Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise
Abstract
1. Introduction
2. AMPK and MTORC1 Pathways
2.1. AMPK/MTORC1 Axis in Organelle Quality Control during Exercise
2.2. MTORC1 Regulators and Exercise: Recent Data on DGKs, FOXO, eIF3f and Cellular Trafficking
3. AMPK and FOXO Transcription Factors
3.1. FOXO Homologues in Energy Metabolism and Post-Translational Modifications
3.2. AMPK/FOXO Axis in Organelle Quality Control during Exercise
3.3. Exercise and Autophagy/Mitophagy Flux
4. The Emerging Roles of Parkin and FOXO3-Dependant Mul1 Pathway in Organelle Turnover and Adaptations to Exercise
5. Exercise in Hypoxia
6. Muscle Contraction Regimens and Cell Component Turnover
7. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Hood, D.A.; Memme, J.M.; Oliveira, A.N.; Triolo, M. Maintenance of Skeletal Muscle Mitochondria in Health, Exercise, and Aging. Annu. Rev. Physiol. 2018, 81, 19–41. [Google Scholar] [CrossRef] [PubMed]
- McGlory, C.; van Vliet, S.; Stokes, T.; Mittendorfer, B.; Phillips, S.M. The impact of exercise and nutrition on the regulation of skeletal muscle mass. J. Physiol. 2018, 597, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Bernardi, H.; Py, G.; Candau, R.B. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R956–R969. [Google Scholar] [CrossRef] [PubMed]
- Kjøbsted, R.; Hingst, J.R.; Fentz, J.; Foretz, M.; Sanz, M.-N.; Pehmøller, C.; Shum, M.; Marette, A.; Mounier, R.; Treebak, J.T.; et al. AMPK in skeletal muscle function and metabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 1741–1777. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Candau, R.B.; Csibi, A.; Pagano, A.F.; Raibon, A.; Bernardi, H. The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am. J. Physiol. Cell Physiol. 2012, 303, C475–C485. [Google Scholar] [CrossRef]
- Marsin, A.-S.; Bouzin, C.; Bertrand, L.; Hue, L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J. Biol. Chem. 2002, 277, 30778–30783. [Google Scholar] [CrossRef]
- Kudo, N.; Barr, A.J.; Barr, R.L.; Desai, S.; Lopaschuk, G.D. High rates of fatty acid oxidation during reperfusion of ischemic hearts are associated with a decrease in malonyl-CoA levels due to an increase in 5′-AMP-activated protein kinase inhibition of acetyl-CoA carboxylase. J. Biol. Chem. 1995, 270, 17513–17520. [Google Scholar] [CrossRef]
- Vavvas, D.; Apazidis, A.; Saha, A.K.; Gamble, J.; Patel, A.; Kemp, B.E.; Witters, L.A.; Ruderman, N.B. Contraction-induced changes in acetyl-CoA carboxylase and 5′-AMP-activated kinase in skeletal muscle. J. Biol. Chem. 1997, 272, 13255–13261. [Google Scholar] [CrossRef]
- Derave, W.; Ai, H.; Ihlemann, J.; Witters, L.A.; Kristiansen, S.; Richter, E.A.; Ploug, T. Dissociation of AMP-activated protein kinase activation and glucose transport in contracting slow-twitch muscle. Diabetes 2000, 49, 1281–1287. [Google Scholar] [CrossRef]
- Salt, I.P.; Connell, J.M.; Gould, G.W. 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) inhibits insulin-stimulated glucose transport in 3T3-L1 adipocytes. Diabetes 2000, 49, 1649–1656. [Google Scholar] [CrossRef]
- Park, H.; Kaushik, V.K.; Constant, S.; Prentki, M.; Przybytkowski, E.; Ruderman, N.B.; Saha, A.K. Coordinate regulation of malonyl-CoA decarboxylase, sn-glycerol-3-phosphate acyltransferase, and acetyl-CoA carboxylase by AMP-activated protein kinase in rat tissues in response to exercise. J. Biol. Chem. 2002, 277, 32571–32577. [Google Scholar] [CrossRef] [PubMed]
- Winder, W.W.; Hardie, D.G. Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. Am. J. Physiol. 1996, 270, E299–E304. [Google Scholar] [CrossRef] [PubMed]
- Wojtaszewski, J.F.; Nielsen, P.; Hansen, B.F.; Richter, E.A.; Kiens, B. Isoform-specific and exercise intensity-dependent activation of 5′-AMP-activated protein kinase in human skeletal muscle. J. Physiol. 2000, 528 Pt 1, 221–226. [Google Scholar] [CrossRef]
- Stein, S.C.; Woods, A.; Jones, N.A.; Davison, M.D.; Carling, D. The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 2000, 345 Pt 3, 437–443. [Google Scholar] [CrossRef]
- Hawley, S.A.; Selbert, M.A.; Goldstein, E.G.; Edelman, A.M.; Carling, D.; Hardie, D.G. 5′-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin activates the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J. Biol. Chem. 1995, 270, 27186–27191. [Google Scholar] [CrossRef] [PubMed]
- Hawley, S.A.; Pan, D.A.; Mustard, K.J.; Ross, L.; Bain, J.; Edelman, A.M.; Frenguelli, B.G.; Hardie, D.G. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005, 2, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-P.; Leiper, F.C.; Woods, A.; Carling, D.; Carlson, M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 2003, 100, 8839–8843. [Google Scholar] [CrossRef]
- Woods, A.; Johnstone, S.R.; Dickerson, K.; Leiper, F.C.; Fryer, L.G.D.; Neumann, D.; Schlattner, U.; Wallimann, T.; Carlson, M.; Carling, D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003, 13, 2004–2008. [Google Scholar] [CrossRef]
- Herrero-Martín, G.; Høyer-Hansen, M.; García-García, C.; Fumarola, C.; Farkas, T.; López-Rivas, A.; Jäättelä, M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009, 28, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Sanders, M.J.; Underwood, E.; Heath, R.; Mayer, F.V.; Carmena, D.; Jing, C.; Walker, P.A.; Eccleston, J.F.; Haire, L.F.; et al. Structure of mammalian AMPK and its regulation by ADP. Nature 2011, 472, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.P.; Helps, N.R.; Cohen, P.T.; Hardie, D.G. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett. 1995, 377, 421–425. [Google Scholar] [PubMed]
- McBride, A.; Ghilagaber, S.; Nikolaev, A.; Hardie, D.G. The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab. 2009, 9, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Csibi, A.; Raibon, A.; Cornille, K.; Gay, S.; Bernardi, H.; Candau, R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J. Cell. Biochem. 2012, 113, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.F.; Kurth-Kraczek, E.J.; Winder, W.W. Chronic activation of 5′-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. J. Appl. Physiol. Bethesda MD 1985 1999, 87, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Marsin, A.S.; Bertrand, L.; Rider, M.H.; Deprez, J.; Beauloye, C.; Vincent, M.F.; Van den Berghe, G.; Carling, D.; Hue, L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr. Biol. 2000, 10, 1247–1255. [Google Scholar] [CrossRef]
- Thomson, D.M.; Herway, S.T.; Fillmore, N.; Kim, H.; Brown, J.D.; Barrow, J.R.; Winder, W.W. AMP-activated protein kinase phosphorylates transcription factors of the CREB family. J. Appl. Physiol. Bethesda MD 1985 2008, 104, 429–438. [Google Scholar] [CrossRef]
- Stoppani, J.; Hildebrandt, A.L.; Sakamoto, K.; Cameron-Smith, D.; Goodyear, L.J.; Neufer, P.D. AMP-activated protein kinase activates transcription of the UCP3 and HKII genes in rat skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E1239–E1248. [Google Scholar] [CrossRef]
- Barnes, T.; Di Sebastiano, K.M.; Vlavcheski, F.; Quadrilatero, J.; Tsiani, E.; Mourtzakis, M. Glutamate increases glucose uptake in L6 myotubes in a concentration- and time-dependent manner that is mediated by AMPK. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2018, 43, 1307–1313. [Google Scholar] [CrossRef]
- Jørgensen, S.B.; Viollet, B.; Andreelli, F.; Frøsig, C.; Birk, J.B.; Schjerling, P.; Vaulont, S.; Richter, E.A.; Wojtaszewski, J.F.P. Knockout of the alpha2 but not alpha1 5′-AMP-activated protein kinase isoform abolishes 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranosidebut not contraction-induced glucose uptake in skeletal muscle. J. Biol. Chem. 2004, 279, 1070–1079. [Google Scholar] [CrossRef]
- Jäger, S.; Handschin, C.; St-Pierre, J.; Spiegelman, B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA 2007, 104, 12017–12022. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Sanchez, A.M.J.; Candau, R.B.; Bernardi, H. FoxO transcription factors: Their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. CMLS 2014, 71, 1657–1671. [Google Scholar] [CrossRef] [PubMed]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Holz, M.K.; Blenis, J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J. Biol. Chem. 2005, 280, 26089–26093. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 2002, 35, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Pullen, N.; Dennis, P.B.; Andjelkovic, M.; Dufner, A.; Kozma, S.C.; Hemmings, B.A.; Thomas, G. Phosphorylation and activation of p70s6k by PDK1. Science 1998, 279, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Youtani, T.; Tomoo, K.; Ishida, T.; Miyoshi, H.; Miura, K. Regulation of human eIF4E by 4E-BP1: Binding analysis using surface plasmon resonance. IUBMB Life 2000, 49, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Youtani, T.; Ide, H.; Hori, H.; Okamoto, K.; Ishikawa, M.; Wakiyama, M.; Nishino, T.; Ishida, T.; Miura, K. Binding analysis of Xenopus laevis translation initiation factor 4E (eIF4E) in initiation complex formation. J. Biochem. (Tokyo) 1999, 126, 897–904. [Google Scholar] [CrossRef]
- Pagano, A.F.; Py, G.; Bernardi, H.; Candau, R.B.; Sanchez, A.M.J. Autophagy and protein turnover signaling in slow-twitch muscle during exercise. Med. Sci. Sports Exerc. 2014, 46, 1314–1325. [Google Scholar] [CrossRef]
- Cheng, S.W.Y.; Fryer, L.G.D.; Carling, D.; Shepherd, P.R. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J. Biol. Chem. 2004, 279, 15719–15722. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Hardie, D.G. AMPK and Raptor: Matching cell growth to energy supply. Mol. Cell 2008, 30, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.M.; Tipton, K.D.; Aarsland, A.; Wolf, S.E.; Wolfe, R.R. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am. J. Physiol. 1997, 273, E99–E107. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.M. Protein requirements and supplementation in strength sports. Nutrition 2004, 20, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.R.; Phillips, S.M.; Babraj, J.A.; Smith, K.; Rennie, M.J. Myofibrillar and collagen protein synthesis in human skeletal muscle in young men after maximal shortening and lengthening contractions. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E1153–E1159. [Google Scholar] [CrossRef] [PubMed]
- Lysenko, E.A.; Popov, D.V.; Vepkhvadze, T.F.; Sharova, A.P.; Vinogradova, O.L. Moderate-Intensity Strength Exercise to Exhaustion Results in More Pronounced Signaling Changes in Skeletal Muscles of Strength-Trained Compared with Untrained Individuals. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mascher, H.; Andersson, H.; Nilsson, P.-A.; Ekblom, B.; Blomstrand, E. Changes in signalling pathways regulating protein synthesis in human muscle in the recovery period after endurance exercise. Acta Physiol. Oxf. Engl. 2007, 191, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Mascher, H.; Ekblom, B.; Rooyackers, O.; Blomstrand, E. Enhanced rates of muscle protein synthesis and elevated mTOR signalling following endurance exercise in human subjects. Acta Physiol. Oxf. Engl. 2011, 202, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Esbjörnsson, M.; Rundqvist, H.C.; Mascher, H.; Österlund, T.; Rooyackers, O.; Blomstrand, E.; Jansson, E. Sprint exercise enhances skeletal muscle p70S6k phosphorylation and more so in women than in men. Acta Physiol. Oxf. Engl. 2012, 205, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Nader, G.A.; Esser, K.A. Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J. Appl. Physiol. Bethesda Md 1985 2001, 90, 1936–1942. [Google Scholar] [CrossRef] [PubMed]
- Luciano, T.F.; Marques, S.O.; Pieri, B.L.; de Souza, D.R.; Araújo, L.V.; Nesi, R.T.; Scheffer, D.L.; Comin, V.H.; Pinho, R.A.; Muller, A.P.; et al. Responses of skeletal muscle hypertrophy in Wistar rats to different resistance exercise models. Physiol. Res. 2017, 66, 317–323. [Google Scholar] [PubMed]
- Ogasawara, R.; Suginohara, T. Rapamycin-insensitive mechanistic target of rapamycin regulates basal and resistance exercise-induced muscle protein synthesis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Atherton, P.J.; Babraj, J.; Smith, K.; Singh, J.; Rennie, M.J.; Wackerhage, H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 786–788. [Google Scholar]
- Dreyer, H.C.; Fujita, S.; Cadenas, J.G.; Chinkes, D.L.; Volpi, E.; Rasmussen, B.B. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J. Physiol. 2006, 576, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Vissing, K.; McGee, S.L.; Farup, J.; Kjølhede, T.; Vendelbo, M.H.; Jessen, N. Differentiated mTOR but not AMPK signaling after strength vs endurance exercise in training-accustomed individuals. Scand. J. Med. Sci. Sports 2013, 23, 355–366. [Google Scholar] [CrossRef]
- Ogasawara, R.; Sato, K.; Matsutani, K.; Nakazato, K.; Fujita, S. The order of concurrent endurance and resistance exercise modifies mTOR signaling and protein synthesis in rat skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1155–E1162. [Google Scholar] [CrossRef]
- Hornberger, T.A.; Chien, S. Mechanical stimuli and nutrients regulate rapamycin-sensitive signaling through distinct mechanisms in skeletal muscle. J. Cell. Biochem. 2006, 97, 1207–1216. [Google Scholar] [CrossRef]
- Hornberger, T.A.; Stuppard, R.; Conley, K.E.; Fedele, M.J.; Fiorotto, M.L.; Chin, E.R.; Esser, K.A. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem. J. 2004, 380, 795–804. [Google Scholar] [CrossRef]
- Hornberger, T.A.; Chu, W.K.; Mak, Y.W.; Hsiung, J.W.; Huang, S.A.; Chien, S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc. Natl. Acad. Sci. USA 2006, 103, 4741–4746. [Google Scholar] [CrossRef] [PubMed]
- West, D.W.D.; Kujbida, G.W.; Moore, D.R.; Atherton, P.; Burd, N.A.; Padzik, J.P.; De Lisio, M.; Tang, J.E.; Parise, G.; Rennie, M.J.; et al. Resistance exercise-induced increases in putative anabolic hormones do not enhance muscle protein synthesis or intracellular signalling in young men. J. Physiol. 2009, 587, 5239–5247. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; McCarthy, J.J.; Fedele, M.J.; Esser, K.A. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J. Physiol. 2011, 589, 1831–1846. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1993, 268, 14553–14556. [Google Scholar] [PubMed]
- Drummond, M.J.; Fry, C.S.; Glynn, E.L.; Dreyer, H.C.; Dhanani, S.; Timmerman, K.L.; Volpi, E.; Rasmussen, B.B. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J. Physiol. 2009, 587, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Frey, J.W.; Hornberger, T.A. Mechanical stimulation induces mTOR signaling via an ERK-independent mechanism: Implications for a direct activation of mTOR by phosphatidic acid. PLoS ONE 2012, 7, e47258. [Google Scholar] [CrossRef] [PubMed]
- Veverka, V.; Crabbe, T.; Bird, I.; Lennie, G.; Muskett, F.W.; Taylor, R.J.; Carr, M.D. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: Compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene 2008, 27, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.-S.; Sun, Y.; Arauz, E.; Jiang, Y.; Chen, J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J. Biol. Chem. 2011, 286, 29568–29574. [Google Scholar] [CrossRef] [PubMed]
- You, J.-S.; Lincoln, H.C.; Kim, C.-R.; Frey, J.W.; Goodman, C.A.; Zhong, X.-P.; Hornberger, T.A. The role of diacylglycerol kinase ζ and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J. Biol. Chem. 2014, 289, 1551–1563. [Google Scholar] [CrossRef]
- You, J.-S.; Dooley, M.S.; Kim, C.-R.; Kim, E.-J.; Xu, W.; Goodman, C.A.; Hornberger, T.A. A DGKζ-FoxO-ubiquitin proteolytic axis controls fiber size during skeletal muscle remodeling. Sci. Signal. 2018, 11, eaao6847. [Google Scholar] [CrossRef]
- Lagirand-Cantaloube, J.; Offner, N.; Csibi, A.; Leibovitch, M.P.; Batonnet-Pichon, S.; Tintignac, L.A.; Segura, C.T.; Leibovitch, S.A. The initiation factor eIF3-f is a major target for Atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008, 27, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Csibi, A.; Raibon, A.; Docquier, A.; Lagirand-Cantaloube, J.; Leibovitch, M.-P.; Leibovitch, S.A.; Bernardi, H. eIF3f: A central regulator of the antagonism atrophy/hypertrophy in skeletal muscle. Int. J. Biochem. Cell Biol. 2013, 45, 2158–2162. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Cornille, K.; Leibovitch, M.-P.; Poupon, A.; Tintignac, L.A.; Sanchez, A.M.J.; Leibovitch, S.A. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE 2010, 5, e8994. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Leibovitch, M.P.; Cornille, K.; Tintignac, L.A.; Leibovitch, S.A. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J. Biol. Chem. 2009, 284, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Docquier, A.; Pavlin, L.; Raibon, A.; Bertrand-Gaday, C.; Sar, C.; Leibovitch, S.; Candau, R.; Bernardi, H. eIF3f depletion impedes mouse embryonic development, reduces adult skeletal muscle mass and amplifies muscle loss during disuse. J. Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Moore, D.R.; Hodson, N.; Ward, C.; Dent, J.R.; O’Leary, M.F.; Shaw, A.M.; Hamilton, D.L.; Sarkar, S.; Gangloff, Y.-G.; et al. Resistance exercise initiates mechanistic target of rapamycin (mTOR) translocation and protein complex co-localisation in human skeletal muscle. Sci. Rep. 2017, 7, 5028. [Google Scholar] [CrossRef]
- Groenewoud, M.J.; Zwartkruis, F.J.T. Rheb and Rags come together at the lysosome to activate mTORC1. Biochem. Soc. Trans. 2013, 41, 951–955. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef]
- Jacobs, B.L.; You, J.-S.; Frey, J.W.; Goodman, C.A.; Gundermann, D.M.; Hornberger, T.A. Eccentric contractions increase the phosphorylation of tuberous sclerosis complex-2 (TSC2) and alter the targeting of TSC2 and the mechanistic target of rapamycin to the lysosome. J. Physiol. 2013, 591, 4611–4620. [Google Scholar] [CrossRef]
- Van Vliet, S.; Shy, E.L.; Abou Sawan, S.; Beals, J.W.; West, D.W.; Skinner, S.K.; Ulanov, A.V.; Li, Z.; Paluska, S.A.; Parsons, C.M.; et al. Consumption of whole eggs promotes greater stimulation of postexercise muscle protein synthesis than consumption of isonitrogenous amounts of egg whites in young men. Am. J. Clin. Nutr. 2017, 106, 1401–1412. [Google Scholar] [CrossRef]
- Abou Sawan, S.; van Vliet, S.; West, D.W.D.; Beals, J.W.; Paluska, S.A.; Burd, N.A.; Moore, D.R. Whole egg, but not egg white ingestion, induces mTOR co-localization with the lysosome after resistance exercise in trained young men. Am. J. Physiol. Cell Physiol. 2018, 315, C537–C543. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Mazumi, T.; Handa, T.; Miyajima, K. Effects of 1,2-diacylglycerol and cholesterol on the hydrolysis activity of phospholipase D in egg-yolk phosphatidylcholine bilayers. Biochim. Biophys. Acta 1993, 1145, 293–297. [Google Scholar] [CrossRef]
- Yamamoto, I.; Konto, A.; Handa, T.; Miyajima, K. Regulation of phospholipase D activity by neutral lipids in egg-yolk phosphatidylcholine small unilamellar vesicles and by calcium ion in aqueous medium. Biochim. Biophys. Acta 1995, 1233, 21–26. [Google Scholar] [CrossRef][Green Version]
- Menon, D.; Salloum, D.; Bernfeld, E.; Gorodetsky, E.; Akselrod, A.; Frias, M.A.; Sudderth, J.; Chen, P.-H.; DeBerardinis, R.; Foster, D.A. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem. 2017, 292, 6303–6311. [Google Scholar] [CrossRef] [PubMed]
- Castellano, B.M.; Thelen, A.M.; Moldavski, O.; Feltes, M.; van der Welle, R.E.N.; Mydock-McGrane, L.; Jiang, X.; van Eijkeren, R.J.; Davis, O.B.; Louie, S.M.; et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 2017, 355, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.Y.; Huang, W.C.; Su, C.W.; Lee, K.W.; Chi, H.C.; Lin, C.T.; Chen, S.-T.; Huang, K.M.; Tsai, M.S.; Yu, H.P.; et al. FoxO6 and PGC-1α form a regulatory loop in myogenic cells. Biosci. Rep. 2013, 33, e00045. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Geles, K.G.; Paik, J.-H.; DePinho, R.A.; Tjian, R. Codependent activators direct myoblast-specific MyoD transcription. Dev. Cell 2008, 15, 534–546. [Google Scholar] [CrossRef]
- Furuyama, T.; Kitayama, K.; Yamashita, H.; Mori, N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem. J. 2003, 375, 365–371. [Google Scholar] [CrossRef]
- Kamei, Y.; Mizukami, J.; Miura, S.; Suzuki, M.; Takahashi, N.; Kawada, T.; Taniguchi, T.; Ezaki, O. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003, 536, 232–236. [Google Scholar] [CrossRef]
- Tsuchida, A.; Yamauchi, T.; Ito, Y.; Hada, Y.; Maki, T.; Takekawa, S.; Kamon, J.; Kobayashi, M.; Suzuki, R.; Hara, K.; et al. Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J. Biol. Chem. 2004, 279, 30817–30822. [Google Scholar] [CrossRef]
- Bastie, C.C.; Nahlé, Z.; McLoughlin, T.; Esser, K.; Zhang, W.; Unterman, T.; Abumrad, N.A. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J. Biol. Chem. 2005, 280, 14222–14229. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Chiacchiera, F.; Grossi, V.; Matrone, A.; Latorre, D.; Simonatto, M.; Fusella, A.; Ryall, J.G.; Finley, L.W.S.; Haigis, M.C.; et al. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell. Mol. Life Sci. CMLS 2013, 70, 2015–2029. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Alt, F.W.; Cheng, H.-L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Kim, J.; Yancopoulos, G.; Valenzuela, D.; Murphy, A.; et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef] [PubMed]
- Slopack, D.; Roudier, E.; Liu, S.T.K.; Nwadozi, E.; Birot, O.; Haas, T.L. Forkhead BoxO transcription factors restrain exercise-induced angiogenesis. J. Physiol. 2014, 592, 4069–4082. [Google Scholar] [CrossRef]
- Sanchez, A.M.J. FoxO transcription factors and endurance training: A role for FoxO1 and FoxO3 in exercise-induced angiogenesis. J. Physiol. 2015, 593, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.F.; Yan, X.; Zhu, M.J.; Du, M. AMP-activated protein kinase enhances the expression of muscle-specific ubiquitin ligases despite its activation of IGF-1/Akt signaling in C2C12 myotubes. J. Cell. Biochem. 2009, 108, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Candau, R.; Bernardi, H. AMP-activated protein kinase stabilizes FOXO3 in primary myotubes. Biochem. Biophys. Res. Commun. 2018, 499, 493–498. [Google Scholar] [CrossRef]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Kops, G.J.; de Ruiter, N.D.; De Vries-Smits, A.M.; Powell, D.R.; Bos, J.L.; Burgering, B.M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 1999, 398, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Tang, E.D.; Zhu, T.; Greenberg, M.E.; Vojtek, A.B.; Guan, K.L. Serum- and glucocorticoid-inducible kinase SGK phosphorylates and negatively regulates B-Raf. J. Biol. Chem. 2001, 276, 31620–31626. [Google Scholar] [CrossRef] [PubMed]
- Bertaggia, E.; Coletto, L.; Sandri, M. Posttranslational modifications control FoxO3 activity during denervation. Am. J. Physiol. Cell Physiol. 2012, 302, C587–C596. [Google Scholar] [CrossRef] [PubMed]
- Senf, S.M.; Sandesara, P.B.; Reed, S.A.; Judge, A.R. p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am. J. Physiol. Cell Physiol. 2011, 300, C1490–C1501. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Mammucari, C.; Milan, G.; Romanello, V.; Masiero, E.; Rudolf, R.; Del Piccolo, P.; Burden, S.J.; Di Lisi, R.; Sandri, C.; Zhao, J.; et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007, 6, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef]
- Lagirand-Cantaloube, J.; Cornille, K.; Csibi, A.; Batonnet-Pichon, S.; Leibovitch, M.P.; Leibovitch, S.A. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS ONE 2009, 4, e4973. [Google Scholar] [CrossRef]
- Tintignac, L.A.; Lagirand, J.; Batonnet, S.; Sirri, V.; Leibovitch, M.P.; Leibovitch, S.A. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 2005, 280, 2847–2856. [Google Scholar] [CrossRef]
- Jogo, M.; Shiraishi, S.; Tamura, T. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 2009, 583, 2715–2719. [Google Scholar] [CrossRef] [PubMed]
- Lokireddy, S.; Wijesoma, I.W.; Sze, S.K.; McFarlane, C.; Kambadur, R.; Sharma, M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am. J. Physiol. Cell Physiol. 2012, 303, C512–C529. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Brault, J.J.; Gygi, S.P.; Glass, D.J.; Valenzuela, D.M.; Gartner, C.; Latres, E.; Goldberg, A.L. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol. 2009, 185, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 2012, 198, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, E.; Kudryashov, D.; Kramerova, I.; Spencer, M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005, 354, 413–424. [Google Scholar] [CrossRef]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010, 11, 45–51. [Google Scholar] [CrossRef]
- Kanki, T. Nix, a receptor protein for mitophagy in mammals. Autophagy 2010, 6, 433–435. [Google Scholar] [CrossRef]
- Novak, I. Mitophagy: A complex mechanism of mitochondrial removal. Antioxid. Redox Signal. 2012, 17, 794–802. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Kamei, Y.; Sugita, S.; Akaike, F.; Kanai, S.; Miura, S.; Hirata, Y.; Troen, B.R.; Kitamura, T.; Nishino, I.; et al. The cathepsin L gene is a direct target of FOXO1 in skeletal muscle. Biochem. J. 2010, 427, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Paré, M.F.; Baechler, B.L.; Fajardo, V.A.; Earl, E.; Wong, E.; Campbell, T.L.; Tupling, A.R.; Quadrilatero, J. Effect of acute and chronic autophagy deficiency on skeletal muscle apoptotic signaling, morphology, and function. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Guan, K.-L. Regulation of the autophagy initiating kinase ULK1 by nutrients: Roles of mTORC1 and AMPK. Cell Cycle Georget. Tex 2011, 10, 1337–1338. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Egan, D.; Kim, J.; Shaw, R.J.; Guan, K.-L. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 2011, 7, 643–644. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Candau, R.; Raibon, A.; Bernardi, H. Autophagy, a Highly Regulated Intracellular System Essential to Skeletal Muscle Homeostasis—Role in Disease, Exercise and Altitude Exposure. Muscle Cell Tissue 2015. [Google Scholar] [CrossRef]
- Mahoney, D.J.; Parise, G.; Melov, S.; Safdar, A.; Tarnopolsky, M.A. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 1498–1500. [Google Scholar] [CrossRef] [PubMed]
- Jamart, C.; Francaux, M.; Millet, G.Y.; Deldicque, L.; Frère, D.; Féasson, L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J. Appl. Physiol. Bethesda MD 1985 2012, 112, 1529–1537. [Google Scholar] [CrossRef]
- Møller, A.B.; Vendelbo, M.H.; Christensen, B.; Clasen, B.F.; Bak, A.M.; Jørgensen, J.O.L.; Møller, N.; Jessen, N. Physical exercise increases autophagic signaling through ULK1 in human skeletal muscle. J. Appl. Physiol. Bethesda MD 1985 2015, 118, 971–979. [Google Scholar] [CrossRef]
- He, C.; Sumpter, R.; Levine, B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 2012, 8, 1548–1551. [Google Scholar] [CrossRef]
- He, C.; Bassik, M.C.; Moresi, V.; Sun, K.; Wei, Y.; Zou, Z.; An, Z.; Loh, J.; Fisher, J.; Sun, Q.; et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012, 481, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Glynn, E.L.; Fry, C.S.; Drummond, M.J.; Dreyer, H.C.; Dhanani, S.; Volpi, E.; Rasmussen, B.B. Muscle protein breakdown has a minor role in the protein anabolic response to essential amino acid and carbohydrate intake following resistance exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R533–R540. [Google Scholar] [CrossRef] [PubMed]
- Jamart, C.; Naslain, D.; Gilson, H.; Francaux, M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E964–E974. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J. Autophagy regulation in human skeletal muscle during exercise. J. Physiol. 2016, 594, 5053–5054. [Google Scholar] [CrossRef] [PubMed]
- Fritzen, A.M.; Madsen, A.B.; Kleinert, M.; Treebak, J.T.; Lundsgaard, A.-M.; Jensen, T.E.; Richter, E.A.; Wojtaszewski, J.; Kiens, B.; Frøsig, C. Regulation of autophagy in human skeletal muscle: Effects of exercise, exercise training and insulin stimulation. J. Physiol. 2016, 594, 745–761. [Google Scholar] [CrossRef] [PubMed]
- Masiero, E.; Agatea, L.; Mammucari, C.; Blaauw, B.; Loro, E.; Komatsu, M.; Metzger, D.; Reggiani, C.; Schiaffino, S.; Sandri, M. Autophagy is required to maintain muscle mass. Cell Metab. 2009, 10, 507–515. [Google Scholar] [CrossRef]
- Lira, V.A.; Okutsu, M.; Zhang, M.; Greene, N.P.; Laker, R.C.; Breen, D.S.; Hoehn, K.L.; Yan, Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 4184–4193. [Google Scholar] [CrossRef]
- Ju, J.-S.; Jeon, S.-I.; Park, J.-Y.; Lee, J.-Y.; Lee, S.-C.; Cho, K.-J.; Jeong, J.-M. Autophagy plays a role in skeletal muscle mitochondrial biogenesis in an endurance exercise-trained condition. J. Physiol. Sci. JPS 2016, 66, 417–430. [Google Scholar] [CrossRef]
- Carter, H.N.; Kim, Y.; Erlich, A.T.; Zarrin-Khat, D.; Hood, D.A. Autophagy and mitophagy flux in young and aged skeletal muscle following chronic contractile activity. J. Physiol. 2018, 596, 3567–3584. [Google Scholar] [CrossRef]
- Sanchez, A.M.J. Mitophagy flux in skeletal muscle during chronic contractile activity and ageing. J. Physiol. 2018, 596, 3461–3462. [Google Scholar] [CrossRef]
- Parousis, A.; Carter, H.N.; Tran, C.; Erlich, A.T.; Mesbah Moosavi, Z.S.; Pauly, M.; Hood, D.A. Contractile activity attenuates autophagy suppression and reverses mitochondrial defects in skeletal muscle cells. Autophagy 2018, 14, 1886–1897. [Google Scholar] [CrossRef] [PubMed]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; de Vries, R.L.A.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S.; et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.-S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deak, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, J.; Kim, S.; Song, S.; Kwon, S.-K.; Lee, S.-H.; Kitada, T.; Kim, J.-M.; Chung, J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 2008, 377, 975–980. [Google Scholar] [CrossRef]
- Tanaka, A.; Cleland, M.M.; Xu, S.; Narendra, D.P.; Suen, D.-F.; Karbowski, M.; Youle, R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010, 191, 1367–1380. [Google Scholar] [CrossRef]
- Chan, N.C.; Salazar, A.M.; Pham, A.H.; Sweredoski, M.J.; Kolawa, N.J.; Graham, R.L.J.; Hess, S.; Chan, D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011, 20, 1726–1737. [Google Scholar] [CrossRef]
- Glauser, L.; Sonnay, S.; Stafa, K.; Moore, D.J. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 2011, 118, 636–645. [Google Scholar] [CrossRef]
- Wang, X.; Winter, D.; Ashrafi, G.; Schlehe, J.; Wong, Y.L.; Selkoe, D.; Rice, S.; Steen, J.; LaVoie, M.J.; Schwarz, T.L. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 2011, 147, 893–906. [Google Scholar] [CrossRef]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013, 496, 372–376. [Google Scholar] [CrossRef]
- Nardin, A.; Schrepfer, E.; Ziviani, E. Counteracting PINK/Parkin Deficiency in the Activation of Mitophagy: A Potential Therapeutic Intervention for Parkinson’s Disease. Curr. Neuropharmacol. 2016, 14, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.-M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Ko, H.S.; Kang, H.; Lee, Y.; Lee, Y.-I.; Pletinkova, O.; Troconso, J.C.; Dawson, V.L.; Dawson, T.M. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 2011, 144, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.F.; Fon, E.A. Presenting mitochondrial antigens: PINK1, Parkin and MDVs steal the show. Cell Res. 2016, 26, 1180–1181. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, A.; McLelland, G.-L.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef] [PubMed]
- Matheoud, D.; Sugiura, A.; Bellemare-Pelletier, A.; Laplante, A.; Rondeau, C.; Chemali, M.; Fazel, A.; Bergeron, J.J.; Trudeau, L.-E.; Burelle, Y.; et al. Parkinson’s Disease-Related Proteins PINK1 and Parkin Repress Mitochondrial Antigen Presentation. Cell 2016, 166, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.F.; Tang, M.Y.; Fon, E.A.; Durcan, T.M. Defending the mitochondria: The pathways of mitophagy and mitochondrial-derived vesicles. Int. J. Biochem. Cell Biol. 2016, 79, 427–436. [Google Scholar] [CrossRef] [PubMed]
- McLelland, G.-L.; Lee, S.A.; McBride, H.M.; Fon, E.A. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 2016, 214, 275–291. [Google Scholar] [CrossRef]
- Peker, N.; Donipadi, V.; Sharma, M.; McFarlane, C.; Kambadur, R. Loss of Parkin Impairs Mitochondrial Function and Leads to Muscle Atrophy. Am. J. Physiol. Cell Physiol. 2018, 315, C164–C185. [Google Scholar] [CrossRef]
- Gouspillou, G.; Godin, R.; Piquereau, J.; Picard, M.; Mofarrahi, M.; Mathew, J.; Purves-Smith, F.M.; Sgarioto, N.; Hepple, R.T.; Burelle, Y.; et al. Protective role of Parkin in skeletal muscle contractile and mitochondrial function. J. Physiol. 2018, 596, 2565–2579. [Google Scholar] [CrossRef]
- Chen, C.C.W.; Erlich, A.T.; Hood, D.A. Role of Parkin and endurance training on mitochondrial turnover in skeletal muscle. Skelet. Muscle 2018, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Neuspiel, M.; Schauss, A.C.; Braschi, E.; Zunino, R.; Rippstein, P.; Rachubinski, R.A.; Andrade-Navarro, M.A.; McBride, H.M. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 2008, 18, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, W.; Chen, G.; Feng, D.; Liu, J.; Ma, B.; Zhou, C.; Mu, C.; Zhang, W.; Chen, Q.; et al. Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy. Autophagy 2015, 11, 1216–1229. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Ren, K.-D.; Yang, J.; Luo, X.-J. Mitochondrial E3 ubiquitin ligase 1: A key enzyme in regulation of mitochondrial dynamics and functions. Mitochondrion 2016, 28, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, J.; Liu, Z.; Liu, H.; Li, H.; Li, Z. IGF-1 potentiates sensory innervation signalling by modulating the mitochondrial fission/fusion balance. Sci. Rep. 2017, 7, 43949. [Google Scholar] [CrossRef]
- Lokireddy, S.; Wijesoma, I.W.; Teng, S.; Bonala, S.; Gluckman, P.D.; McFarlane, C.; Sharma, M.; Kambadur, R. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012, 16, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Braschi, E.; Zunino, R.; McBride, H.M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009, 10, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, C.; Przyklenk, A.; Metten, A.; Schiffer, T.; Bloch, W.; Brixius, K.; Gehlert, S. Influence of endurance training on skeletal muscle mitophagy regulatory proteins in type 2 diabetic men. Endocr. Res. 2017, 42, 325–330. [Google Scholar] [CrossRef]
- Katayama, K.; Matsuo, H.; Ishida, K.; Mori, S.; Miyamura, M. Intermittent hypoxia improves endurance performance and submaximal exercise efficiency. High Alt. Med. Biol. 2003, 4, 291–304. [Google Scholar] [CrossRef]
- Flaherty, G.; O’Connor, R.; Johnston, N. Altitude training for elite endurance athletes: A review for the travel medicine practitioner. Travel Med. Infect. Dis. 2016, 14, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Brocherie, F.; Girard, O.; Faiss, R.; Millet, G.P. Effects of Repeated-Sprint Training in Hypoxia on Sea-Level Performance: A Meta-Analysis. Sports Med. 2017, 47, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Girard, O.; Brocherie, F.; Millet, G.P. Effects of Altitude/Hypoxia on Single- and Multiple-Sprint Performance: A Comprehensive Review. Sports Med. 2017, 47, 1931–1949. [Google Scholar] [CrossRef] [PubMed]
- Van der Zwaard, S.; Brocherie, F.; Kom, B.L.G.; Millet, G.P.; Deldicque, L.; van der Laarse, W.J.; Girard, O.; Jaspers, R.T. Adaptations in muscle oxidative capacity, fiber size, and oxygen supply capacity after repeated-sprint training in hypoxia combined with chronic hypoxic exposure. J. Appl. Physiol. Bethesda MD 1985 2018, 124, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Borrani, F. Effects of intermittent hypoxic training performed at high hypoxia level on exercise performance in highly trained runners. J. Sports Sci. 2018, 36, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.M.; Davies, B. Physiological implications of altitude training for endurance performance at sea level: A review. Br. J. Sports Med. 1997, 31, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Böning, D. Altitude and hypoxia training—A short review. Int. J. Sports Med. 1997, 18, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-Y.; Hwang, H.; Park, J.; Lee, S.; Lim, K. The effects of altitude/hypoxic training on oxygen delivery capacity of the blood and aerobic exercise capacity in elite athletes—A meta-analysis. J. Exerc. Nutr. Biochem. 2016, 20, 15–22. [Google Scholar] [CrossRef]
- Roels, B.; Millet, G.P.; Marcoux, C.J.L.; Coste, O.; Bentley, D.J.; Candau, R.B. Effects of hypoxic interval training on cycling performance. Med. Sci. Sports Exerc. 2005, 37, 138–146. [Google Scholar] [CrossRef]
- Czuba, M.; Waskiewicz, Z.; Zajac, A.; Poprzecki, S.; Cholewa, J.; Roczniok, R. The effects of intermittent hypoxic training on aerobic capacity and endurance performance in cyclists. J. Sports Sci. Med. 2011, 10, 175–183. [Google Scholar] [PubMed]
- Nakamoto, F.P.; Ivamoto, R.K.; Andrade, M.D.S.; de Lira, C.A.B.; Silva, B.M.; da Silva, A.C. Effect of Intermittent Hypoxic Training Followed by Intermittent Hypoxic Exposure on Aerobic Capacity of Long Distance Runners. J. Strength Cond. Res. 2016, 30, 1708–1720. [Google Scholar] [CrossRef]
- Hoppeler, H.; Kleinert, E.; Schlegel, C.; Claassen, H.; Howald, H.; Kayar, S.R.; Cerretelli, P. Morphological adaptations of human skeletal muscle to chronic hypoxia. Int. J. Sports Med. 1990, 11 (Suppl. 1), S3–S9. [Google Scholar] [CrossRef]
- MacDougall, J.D.; Green, H.J.; Sutton, J.R.; Coates, G.; Cymerman, A.; Young, P.; Houston, C.S. Operation Everest II: Structural adaptations in skeletal muscle in response to extreme simulated altitude. Acta Physiol. Scand. 1991, 142, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, M.; Savard, G.K.; Areskog, N.-H.; Lundby, C.; Saltin, B. Skeletal muscle adaptations to prolonged exposure to extreme altitude: A role of physical activity? High Alt. Med. Biol. 2008, 9, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Watier, T.; Sanchez, A.M. Micro-RNAs, Exercise and Cellular Plasticity in Humans: The Impact of Dietary Factors and Hypoxia. MicroRNA 2017, 6, 110–124. [Google Scholar] [CrossRef] [PubMed]
- D’Hulst, G.; Jamart, C.; Van Thienen, R.; Hespel, P.; Francaux, M.; Deldicque, L. Effect of acute environmental hypoxia on protein metabolism in human skeletal muscle. Acta Physiol. 2013, 208, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Masschelein, E.; Van Thienen, R.; D’Hulst, G.; Hespel, P.; Thomis, M.; Deldicque, L. Acute environmental hypoxia induces LC3 lipidation in a genotype-dependent manner. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2014, 28, 1022–1034. [Google Scholar] [CrossRef] [PubMed]
- Gnimassou, O.; Fernández-Verdejo, R.; Brook, M.; Naslain, D.; Balan, E.; Sayda, M.; Cegielski, J.; Nielens, H.; Decottignies, A.; Demoulin, J.-B.; et al. Environmental hypoxia favors myoblast differentiation and fast phenotype but blunts activation of protein synthesis after resistance exercise in human skeletal muscle. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 5272–5284. [Google Scholar] [CrossRef] [PubMed]
- Aragonés, J.; Schneider, M.; Van Geyte, K.; Fraisl, P.; Dresselaers, T.; Mazzone, M.; Dirkx, R.; Zacchigna, S.; Lemieux, H.; Jeoung, N.H.; et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat. Genet. 2008, 40, 170–180. [Google Scholar] [CrossRef]
- Kelly, D.P. Hypoxic reprogramming. Nat. Genet. 2008, 40, 132–134. [Google Scholar] [CrossRef]
- Chaillou, T.; Koulmann, N.; Simler, N.; Meunier, A.; Serrurier, B.; Chapot, R.; Peinnequin, A.; Beaudry, M.; Bigard, X. Hypoxia transiently affects skeletal muscle hypertrophy in a functional overload model. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R643–R654. [Google Scholar] [CrossRef]
- Viganò, A.; Ripamonti, M.; De Palma, S.; Capitanio, D.; Vasso, M.; Wait, R.; Lundby, C.; Cerretelli, P.; Gelfi, C. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics 2008, 8, 4668–4679. [Google Scholar] [CrossRef] [PubMed]
- Bigard, X. Molecular factors involved in the control of muscle mass during hypoxia-exposure: The main hypotheses are revisited. Acta Physiol. 2013, 208, 222–223. [Google Scholar] [CrossRef] [PubMed]
- Roig, M.; O’Brien, K.; Kirk, G.; Murray, R.; McKinnon, P.; Shadgan, B.; Reid, W.D. The effects of eccentric versus concentric resistance training on muscle strength and mass in healthy adults: A systematic review with meta-analysis. Br. J. Sports Med. 2009, 43, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Norrbrand, L.; Fluckey, J.D.; Pozzo, M.; Tesch, P.A. Resistance training using eccentric overload induces early adaptations in skeletal muscle size. Eur. J. Appl. Physiol. 2008, 102, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, S.K.; Farup, J.; Møller, A.B.; Vendelbo, M.H.; Holm, L.; Jessen, N.; Vissing, K. Effects of divergent resistance exercise contraction mode and dietary supplementation type on anabolic signalling, muscle protein synthesis and muscle hypertrophy. Amino Acids 2014, 46, 2377–2392. [Google Scholar] [CrossRef] [PubMed]
- Ato, S.; Makanae, Y.; Kido, K.; Fujita, S. Contraction mode itself does not determine the level of mTORC1 activity in rat skeletal muscle. Physiol. Rep. 2016, 4, e12976. [Google Scholar] [CrossRef] [PubMed]
- Garma, T.; Kobayashi, C.; Haddad, F.; Adams, G.R.; Bodell, P.W.; Baldwin, K.M. Similar acute molecular responses to equivalent volumes of isometric, lengthening, or shortening mode resistance exercise. J. Appl. Physiol. Bethesda MD 1985 2007, 102, 135–143. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, T.K.; Duffy, L.R.; Frey, J.W.; Hornberger, T.A. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J. Physiol. 2009, 587, 3691–3701. [Google Scholar] [CrossRef] [PubMed]
- Ochi, E.; Ishii, N.; Nakazato, K. Time Course Change of IGF1/Akt/mTOR/p70s6k Pathway Activation in Rat Gastrocnemius Muscle During Repeated Bouts of Eccentric Exercise. J. Sports Sci. Med. 2010, 9, 170–175. [Google Scholar] [PubMed]
- Ato, S.; Makanae, Y.; Kido, K.; Sase, K.; Yoshii, N.; Fujita, S. The effect of different acute muscle contraction regimens on the expression of muscle proteolytic signaling proteins and genes. Physiol. Rep. 2017, 5, e13364. [Google Scholar] [CrossRef] [PubMed]
- Lo Verso, F.; Carnio, S.; Vainshtein, A.; Sandri, M. Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy 2014, 10, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Klarod, K.; Philippe, M.; Gatterer, H.; Burtscher, M. Different training responses to eccentric endurance exercise at low and moderate altitudes in pre-diabetic men: A pilot study. Sport Sci. Health 2017, 13, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Rizo-Roca, D.; Ríos-Kristjánsson, J.G.; Núñez-Espinosa, C.; Santos-Alves, E.; Gonçalves, I.O.; Magalhães, J.; Ascensão, A.; Pagès, T.; Viscor, G.; Torrella, J.R. Intermittent hypobaric hypoxia combined with aerobic exercise improves muscle morphofunctional recovery after eccentric exercise to exhaustion in trained rats. J. Appl. Physiol. Bethesda MD 1985 2017, 122, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Rizo-Roca, D.; Ríos-Kristjánsson, J.G.; Núñez-Espinosa, C.; Santos-Alves, E.; Magalhães, J.; Ascensão, A.; Pagès, T.; Viscor, G.; Torrella, J.R. Modulation of mitochondrial biomarkers by intermittent hypobaric hypoxia and aerobic exercise after eccentric exercise in trained rats. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2017, 42, 683–693. [Google Scholar] [CrossRef] [PubMed]
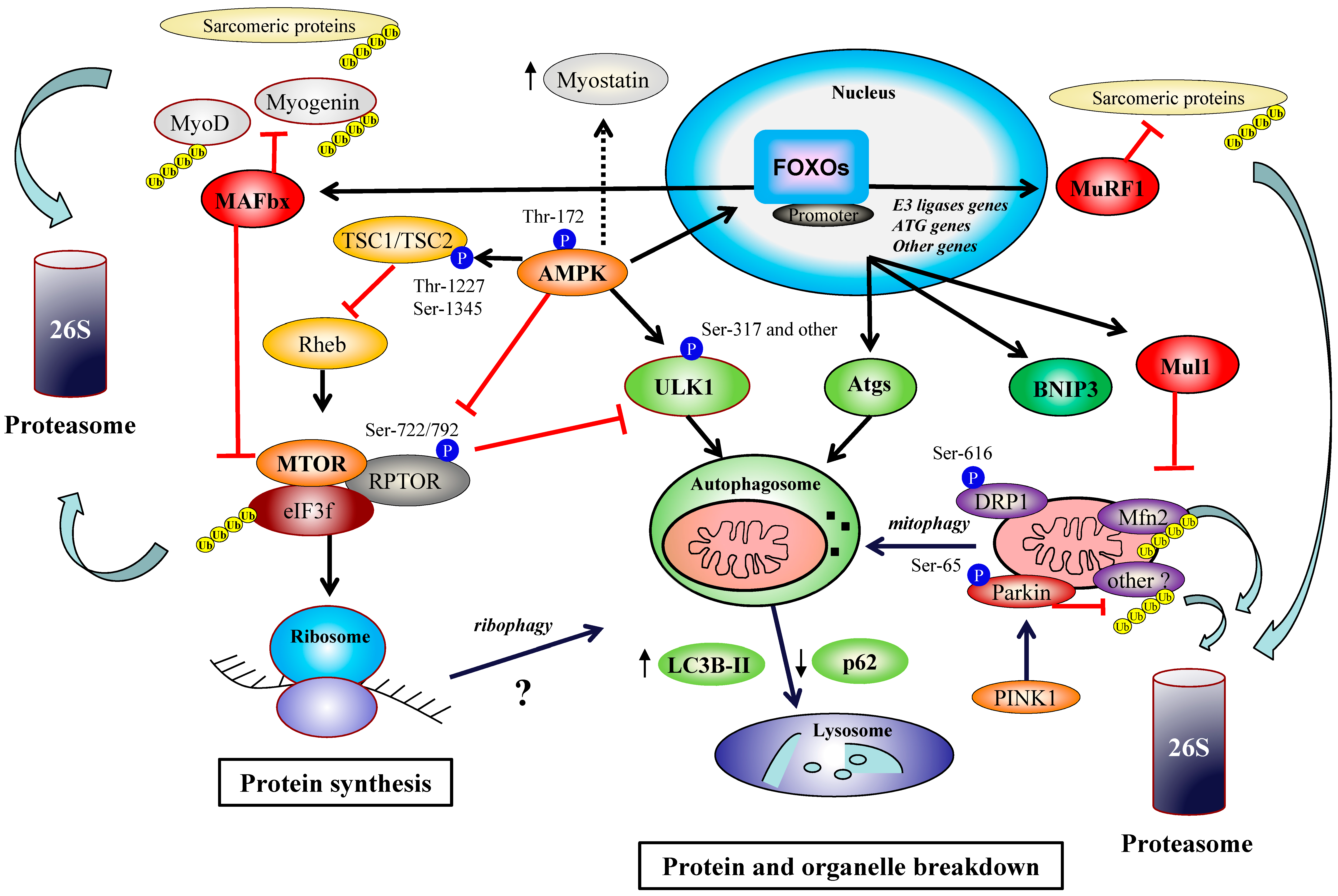
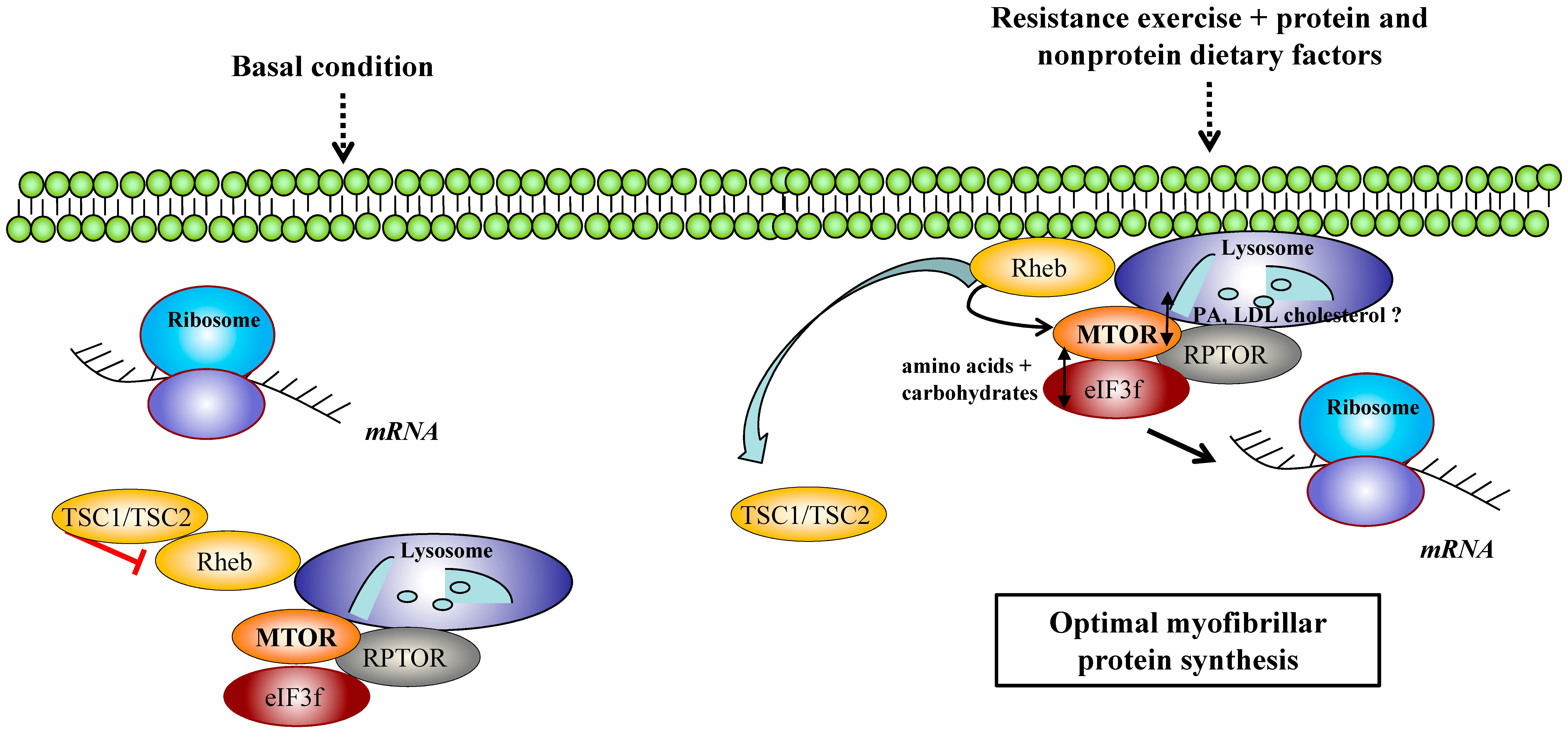

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez, A.M.; Candau, R.; Bernardi, H. Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise. Cells 2019, 8, 542. https://doi.org/10.3390/cells8060542
Sanchez AM, Candau R, Bernardi H. Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise. Cells. 2019; 8(6):542. https://doi.org/10.3390/cells8060542
Chicago/Turabian StyleSanchez, Anthony MJ, Robin Candau, and Henri Bernardi. 2019. "Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise" Cells 8, no. 6: 542. https://doi.org/10.3390/cells8060542
APA StyleSanchez, A. M., Candau, R., & Bernardi, H. (2019). Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise. Cells, 8(6), 542. https://doi.org/10.3390/cells8060542






