dynamin and cytokinesis. Traffic 2006, 7, 239–247. [Google Scholar] [CrossRef]
- Smith, C.M.; Chircop, M. Clathrin-Mediated Endocytic Proteins are Involved in Regulating Mitotic Progression and Completion. Traffic 2012, 13, 1628–1641. [Google Scholar] [CrossRef]
- Gu, C.; Yaddanapudi, S.; Weins, A.; Osborn, T.; Reiser, J.; Pollak, M.; Hartwig, J.; Sever, S. Direct dynamin-actin interactions regulate the actin cytoskeleton. Embo J. 2010, 29, 3593–3606. [Google Scholar] [CrossRef]
- Gu, C.; Chang, J.; Shchedrina, V.A.; Pham, V.A.; Hartwig, J.H.; Suphamungmee, W.; Lehman, W.; Hyman, B.T.; Bacskai, B.J.; Sever, S. Regulation of dynamin oligomerization in cells: The role of dynamin-actin interactions and its GTPase activity. Traffic 2014, 15, 819–838. [Google Scholar] [CrossRef]
- Marie-Anais, F.; Mazzolini, J.; Herit, F.; Niedergang, F. Dynamin-Actin Cross Talk Contributes to Phagosome Formation and Closure. Traffic 2016, 17, 487–499. [Google Scholar] [CrossRef] [Green Version]
- Sever, S.; Chang, J.; Gu, C. Dynamin rings: Not just for fission. Traffic 2013, 14, 1194–1199. [Google Scholar] [CrossRef]
- Witke, W.; Podtelejnikov, A.V.; Di Nardo, A.; Sutherland, J.D.; Gurniak, C.B.; Dotti, C.; Mann, M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. Embo J. 1998, 17, 967–976. [Google Scholar] [CrossRef]
- Yamada, H.; Abe, T.; Satoh, A.; Okazaki, N.; Tago, S.; Kobayashi, K.; Yoshida, Y.; Oda, Y.; Watanabe, M.; Tomizawa, K.; et al. Stabilization of actin bundles by a dynamin 1/cortactin ring complex is necessary for growth cone filopodia. J. Neurosci. 2013, 33, 4514–4526. [Google Scholar] [CrossRef]
- Schafer, D.A.; Weed, S.A.; Binns, D.; Karginov, A.V.; Parsons, J.T.; Cooper, J.A. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 2002, 12, 1852–1857. [Google Scholar] [CrossRef]
- Schlunck, G.; Damke, H.; Kiosses, W.B.; Rusk, N.; Symons, M.H.; Waterman-Storer, C.M.; Schmid, S.L.; Schwartz, M.A. Modulation of Rac localization and function by dynamin. Mol. Biol Cell 2004, 15, 256–267. [Google Scholar] [CrossRef]
- Chen, Q.; Pollard, T.D. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 2011, 195, 485–498. [Google Scholar] [CrossRef] [Green Version]
- Schramm, A.C.; Hocky, G.M.; Voth, G.A.; Blanchoin, L.; Martiel, J.L.; De La Cruz, E.M. Actin Filament Strain Promotes Severing and Cofilin Dissociation. Biophys. J. 2017, 112, 2624–2633. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Ando, Y.; Yasuda, S.; Hosoya, H.; Watanabe, N.; Ishizaki, T.; Narumiya, S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell 2008, 19, 2328–2338. [Google Scholar] [CrossRef]
- Uchida, K.S.; Yumura, S. Dynamics of novel feet of Dictyostelium cells during migration. J. Cell Sci. 2004, 117, 1443–1455. [Google Scholar] [CrossRef] [Green Version]
- Kruchten, A.E.; McNiven, M.A. Dynamin as a mover and pincher during cell migration and invasion. J. Cell Sci. 2006, 119, 1683–1690. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, G.C.; Slepnev, V.I.; Neff, L.; Ringstad, N.; Takei, K.; Daniell, L.; Kim, W.; Cao, H.; McNiven, M.; Baron, R.; et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 2000, 150, 377–389. [Google Scholar] [CrossRef]
- Gueho, A.; Bosmani, C.; Gopaldass, N.; Molle, V.; Soldati, T.; Letourneur, F. Dictyostelium EHD associates with Dynamin and participates in phagosome maturation. J. Cell Sci. 2016, 129, 2354–2367. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujimoto, K.; Tanaka, M.; Rana, A.Y.K.M.M.; Jahan, M.G.S.; Itoh, G.; Tsujioka, M.; Uyeda, T.Q.P.; Miyagishima, S.-y.; Yumura, S. Dynamin-Like Protein B of Dictyostelium Contributes to Cytokinesis Cooperatively with Other Dynamins. Cells 2019, 8, 781. https://doi.org/10.3390/cells8080781
Fujimoto K, Tanaka M, Rana AYKMM, Jahan MGS, Itoh G, Tsujioka M, Uyeda TQP, Miyagishima S-y, Yumura S. Dynamin-Like Protein B of Dictyostelium Contributes to Cytokinesis Cooperatively with Other Dynamins. Cells. 2019; 8(8):781. https://doi.org/10.3390/cells8080781
Chicago/Turabian StyleFujimoto, Koushiro, Masahito Tanaka, A.Y. K. Md. Masud Rana, Md. Golam Sarowar Jahan, Go Itoh, Masatsune Tsujioka, Taro Q. P. Uyeda, Shin-ya Miyagishima, and Shigehiko Yumura. 2019. "Dynamin-Like Protein B of Dictyostelium Contributes to Cytokinesis Cooperatively with Other Dynamins" Cells 8, no. 8: 781. https://doi.org/10.3390/cells8080781
APA StyleFujimoto, K., Tanaka, M., Rana, A. Y. K. M. M., Jahan, M. G. S., Itoh, G., Tsujioka, M., Uyeda, T. Q. P., Miyagishima, S.-y., & Yumura, S. (2019). Dynamin-Like Protein B of Dictyostelium Contributes to Cytokinesis Cooperatively with Other Dynamins. Cells, 8(8), 781. https://doi.org/10.3390/cells8080781





