pHluorin-BACE1-mCherry Acts as a Reporter for the Intracellular Distribution of Active BACE1 In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Antibodies
2.3. Expression Plasmids
2.4. Western Blot Analysis
2.5. Cell Culture and Transient Transfection
2.6. BACE1 Activity Measurement
2.7. ELISA Analysis of Aβ (1–40)
2.8. Immunostaining
2.9. Live Cell Imaging and Drug Treatment
2.10. In Utero Electroporation
2.11. Statistical Analysis
3. Results
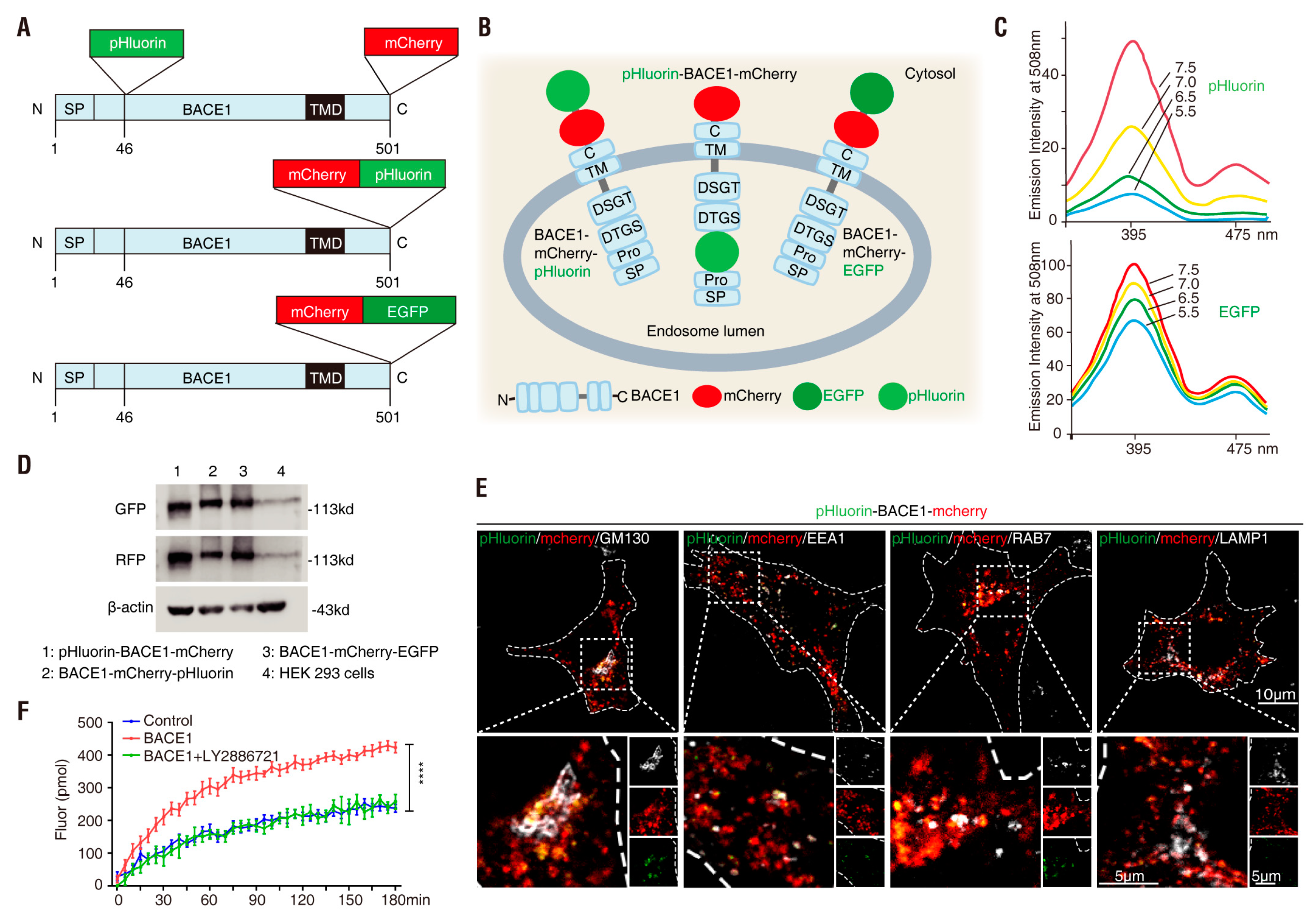
3.1. Generation of BACE1 Fusion Proteins
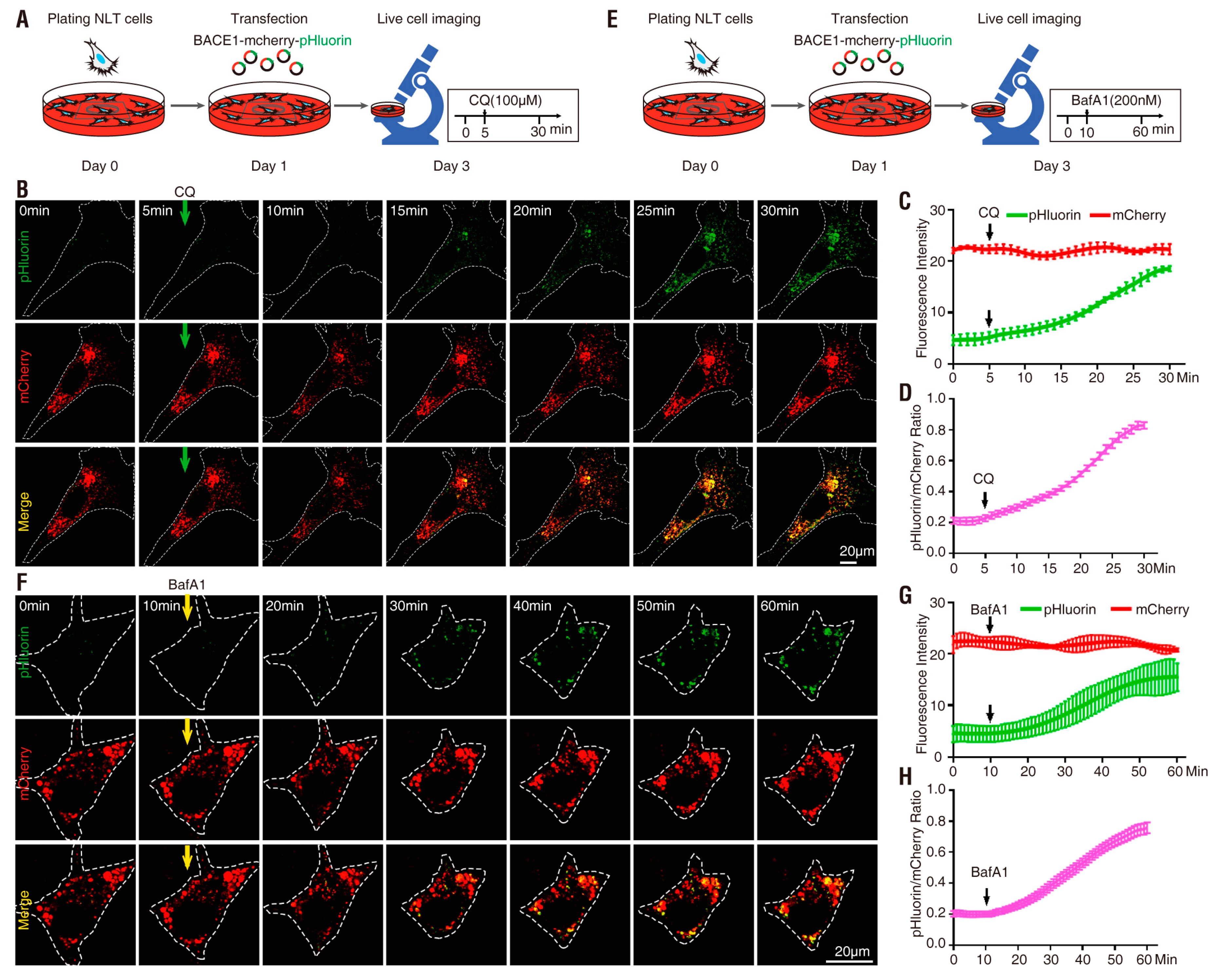
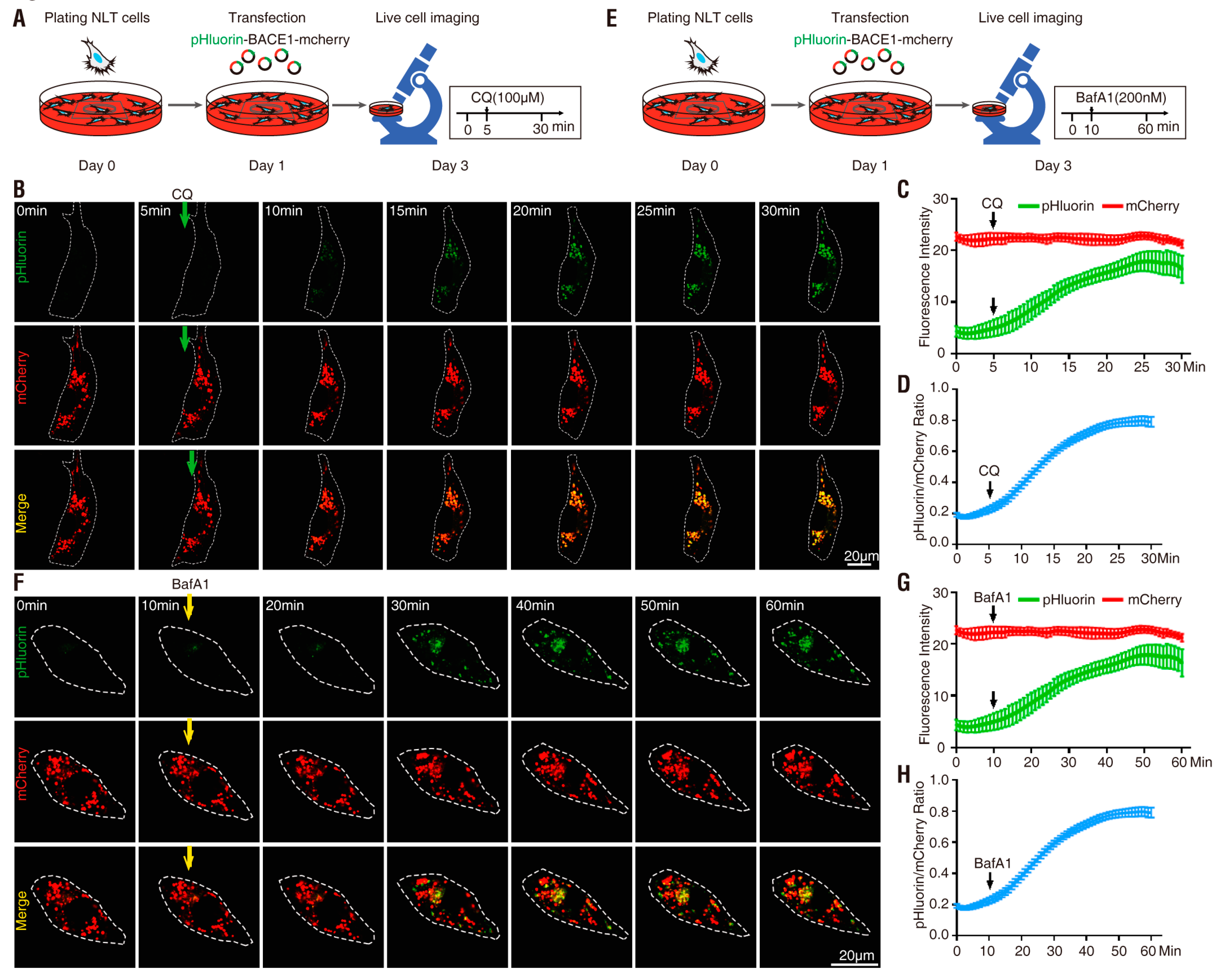
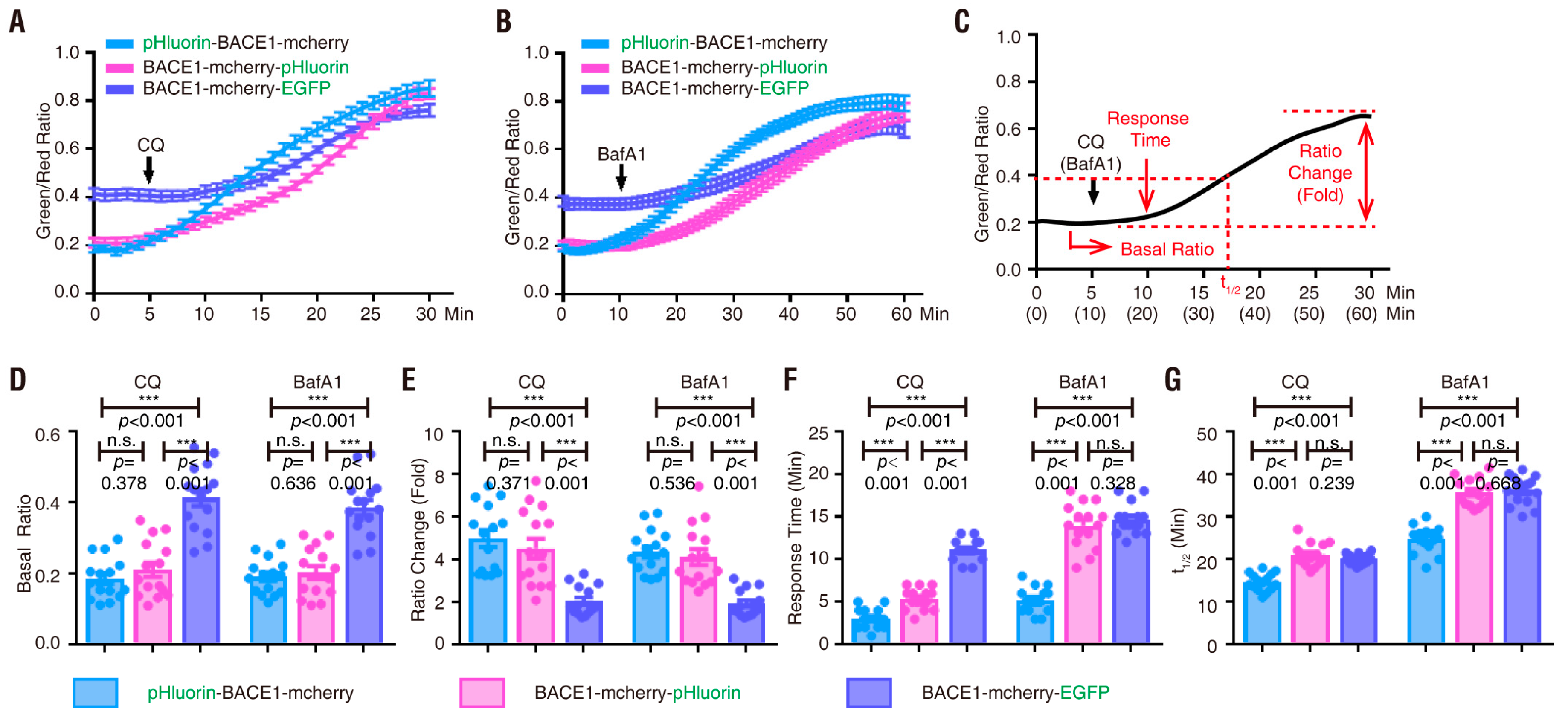
3.2. pHluorin-BACE1-mCherry, a Better pH Sensor for BACE1 in NLT Cells
3.3. pHluorin-BACE1-mCherry, a Better pH Sensor for BACE1 in Cultured Neurons
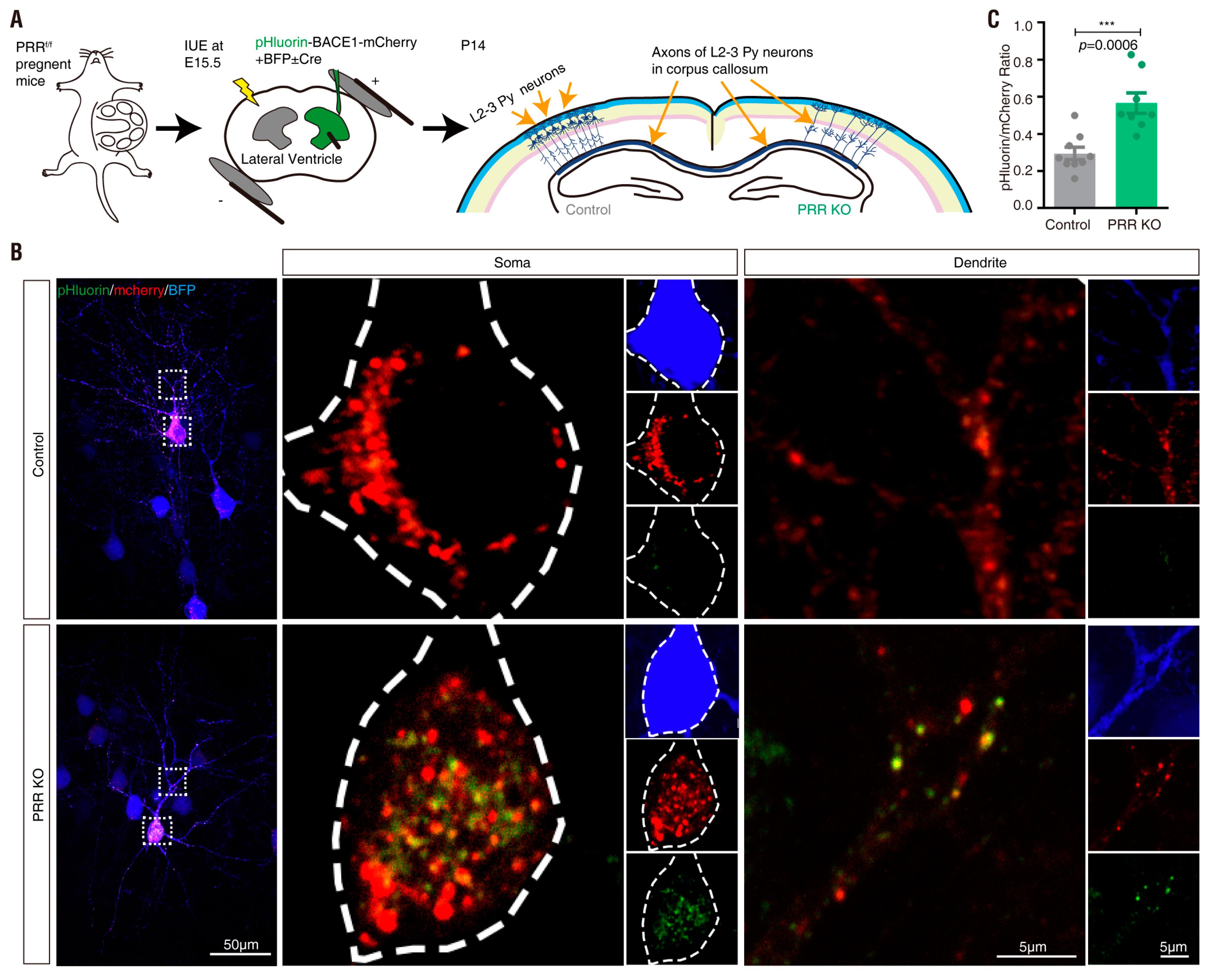
3.4. PRR Regulation of phluorin-BACE1-mCherry and BACE1 Activity in Culture and In Vivo
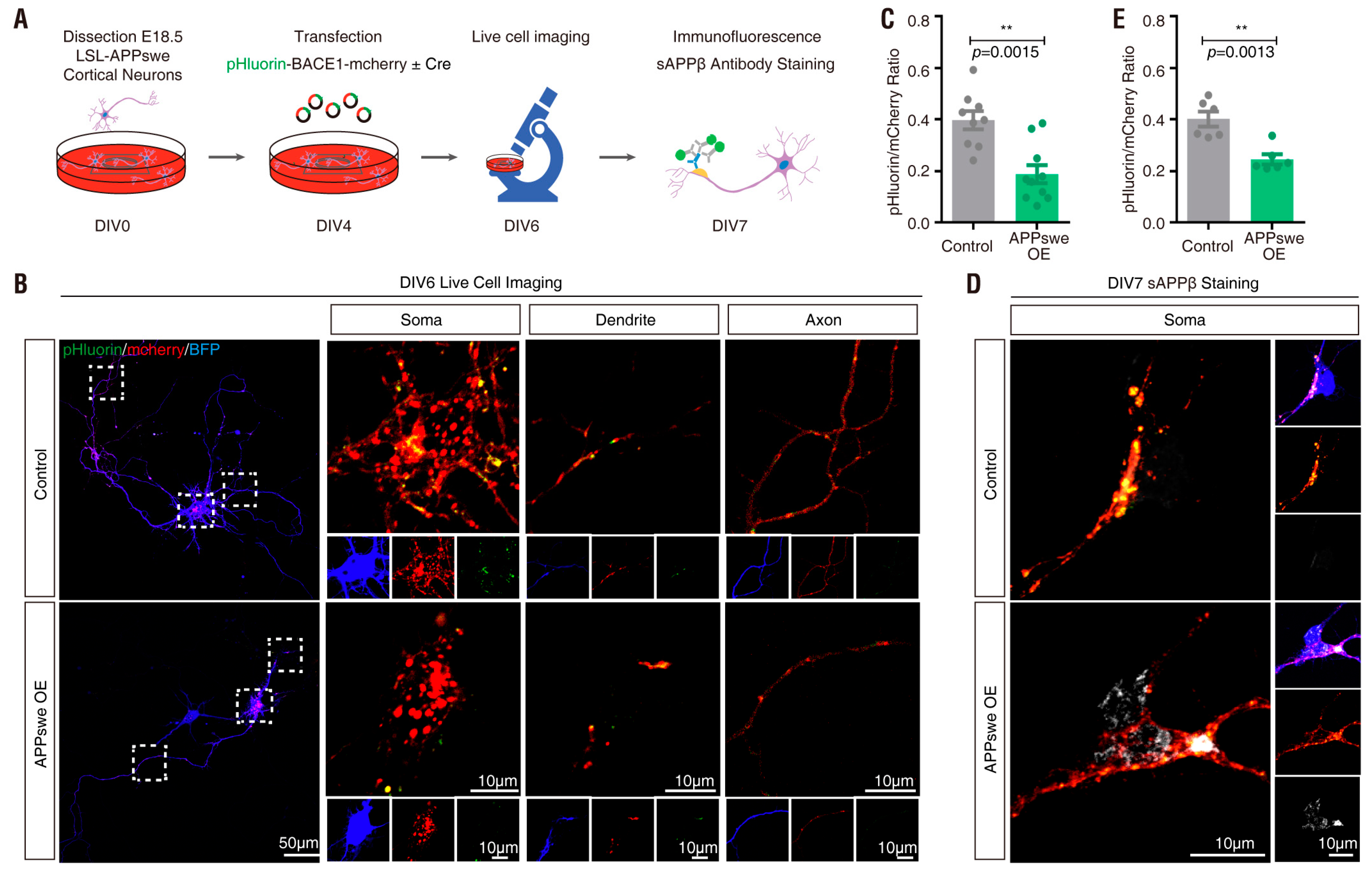
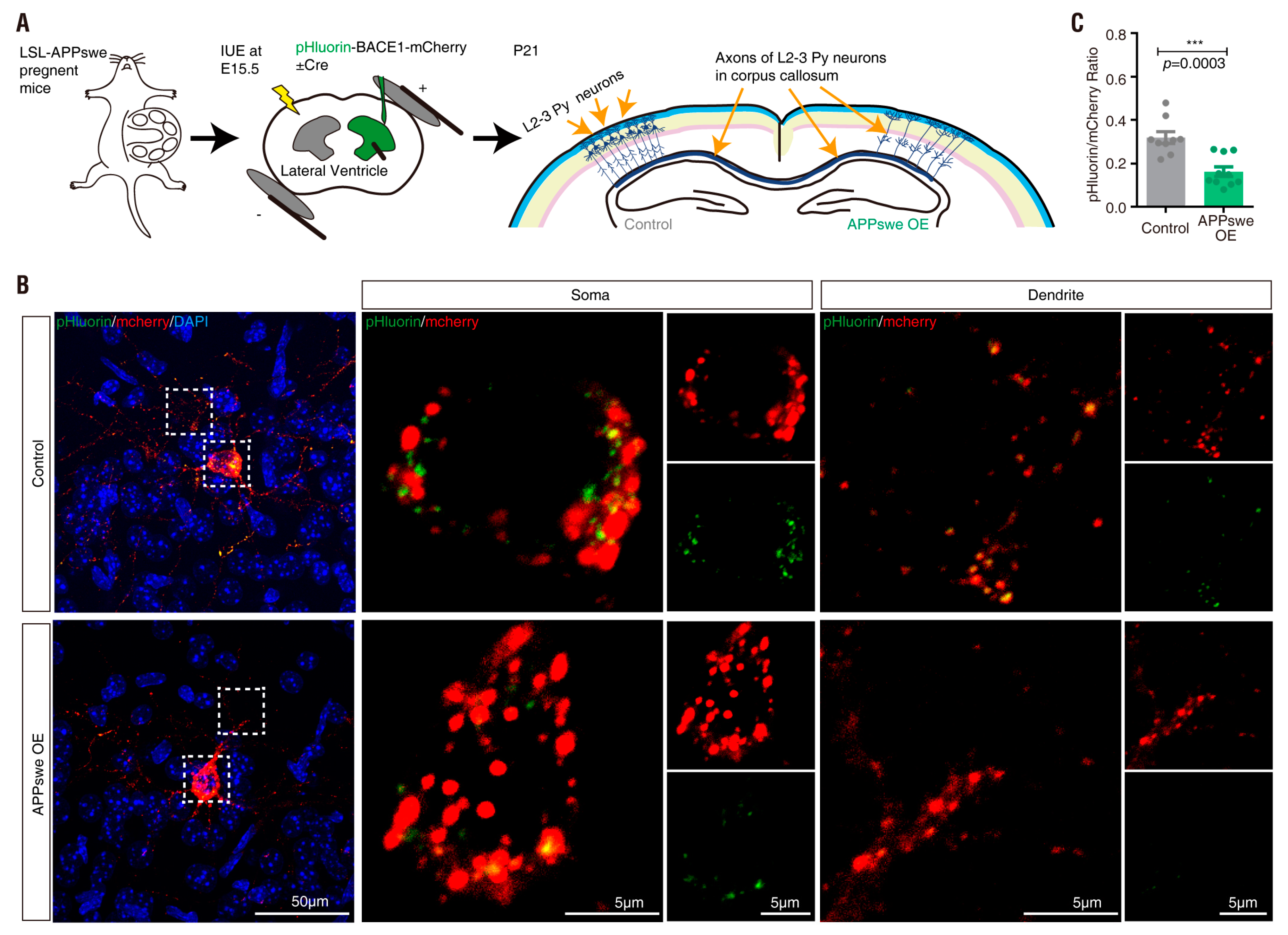
3.5. APPswe Regulation of pHluorin-BACE1-mCherry in Culture and In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | β-amyloid |
| AD | Alzheimer’s disease |
| APP | amyloid precursor protein |
| APPswe | Swedish mutant APP |
| BACE1 | β-site APP cleaving enzyme 1 |
| BafA1 | bafilomycin A1 |
| BBS | α-bungarotoxin binding site |
| BFP | blue fluorescent protein |
| CMV | cytomegalovirus |
| CQ | chloroquine |
| DABCO | 1,4-Diazabicyclo-octane |
| EGFP | enhanced green fluorescent protein |
| ER | Endoplasmic reticulum |
| FBS | fetal bovine serum |
| GFP | green fluorescent protein |
| HRP | horseradish peroxidase |
| IRES | internal ribosome entry site |
| IUE | in utero electroporation |
| NPCs | neural progenitor cells |
| NSCs | neural stem cells |
| PCR | polymerase chain reaction |
| PEI | polyethylenimine |
| PFA | paraformaldehyde |
| Pro | pro-peptide |
| proBACE1 | immature precursor protein BACE1 |
| PRR/ATP6AP2 | (pro)renin receptor |
| PVA | poly(vinyl alcohol) |
| SP | signal peptide |
| TGN | trans-Golgi Networks |
| TMD | transmembrane domain |
| V-ATPase | vacuolar-type ATPase |
References
- Cole, S.L.; Vassar, R. BACE1 structure and function in health and Alzheimer’s disease. Curr. Alzheimer Res. 2008, 5, 100–120. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; Karran, E. The cellular phase of Alzheimer’s disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, R.E.; Bertram, L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell 2005, 120, 545–555. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2012 Alzheimer’s disease facts and figures. Alzheimers Dement. 2012, 8, 131–168. [Google Scholar]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Leon, M.J.D.; Zetterberg, H. Alzheimer’s disease. Lancet 2006, 368, 387–403. [Google Scholar] [CrossRef]
- Hussain, I.; Powell, D.; Howlett, D.R.; Tew, D.G.; Meek, T.D.; Chapman, C.; Gloger, I.S.; Murphy, K.E.; Southan, C.D.; Ryan, D.M. Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol. Cell. Neurosci. 1999, 14, 419–427. [Google Scholar] [CrossRef]
- Vassar, R.; Bennett, B.D.; Babu-Khan, S.; Kahn, S.; Mendiaz, E.A.; Denis, P.; Teplow, D.B.; Ross, S.; Amarante, P.; Loeloff, R. β-Secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999, 286, 735–741. [Google Scholar] [CrossRef]
- Yan, R.; Bienkowski, M.J.; Shuck, M.E.; Miao, H.; Tory, M.C.; Pauley, A.M.; Brashler, J.R.; Stratman, N.C.; Mathews, W.R.; Buhl, A.E. Membrane-anchored aspartyl protease with Alzheimer’s disease β-secretase activity. Nature 1999, 402, 533. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Wang, Y.; McCarthy, D.; Wen, H.; Borchelt, D.R.; Price, D.L.; Wong, P.C. BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat. Neurosci. 2001, 4, 233. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Bolon, B.; Kahn, S.; Bennett, B.D.; Babu-Khan, S.; Denis, P.; Fan, W.; Kha, H.; Zhang, J.; Gong, Y. Mice deficient in BACE1, the Alzheimer’s β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 2001, 4, 231. [Google Scholar] [CrossRef]
- Roberds, S.L.; Anderson, J.; Basi, G.; Bienkowski, M.J.; Branstetter, D.G.; Chen, K.S.; Freedman, S.; Frigon, N.L.; Games, D.; Hu, K. BACE knockout mice are healthy despite lacking the primary β-secretase activity in brain: Implications for Alzheimer’s disease therapeutics. Hum. Mol. Genet. 2001, 10, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.D.; Denis, P.; Haniu, M.; Teplow, D.B.; Kahn, S.; Louis, J.C.; Citron, M.; Vassar, R. A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer’s beta -secretase. J. Biol. Chem. 2000, 275, 37712–37717. [Google Scholar] [CrossRef]
- Benjannet, S.; Elagoz, A.; Wickham, L.; Mamarbachi, M.; Munzer, J.S.; Basak, A.; Lazure, C.; Cromlish, J.A.; Sisodia, S.; Checler, F. Post-translational processing of β-secretase (BACE) and its ectodomain shedding: The pro-and transmembrane/cytosolic domains affect its cellular activity and amyloid Aβ production. J. Biol. Chem. 2001. [Google Scholar] [CrossRef] [PubMed]
- Andrew, R.J.; Fernandez, C.G.; Stanley, M.; Jiang, H.; Nguyen, P.; Rice, R.C.; Buggia-Prévot, V.; De Rossi, P.; Vetrivel, K.S.; Lamb, R.; et al. Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E9665–E9674. [Google Scholar] [CrossRef] [PubMed]
- Vetrivel, K.S.; Meckler, X.; Chen, Y.; Nguyen, P.D.; Seidah, N.G.; Vassar, R.; Wong, P.C.; Fukata, M.; Kounnas, M.Z.; Thinakaran, G. Alzheimer disease Abeta production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J. Biol. Chem. 2009, 284, 3793–3803. [Google Scholar] [CrossRef]
- Haass, C.; Capell, A.; Citron, M.; Teplow, D.B.; Selkoe, D.J. The vacuolar H-ATPase inhibitor bafilomycin A1 differentially affects proteolytic processing of mutant and wild-type-amyloid precursor protein. J. Biol. Chem. 1995, 270, 6186–6192. [Google Scholar] [CrossRef]
- Haass, C.; Hung, A.Y.; Schlossmacher, M.; Teplow, D.B.; Selkoe, D.J. beta-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J. Biol. Chem. 1993, 268, 3021–3024. [Google Scholar] [PubMed]
- Knops, J.; Suomensaari, S.; Lee, M.; McConlogue, L.; Seubert, P.; Sinha, S. Cell-type and amyloid precursor protein-type specific inhibition of Aβ release by bafilomycin A1, a selective inhibitor of vacuolar ATPases. J. Biol. Chem. 1995, 270, 2419–2422. [Google Scholar] [CrossRef]
- Haass, C.; Lemere, C.A.; Capell, A.; Citron, M.; Seubert, P.; Schenk, D.; Lannfelt, L.; Selkoe, D.J. The Swedish mutation causes early-onset Alzheimer’s disease by β-secretase cleavage within the secretory pathway. Nat. Med. 1995, 1, 1291. [Google Scholar] [CrossRef]
- Koo, E.H.; Squazzo, S.L. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J. Biol. Chem. 1994, 269, 17386–17389. [Google Scholar] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Perez-Gonzalez, R.; Gauthier, S.A.; Kumar, A.; Levy, E. The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J. Biol. Chem. 2012, 287, 43108–43115. [Google Scholar] [CrossRef]
- Yuyama, K.; Igarashi, Y. Exosomes as Carriers of Alzheimer’s Amyloid-β. Front. Neurosci. 2017, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Capell, A.; Steiner, H.; Willem, M.; Kaiser, H.; Meyer, C.; Walter, J.; Lammich, S.; Multhaup, G.; Haass, C. Maturation and pro-peptide cleavage of β-secretase. J. Biol. Chem. 2000, 275, 30849–30854. [Google Scholar] [CrossRef] [PubMed]
- Creemers, J.W.; Dominguez, D.I.; Plets, E.; Serneels, L.; Taylor, N.A.; Multhaup, G.; Craessaerts, K.; Annaert, W.; De Strooper, B. Processing of β-secretase by furin and other members of the proprotein convertase family. J. Biol. Chem. 2001, 276, 4211–4217. [Google Scholar] [CrossRef] [PubMed]
- Haniu, M.; Denis, P.; Young, Y.; Mendiaz, E.A.; Fuller, J.; Hui, J.O.; Bennett, B.D.; Kahn, S.; Ross, S.; Burgess, T. Characterization of Alzheimer’s β-secretase protein BACE a pepsin family member with unusual properties. J. Biol. Chem. 2000, 275, 21099–21106. [Google Scholar] [CrossRef]
- Huse, J.T.; Pijak, D.S.; Leslie, G.J.; Lee, V.M.-Y.; Doms, R.W. Maturation and endosomal targeting of BACE: The Alzheimer’s disease β-Secretase. J. Biol. Chem. 2000. [Google Scholar] [CrossRef]
- Bennett, B.D.; Babu-Khan, S.; Loeloff, R.; Louis, J.-C.; Curran, E.; Citron, M.; Vassar, R. Expression analysis of BACE2 in brain and peripheral tissues. J. Biol. Chem. 2000. [Google Scholar] [CrossRef]
- Fluhrer, R.; Capell, A.; Westmeyer, G.; Willem, M.; Hartung, B.; Condron, M.M.; Teplow, D.B.; Haass, C.; Walter, J. A non-amyloidogenic function of BACE-2 in the secretory pathway. J. Neurochem. 2002, 81, 1011–1020. [Google Scholar] [CrossRef]
- Yan, R.; Han, P.; Miao, H.; Greengard, P.; Xu, H. The transmembrane domain of the Alzheimer’s beta-secretase (BACE1) determines its late Golgi localization and access to APP substrate. J. Biol. Chem. 2001. [Google Scholar] [CrossRef]
- Wahle, T.; Prager, K.; Raffler, N.; Haass, C.; Famulok, M.; Walter, J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol. Cell. Neurosci. 2005, 29, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Fluhrer, R.; Hartung, B.; Willem, M.; Kaether, C.; Capell, A.; Lammich, S.; Multhaup, G.; Haass, C. Phosphorylation regulates intracellular trafficking of β-secretase. J. Biol. Chem. 2001, 276, 14634–14641. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.H.; von Arnim, C.A.; Hyman, B.T.; Tanzi, R.E.; Tesco, G. BACE is degraded via the lysosomal pathway. J. Biol. Chem. 2005, 280, 32499–32504. [Google Scholar] [CrossRef]
- He, X.; Li, F.; Chang, W.-P.; Tang, J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE). J. Biol. Chem. 2005, 280, 11696–11703. [Google Scholar] [CrossRef]
- Wahle, T.; Thal, D.R.; Sastre, M.; Rentmeister, A.; Bogdanovic, N.; Famulok, M.; Heneka, M.T.; Walter, J. GGA1 is expressed in the human brain and affects the generation of amyloid β-peptide. J. Neurosci. 2006, 26, 12838–12846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, C.-L.; Tang, F.-L.; Peng, Y.; Shen, C.-Y.; Mei, L.; Xiong, W.-C. VPS35 regulates developing mouse hippocampal neuronal morphogenesis by promoting retrograde trafficking of BACE1. Biol. Open 2012, 1, 1248–1257. [Google Scholar] [CrossRef]
- Geisberger, S.; Maschke, U.; Gebhardt, M.; Kleinewietfeld, M.; Manzel, A.; Linker, R.A.; Chidgey, A.; Dechend, R.; Nguyen, G.; Daumke, O. New role for the (pro)renin receptor in T-cell development. Blood 2015, 126, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.-F.; Jung, J.-U.; Shun, C.; Xiong, S.; Xiong, L.; Shi, X.-M.; Mei, L.; Xiong, W.-C. Swedish mutant APP suppresses osteoblast differentiation and causes osteoporotic deficit, which are ameliorated by N-acetyl-L-cysteine. J. Bone Miner. Res. 2013, 28, 2122–2135. [Google Scholar] [CrossRef]
- Tang, F.-L.; Liu, W.; Hu, J.-X.; Erion, J.R.; Ye, J.; Mei, L.; Xiong, W.-C. VPS35 Deficiency or Mutation Causes Dopaminergic Neuronal Loss by Impairing Mitochondrial Fusion and Function. Cell Rep. 2015, 12, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-J.; Wang, C.-Z.; Dai, P.-G.; Xie, Y.; Song, N.-N.; Liu, Y.; Du, Q.-S.; Mei, L.; Ding, Y.-Q.; Xiong, W.-C. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat. Cell Biol. 2007, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, G.M., III; Lee, S.-H.; Singh, D.; Yuan, Y.; Ng, Y.-G.; Reichardt, L.F.; Arikkath, J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 2012, 7, 1741. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, G. High Ca 2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc. 2006, 1, 695. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Jung, J.-U.; Guo, H.-H.; Pan, J.-X.; Sun, X.-D.; Mei, L.; Xiong, W.-C. Osteoblastic Lrp4 promotes osteoclastogenesis by regulating ATP release and adenosine-A2AR signaling. J. Cell Biol. 2017, 216, 761–778. [Google Scholar] [CrossRef]
- Takeshi, K.; Kaori, C.; Yo-Ichi, N.; Mikio, H. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. Embo J. EMBO J. 2014, 22, 4190–4201. [Google Scholar]
- Guo, Y.; He, X.; Zhao, L.; Liu, L.; Song, H.; Wang, X.; Xu, J.; Ju, X.; Guo, W.; Zhu, X. Gβ2 Regulates the Multipolar-Bipolar Transition of Newborn Neurons in the Developing Neocortex; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192. [Google Scholar] [CrossRef]
- Das, U.; Scott, D.A.; Ganguly, A.; Koo, E.H.; Tang, Y.; Roy, S. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 2013, 79, 447–460. [Google Scholar] [CrossRef]
- He, W.; Lu, Y.; Qahwash, I.; Hu, X.-Y.; Chang, A.; Yan, R. Reticulon family members modulate BACE1 activity and amyloid-β peptide generation. Nat. Med. 2004, 10, 959. [Google Scholar] [CrossRef]
- Wen, L.; Tang, F.-L.; Hong, Y.; Luo, S.-W.; Wang, C.-L.; He, W.; Shen, C.; Jung, J.-U.; Xiong, F.; Lee, D.-h. VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J. Cell Biol. 2011, 195, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Kandalepas, P.C.; Sadleir, K.R.; Eimer, W.A.; Zhao, J.; Nicholson, D.A.; Vassar, R. The Alzheimer’s β-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013, 126, 329–352. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.L.; Biscaro, B.; Piazza, F.; Tesco, G. BACE1 endocytosis and trafficking are differentially regulated by ubiquitination at lysine 501 and the di-leucine motif in the C-terminus. J. Biol. Chem. 2012, 287, 42867–42880. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Dunn, I.C.; Wray, S.; Liu, Y.; Chappell, P.E.; Levine, J.E.; Radovick, S. An alternative gonadotropin-releasing hormone (GnRH) RNA splicing product found in cultured GnRH neurons and mouse hypothalamus. J. Biol. Chem. 1997, 272, 12620–12625. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Noriega, A.; Grubb, J.H.; Talkad, V.; Sly, W.S. Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J. Cell Biol. 1980, 85, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Homewood, C.; Warhurst, D.; Peters, W.; Baggaley, V. Lysosomes, pH and the anti-malarial action of chloroquine. Nature 1972, 235, 50. [Google Scholar] [CrossRef] [PubMed]
- Bowman, E.J.; Siebers, A.; Altendorf, K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells and plant cells. Proc. Natl. Acad. Sci. USA 1988, 85, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Altendorf, K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 1997, 200, 1–8. [Google Scholar] [PubMed]
- Yoshimori, T.; Yamamoto, A.; Moriyama, Y.; Futai, M.; Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H (+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991, 266, 17707–17712. [Google Scholar]
- Anthony, R.; Gabin, S.; Martijn, R.; Michael, B. (Pro)renin receptor and V-ATPase: From Drosophila to humans. Clin. Sci. 2014, 126, 529–536. [Google Scholar]
- Ludwig, J.; Kerscher, S.; Brandt, U.; Pfeiffer, K.; Getlawi, F.; Apps, D.K.; Schagger, H. Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J. Biol. Chem. 2015, 273, 10939–10947. [Google Scholar] [CrossRef]
- Trepiccione, F.; Gerber, S.D.; Grahammer, F.; López-Cayuqueo, K.I.; Baudrie, V.; Păunescu, T.G.; Capen, D.E.; Picard, N.; Alexander, R.T.; Huber, T.B. Renal Atp6ap2/(Pro)renin Receptor Is Required for Normal Vacuolar H+-ATPase Function but Not for the Renin-Angiotensin System. J. Am. Soc. Nephrol. 2016, 27, 3320–3330. [Google Scholar] [CrossRef]
- Yoichi, O.; Kenichiro, K.; Atsuhiro, I.; Mariyo, S.; Asako, K.M.; Kanako, B.; Tatsuya, N.; Hideaki, K.; Ge-Hong, S.W.; Yoh, W. Prorenin receptor is essential for normal podocyte structure and function. J. Am. Soc. Nephrol. 2012, 22, 2203–2212. [Google Scholar]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases — nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 2002, 3, 94. [Google Scholar] [CrossRef]
- Kenichiro, K.; Atsuhiro, I.; Motoaki, S.; Ge-Hong, S.W.; Yoh, W.; Asako, K.M.; Kanako, B.; Tatsuya, N.; Yoichi, O.; Mariyo, S. The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circul. Res. 2010, 107, 30–34. [Google Scholar]
- Riediger, F.; Quack, I.; Qadri, F.; Hartleben, B.; Park, J.K.; Potthoff, S.A.; Sohn, D.; Sihn, G.; Rousselle, A.; Fokuhl, V. Prorenin receptor is essential for podocyte autophagy and survival. J. Am. Soc. Nephrol. 2011, 22, 2193. [Google Scholar] [CrossRef]
- Pan, J.-X.; Tang, F.; Xiong, F.; Xiong, L.; Zeng, P.; Wang, B.; Zhao, K.; Guo, H.; Shun, C.; Xia, W.-F. APP promotes osteoblast survival and bone formation by regulating mitochondrial function and preventing oxidative stress. Cell Death Dis. 2018, 9, 1077. [Google Scholar] [CrossRef]
- Patterson, G.H.; Knobel, S.M.; Sharif, W.D.; Kain, S.R.; Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997, 73, 2782–2790. [Google Scholar] [CrossRef]
- Llopis, J.; McCaffery, J.M.; Miyawaki, A.; Farquhar, M.G.; Tsien, R.Y. Measurement of cytosolic, mitochondrial and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 6803–6808. [Google Scholar] [CrossRef]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2009, 11, 50. [Google Scholar] [CrossRef]
- Demaurex, N. pH Homeostasis of Cellular Organelles. Physiology 2002, 17, 1–5. [Google Scholar] [CrossRef]
- Jankowski, A.; Kim, J.H.; Collins, R.F.; Daneman, R.; Walton, P.; Grinstein, S. In situ measurements of the pH of mammalian peroxisomes using the fluorescent protein pHluorin. J. Biol. Chem. 2001, 276, 48748–48753. [Google Scholar] [CrossRef]
- Machen, T.E.; Leigh, M.J.; Taylor, C.; Kimura, T.; Asano, S.; Moore, H.-P.H. pH of TGN and recycling endosomes of H+/K+-ATPase-transfected HEK-293 cells: Implications for pH regulation in the secretory pathway. Am. J. Physiol. Cell Physiol. 2003, 285, C205–C214. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rost, B.R.; Schneider, F.; Grauel, M.K.; Wozny, C.; Bentz, C.G.; Blessing, A.; Rosenmund, T.; Jentsch, T.J.; Schmitz, D.; Hegemann, P. Optogenetic acidification of synaptic vesicles and lysosomes. Nat. Neurosci. 2015, 18, 1845. [Google Scholar] [CrossRef] [PubMed]
- Ohara-Imaizumi, M.; Nakamichi, Y.; Tanaka, T.; Katsuta, H.; Ishida, H.; Nagamatsu, S. Monitoring of exocytosis and endocytosis of insulin secretory granules in the pancreatic β-cell line MIN6 using pH-sensitive green fluorescent protein (pHluorin) and confocal laser microscopy. Biochem. J. 2002, 363, 73–80. [Google Scholar] [CrossRef]
- Prosser, D.C.; Whitworth, K.; Wendland, B. Quantitative analysis of endocytosis with cytoplasmic pHluorin chimeras. Traffic 2010, 11, 1141–1150. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Uchiyama, Y. A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS ONE 2014, 9, e110600. [Google Scholar] [CrossRef]
- Karagiannis, J.; Young, P.G. Intracellular pH homeostasis during cell-cycle progression and growth state transition in Schizosaccharomyces pombe. J. Cell Sci. 2001, 114, 2929–2941. [Google Scholar] [PubMed]
- Kopec, C.D.; Li, B.; Wei, W.; Boehm, J.; Malinow, R. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J. Neurosci. 2006, 26, 2000–2009. [Google Scholar] [CrossRef]
- Lin, D.-T.; Huganir, R.L. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J. Neurosci. 2007, 27, 13903–13908. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Chi, Z.; Keuss, M.J.; Pai, Y.-M.E.; Kang, H.C.; Shin, J.-h.; Bugayenko, A.; Wang, H.; Xiong, Y. The AAA+ ATPase Thorase regulates AMPA receptor-dependent synaptic plasticity and behavior. Cell 2011, 145, 284–299. [Google Scholar] [CrossRef]
- Bauereiss, A.; Welzel, O.; Jung, J.; Grosse-Holz, S.; Lelental, N.; Lewczuk, P.; Wenzel, E.M.; Kornhuber, J.; Groemer, T.W. Surface trafficking of APP and BACE in live cells. Traffic 2015, 16, 655–675. [Google Scholar] [CrossRef][Green Version]
- Buggia-Prévot, V.; Fernandez, C.G.; Udayar, V.; Vetrivel, K.S.; Elie, A.; Roseman, J.; Sasse, V.A.; Lefkow, M.; Meckler, X.; Bhattacharyya, S. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer’s disease Aβ production. Cell Rep. 2013, 5, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Das, U.; Wang, L.; Ganguly, A.; Saikia, J.M.; Wagner, S.L.; Koo, E.H.; Roy, S. Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat. Neurosci. 2016, 19, 55. [Google Scholar] [CrossRef]
- Maxson, M.E.; Grinstein, S. The vacuolar-type H+-ATPase at a glance – more than a proton pump. J. Cell Sci. 2014, 127, 4987–4993. [Google Scholar] [CrossRef] [PubMed]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917. [Google Scholar] [CrossRef]
- Valenzuela, R.; Barroso-Chinea, P.; Villar-Cheda, B.; Joglar, B.; Munoz, A.; Lanciego, J.L.; Labandeira-Garcia, J.L. Location of prorenin receptors in primate substantia nigra: Effects on dopaminergic cell death. J. Neuropathol. Exp. Neurol. 2010, 69, 1130–1142. [Google Scholar] [CrossRef]
- Korvatska, O.; Strand, N.S.; Berndt, J.D.; Strovas, T.; Chen, D.-H.; Leverenz, J.B.; Kiianitsa, K.; Mata, I.F.; Karakoc, E.; Greenup, J.L.; et al. Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum. Mol. Genet. 2013, 22, 3259–3268. [Google Scholar] [CrossRef] [PubMed]
- Gupta, H.V.; Vengoechea, J.; Sahaya, K.; Virmani, T. A splice site mutation in ATP6AP2 causes X-linked intellectual disability, epilepsy and parkinsonism. Parkinsonism Relat. Disord. 2015, 21, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Vassar, R.; Kovacs, D.M.; Yan, R.; Wong, P.C. The β-Secretase Enzyme BACE in Health and Alzheimer’s Disease: Regulation, Cell Biology, Function and Therapeutic Potential. J. Neurosci. 2009, 29, 12787–12794. [Google Scholar] [CrossRef]
- Vassar, R.; Kandalepas, P.C. The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s disease. Alzheimers Res. Ther. 2011, 3, 20. [Google Scholar] [CrossRef]
- Sinha, S.; Anderson, J.P.; Barbour, R.; Basi, G.S.; Caccavello, R.; Davis, D.; Doan, M.; Dovey, H.F.; Frigon, N.; Hong, J.; et al. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 1999, 402, 537. [Google Scholar] [CrossRef] [PubMed]
- Citron, M.; Oltersdorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the β-amyloid precursor protein in familial Alzheimer’s disease increases β-protein production. Nature 1992, 360, 672. [Google Scholar] [CrossRef] [PubMed]
- Kitazume, S.; Tachida, Y.; Oka, R.; Shirotani, K.; Saido, T.C.; Hashimoto, Y. Alzheimer’s β-secretase, β-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc. Natl. Acad. Sci. USA 2001, 98, 13554–13559. [Google Scholar] [CrossRef]
- Lichtenthaler, S.F.; Dominguez, D.I.; Westmeyer, G.G.; Reiss, K.; Haass, C.; Saftig, P.; De Strooper, B.; Seed, B. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J. Biol. Chem. 2003, 278, 48713–48719. [Google Scholar] [CrossRef] [PubMed]
- Eggert, S.; Paliga, K.; Soba, P.; Evin, G.; Masters, C.L.; Weidemann, A.; Beyreuther, K. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves α-, β-, γ- and epsilon-like cleavages. Modulation of APLP-1 processing by N-glycosylation. J. Biol. Chem. 2004, 279, 18146–18156. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Südhof, T.C. Cleavage of amyloid-β precursor protein and amyloid-β precursor-like protein by BACE 1. J. Biol. Chem. 2004, 279, 10542–10550. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, L.; Ikin, A.; Lamprianou, S.; Vacaresse, N.; Revelli, J.; Platt, K.; Paganetti, P.; Mathews, P.; Harroch, S.; Buxbaum, J. BACE (β-secretase) modulates the processing of APLP2 in vivo. Mol. Cell. Neurosci. 2004, 25, 642–649. [Google Scholar] [CrossRef]
- Von Arnim, C.A.; Kinoshita, A.; Peltan, I.D.; Tangredi, M.M.; Herl, L.; Lee, B.M.; Spoelgen, R.; Hshieh, T.T.; Ranganathan, S.; Battey, F.D. The low density lipoprotein receptor-related protein (LRP) is a novel β-secretase (BACE1) substrate. J. Biol. Chem. 2005, 280, 17777–17785. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Ingano, L.A.M.; Carey, B.W.; Pettingell, W.P.; Kovacs, D.M. Presenilin/γ-secretase-mediated cleavage of the voltage-gated sodium channel β2 subunit regulates cell adhesion and migration. J. Biol. Chem. 2005, 280, 23251–23261. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.-K.; Sakurai, T.; Oyama, F.; Kaneko, K.; Wada, K.; Miyazaki, H.; Kurosawa, M.; De Strooper, B.; Saftig, P.; Nukina, N. β Subunits of voltage-gated sodium channels are novel substrates of β-site amyloid precursor protein-cleaving enzyme (BACE1) and γ-secretase. J. Biol. Chem. 2005, 280, 23009–23017. [Google Scholar] [CrossRef]
- Hu, X.; Hicks, C.W.; He, W.; Wong, P.; Macklin, W.B.; Trapp, B.D.; Yan, R. Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 2006, 9, 1520. [Google Scholar] [CrossRef] [PubMed]
- Willem, M.; Garratt, A.N.; Novak, B.; Citron, M.; Kaufmann, S.; Rittger, A.; DeStrooper, B.; Saftig, P.; Birchmeier, C.; Haass, C. Control of peripheral nerve myelination by the β-secretase BACE1. Science 2006, 314, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; He, W.; Diaconu, C.; Tang, X.; Kidd, G.J.; Macklin, W.B.; Trapp, B.D.; Yan, R. Genetic deletion of BACE1 in mice affects remyelination of sciatic nerves. FASEB J. 2008, 22, 2970–2980. [Google Scholar] [CrossRef] [PubMed]
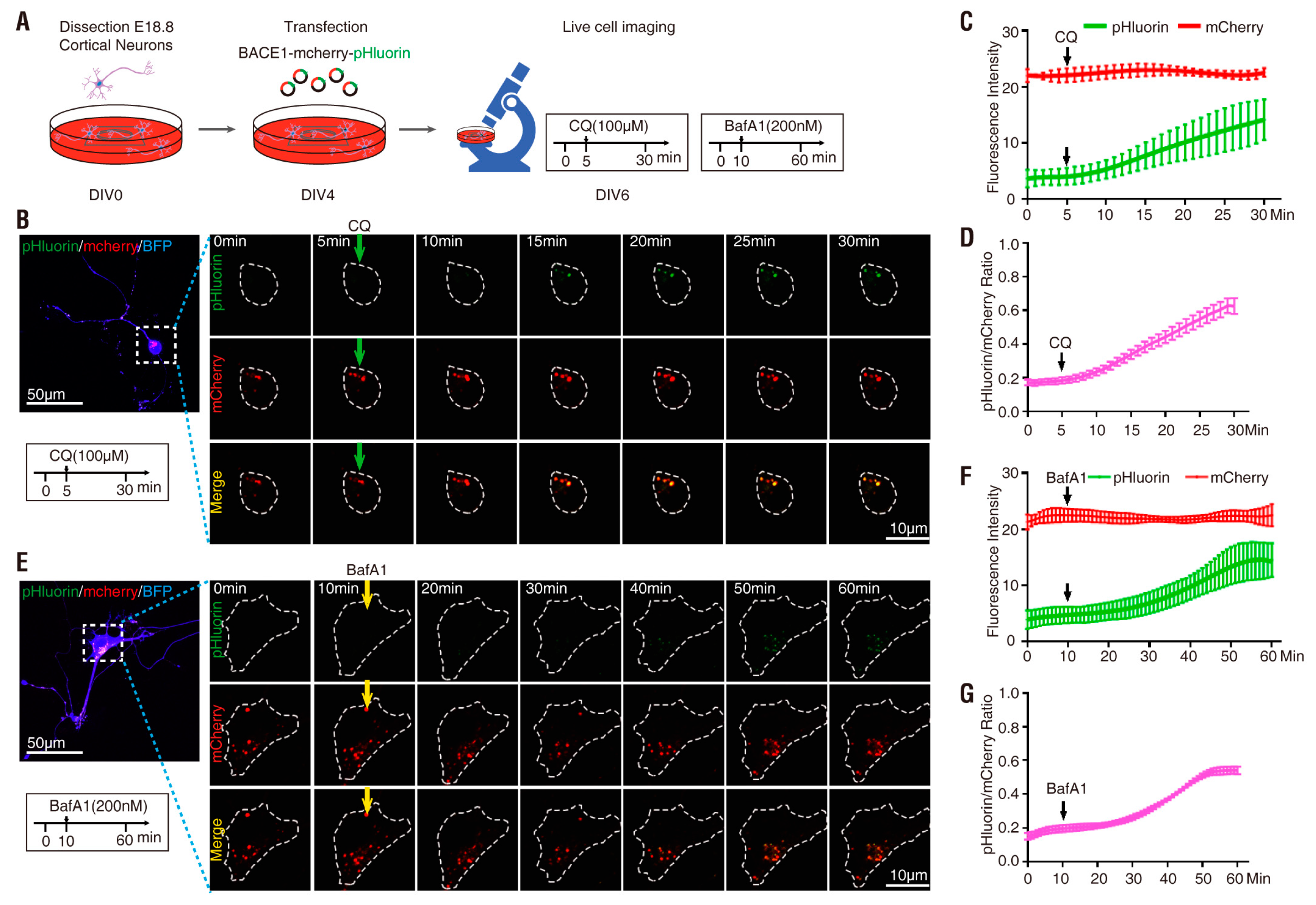
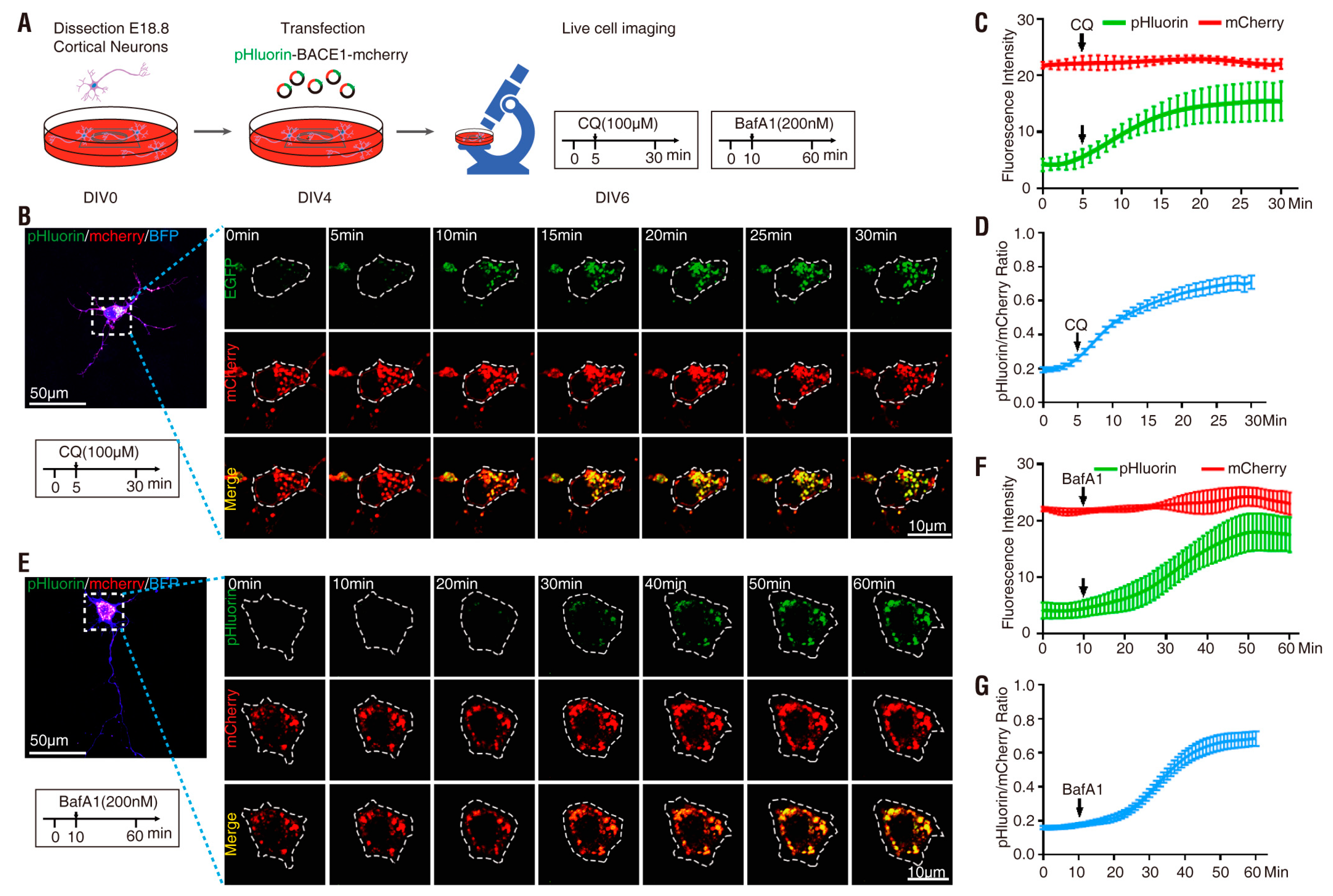
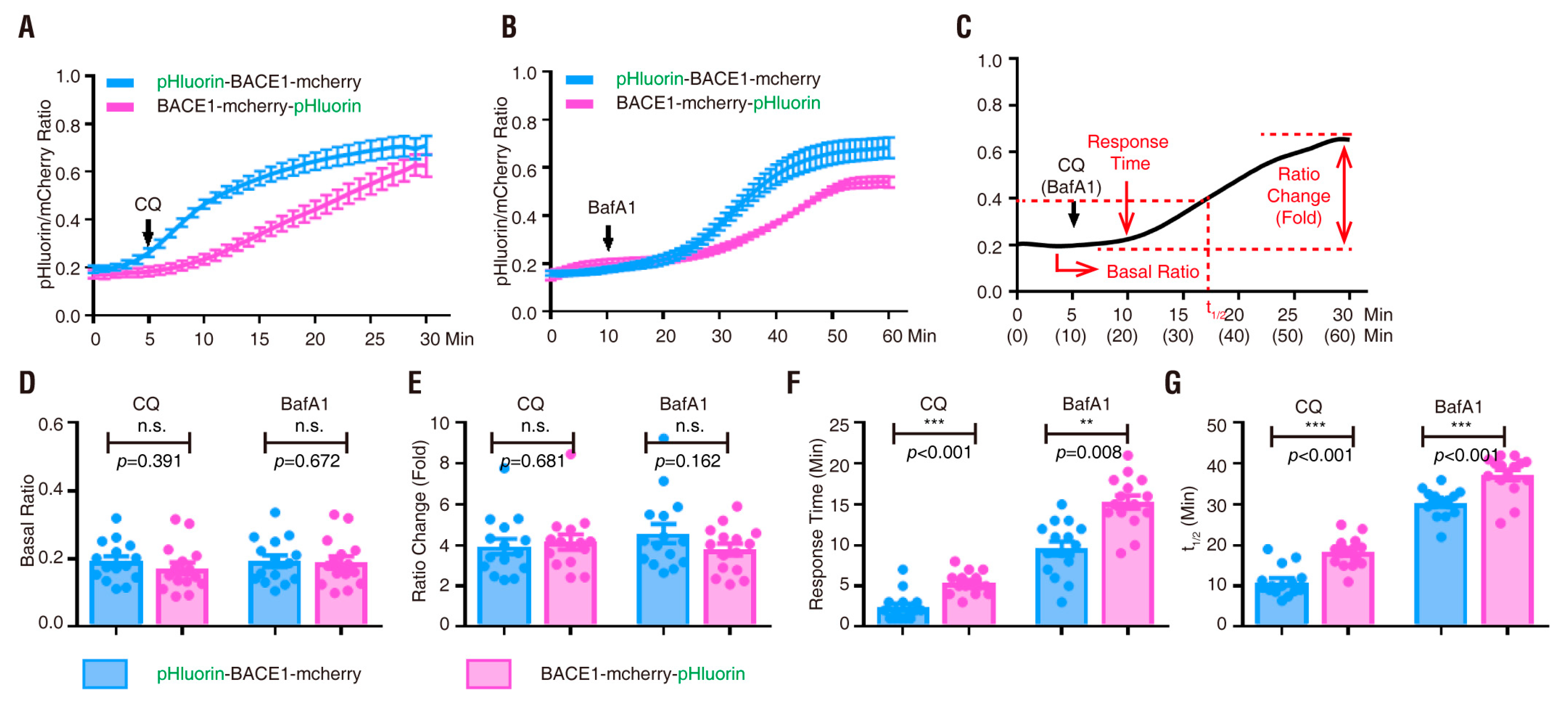
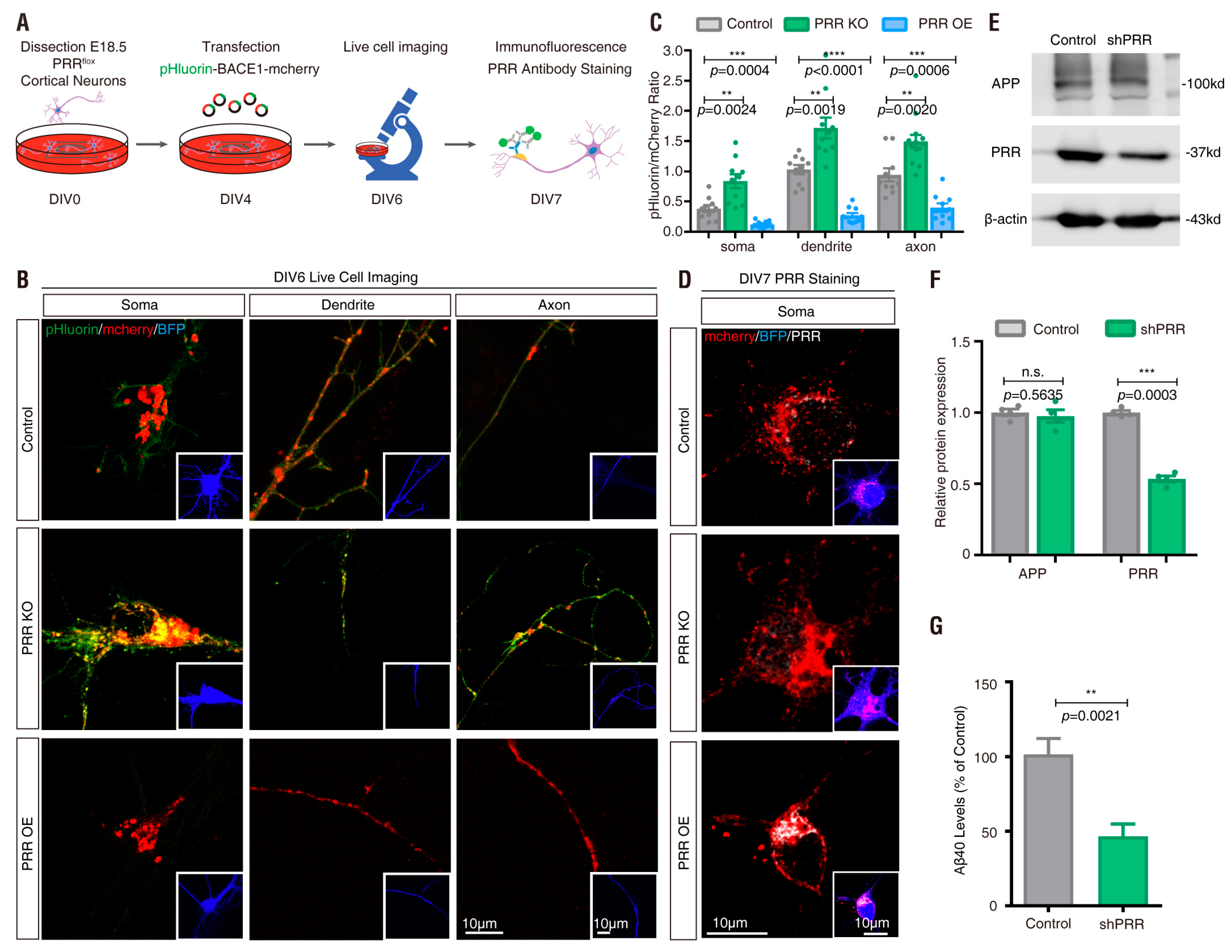
| Plasmid | Drug Treatment | Basal Ratio | Ratio Change (fold) | Response Time (min) | t1/2 (min) |
|---|---|---|---|---|---|
| pHluorin-BACE1-mCherry | CQ | 0.185 ± 0.059 | 4.971 ± 1.548 | 3.067 ± 1.163 | 14.673 ± 2.038 |
| BACE1-mCherry-pHluorin | CQ | 0.211 ± 0.079 | 4.505 ± 1.774 | 5.333 ± 1.234 | 20.960 ± 2.681 |
| BACE1-mCherry-EGFP | CQ | 0.414 ± 0.092 | 2.031 ± 0.664 | 11.067 ± 1.223 | 20.060 ± 1.206 |
| pHluorin-BACE1-mCherry | BafA1 | 0.191 ± 0.050 | 4.344 ± 1.022 | 5.200 ± 1.474 | 24.680 ± 3.153 |
| BACE1-mCherry-pHluorin | BafA1 | 0.203 ± 0.069 | 4.099 ± 1.436 | 13.933 ± 2.631 | 35.640 ± 3.193 |
| BACE1-mCherry-EGFP | BafA1 | 0.385 ± 0.082 | 1.918 ± 0.592 | 14.667 ± 1.799 | 36.153 ± 3.408 |
| Plasmid | Drug Treatment | Basal Ratio | Ratio Change (fold) | Response Time (min) | t1/2 (min) |
|---|---|---|---|---|---|
| pHluorin-BACE1-mCherry | CQ | 0.192 ± 0.059 | 3.937 ± 1.455 | 2.400 ± 0.682 | 10.953 ± 3.596 |
| BACE1-mCherry-pHluorin | CQ | 0.172 ± 0.068 | 4.157 ± 1.443 | 5.333 ± 1.345 | 18.267 ± 3.625 |
| pHluorin-BACE1-mCherry | BafA1 | 0.193 ± 0.066 | 4.570 ± 1.810 | 9.600 ± 3.334 | 30.333 ± 3.400 |
| BACE1-mCherry-pHluorin | BafA1 | 0.189 ± 0.070 | 3.778 ± 1.137 | 15.267 ± 3.195 | 37.207 ± 4.881 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Zhao, Y.; Tang, F.-L.; Xiong, L.; Su, C.; Mei, L.; Zhu, X.-J.; Xiong, W.-C. pHluorin-BACE1-mCherry Acts as a Reporter for the Intracellular Distribution of Active BACE1 In Vitro and In Vivo. Cells 2019, 8, 474. https://doi.org/10.3390/cells8050474
Zhao L, Zhao Y, Tang F-L, Xiong L, Su C, Mei L, Zhu X-J, Xiong W-C. pHluorin-BACE1-mCherry Acts as a Reporter for the Intracellular Distribution of Active BACE1 In Vitro and In Vivo. Cells. 2019; 8(5):474. https://doi.org/10.3390/cells8050474
Chicago/Turabian StyleZhao, Lu, Yang Zhao, Fu-Lei Tang, Lei Xiong, Ce Su, Lin Mei, Xiao-Juan Zhu, and Wen-Cheng Xiong. 2019. "pHluorin-BACE1-mCherry Acts as a Reporter for the Intracellular Distribution of Active BACE1 In Vitro and In Vivo" Cells 8, no. 5: 474. https://doi.org/10.3390/cells8050474
APA StyleZhao, L., Zhao, Y., Tang, F.-L., Xiong, L., Su, C., Mei, L., Zhu, X.-J., & Xiong, W.-C. (2019). pHluorin-BACE1-mCherry Acts as a Reporter for the Intracellular Distribution of Active BACE1 In Vitro and In Vivo. Cells, 8(5), 474. https://doi.org/10.3390/cells8050474




