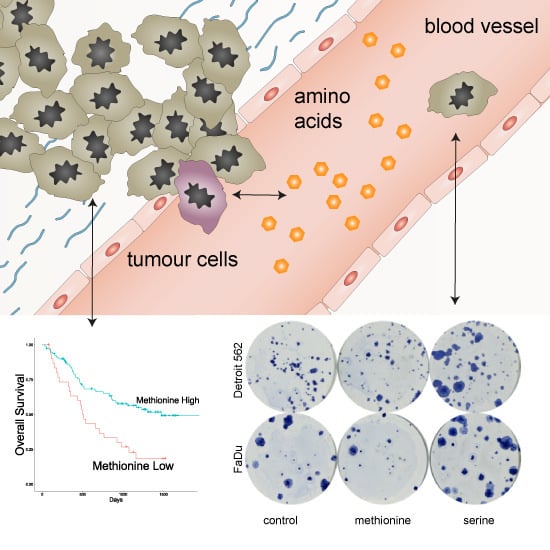
Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection and Samples Preparation
2.2. Amino Acids Profiling
2.3. Colony Forming Assay
2.4. Statistical Analysis
3. Results
3.1. Clinicopathological Characterization of HNSCC Patients
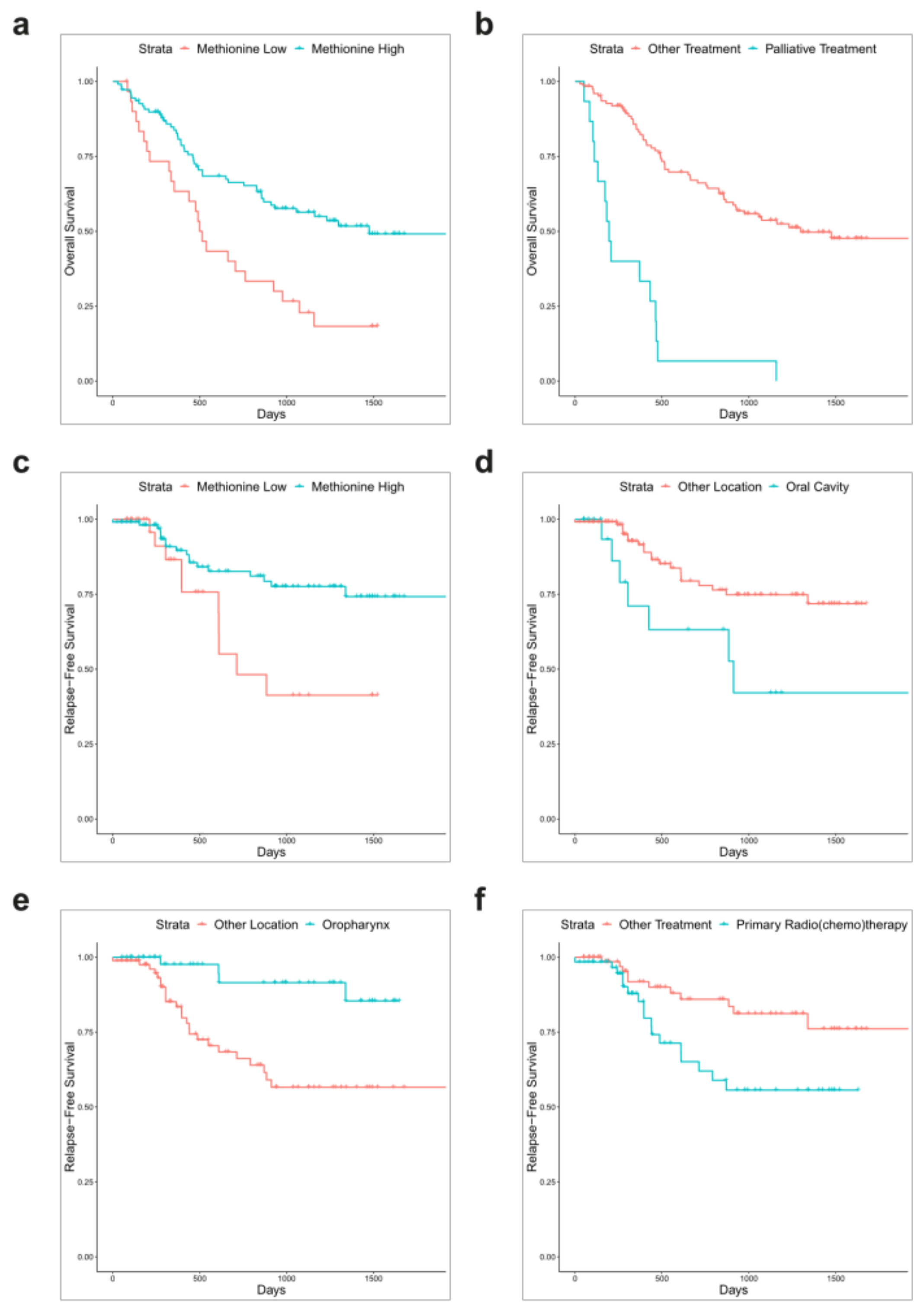
3.2. Association between Amino Acid Serum Levels and Survival
3.3. Multivariate Analysis
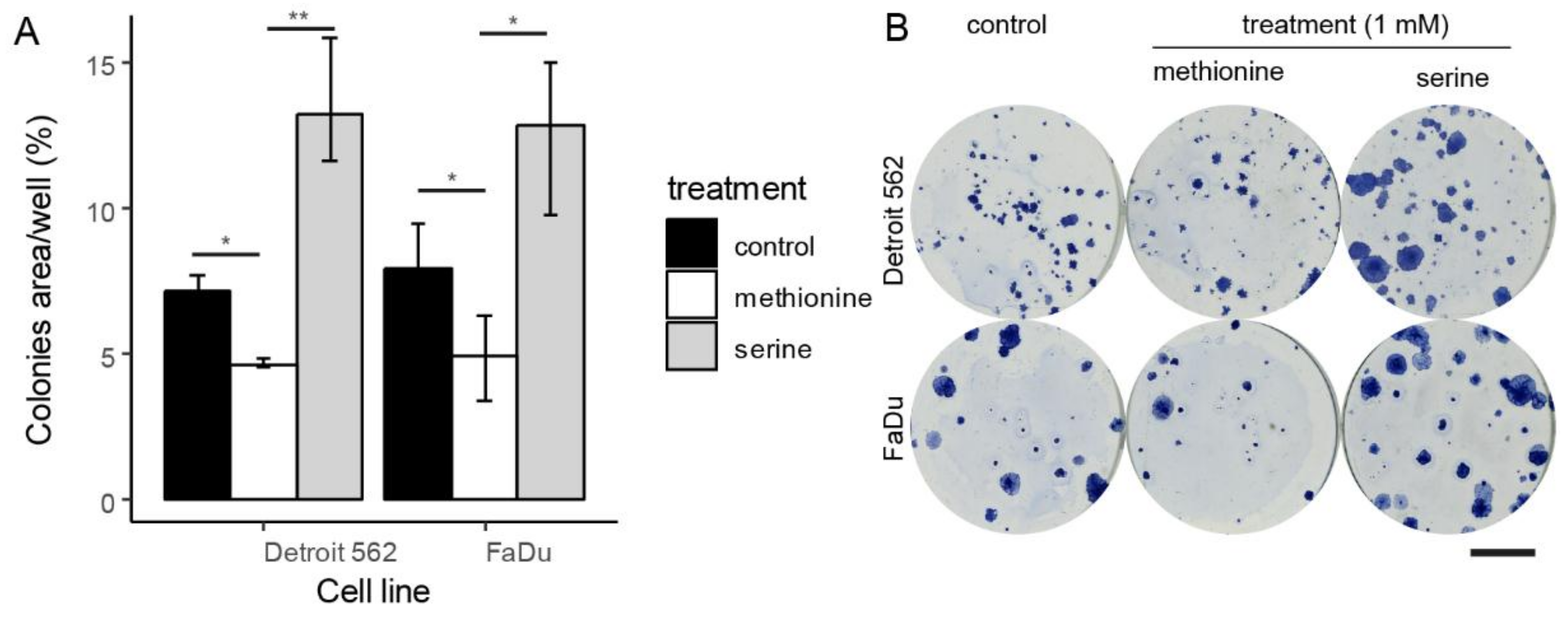
3.4. Colony-Forming Capacity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Goldson, T.M.; Han, Y.; Knight, K.B.; Weiss, H.L.; Resto, V.A. Clinicopathological predictors of lymphatic metastasis in HNSCC: Implications for molecular mechanisms of metastatic disease. J. Exp Ther. Oncol. 2010, 8, 211–221. [Google Scholar]
- Polanska, H.; Raudenska, M.; Gumulec, J.; Sztalmachova, M.; Adam, V.; Kizek, R.; Masarik, M. Clinical significance of head and neck squamous cell cancer biomarkers. Oral. Oncol. 2014, 50, 168–177. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef]
- Li, X.; Wenes, M.; Romero, P.; Huang, S.C.-C.; Fendt, S.-M.; Ho, P.-C. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 2019. [Google Scholar] [CrossRef]
- Vučetić, M.; Cormerais, Y.; Parks, S.K.; Pouysségur, J. The Central Role of Amino Acids in Cancer Redox Homeostasis: Vulnerability Points of the Cancer Redox Code. Front. Oncol. 2017, 7, 319. [Google Scholar] [CrossRef]
- Klupczynska, A.; Derezinski, P.; Dyszkiewicz, W.; Pawlak, K.; Kasprzyk, M.; Kokot, Z.J. Evaluation of serum amino acid profiles’ utility in non-small cell lung cancer detection in Polish population. Lung Cancer 2016, 100, 71–76. [Google Scholar] [CrossRef]
- Gu, Y.; Chen, T.; Fu, S.; Sun, X.; Wang, L.; Wang, J.; Lu, Y.; Ding, S.; Ruan, G.; Teng, L.; et al. Perioperative dynamics and significance of amino acid profiles in patients with cancer. J. Transl. Med. 2015, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Plewa, S.; Horala, A.; Derezinski, P.; Klupczynska, A.; Nowak-Markwitz, E.; Matysiak, J.; Kokot, Z.J. Usefulness of Amino Acid Profiling in Ovarian Cancer Screening with Special Emphasis on Their Role in Cancerogenesis. Int. J. Mol. Sci. 2017, 18, 2727. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, X.; Yin, M.; Fan, L.; Zhang, H.; Zhao, F.; Zhang, W.; Ke, C.; Zhang, G.; Hou, Y.; et al. Discrimination between malignant and benign ovarian tumors by plasma metabolomic profiling using ultra performance liquid chromatography/mass spectrometry. Clin. Chim. Acta 2012, 413, 861–868. [Google Scholar] [CrossRef]
- Wirtz, E.D.; Hoshino, D.; Maldonado, A.T.; Tyson, D.R.; Weaver, A.M. Response of head and neck squamous cell carcinoma cells carrying PIK3CA mutations to selected targeted therapies. JAMA Otolaryngol Head Neck Surg. 2015, 141, 543–549. [Google Scholar] [CrossRef]
- Lausen, B.; Schumacher, M. Maximally Selected Rank Statistics. Biometrics 1992, 48, 73–85. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Scioscia, K.A.; Snyderman, C.H.; Wagner, R. Altered serum amino acid profiles in head and neck cancer. Nutr. Cancer 1998, 30, 144–147. [Google Scholar] [CrossRef]
- Toyoda, M.; Kaira, K.; Ohshima, Y.; Ishioka, N.S.; Shino, M.; Sakakura, K.; Takayasu, Y.; Takahashi, K.; Tominaga, H.; Oriuchi, N.; et al. Prognostic significance of amino-acid transporter expression (LAT1, ASCT2, and xCT) in surgically resected tongue cancer. Br. J. Cancer 2014, 110, 2506–2513. [Google Scholar] [CrossRef]
- Ruth, M.R.; Field, C.J. The immune modifying effects of amino acids on gut-associated lymphoid tissue. J. Anim. Sci. Biotechnol. 2013, 4, 27. [Google Scholar] [CrossRef]
- Curis, E.; Nicolis, I.; Moinard, C.; Osowska, S.; Zerrouk, N.; Bénazeth, S.; Cynober, L. Almost all about citrulline in mammals. Amino Acids 2005, 29, 177. [Google Scholar] [CrossRef]
- Crenn, P.; Messing, B.; Cynober, L. Citrulline as a biomarker of intestinal failure due to enterocyte mass reduction. Clin. Nutr. 2008, 27, 328–339. [Google Scholar] [CrossRef]
- van Vliet, M.J.; Tissing, W.J.; Rings, E.H.; Koetse, H.A.; Stellaard, F.; Kamps, W.A.; de Bont, E.S. Citrulline as a marker for chemotherapy induced mucosal barrier injury in pediatric patients. Pediatr Blood Cancer 2009, 53, 1188–1194. [Google Scholar] [CrossRef]
- van Monsjou, H.S.; Wreesmann, V.B.; van den Brekel, M.W.; Balm, A.J. Head and neck squamous cell carcinoma in young patients. Oral. Oncol. 2013, 49, 1097–1102. [Google Scholar] [CrossRef]
- Du, E.; Mazul, A.L.; Farquhar, D.; Brennan, P.; Anantharaman, D.; Abedi-Ardekani, B.; Weissler, M.C.; Hayes, D.N.; Olshan, A.F.; Zevallos, J.P. Long-term Survival in Head and Neck Cancer: Impact of Site, Stage, Smoking, and Human Papillomavirus Status. The Laryngoscope 2019, 0. [Google Scholar] [CrossRef]
- Hoffman, R.M. Altered methionine metabolism and transmethylation in cancer. Anticancer Res. 1985, 5, 1–30. [Google Scholar]
- Kune, G.; Watson, L. Colorectal cancer protective effects and the dietary micronutrients folate, methionine, vitamins B6, B12, C, E, selenium, and lycopene. Nutr. Cancer 2006, 56, 11–21. [Google Scholar] [CrossRef]
- Giovannucci, E.; Stampfer, M.J.; Colditz, G.A.; Rimm, E.B.; Trichopoulos, D.; Rosner, B.A.; Speizer, F.E.; Willett, W.C. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J. Natl. Cancer Inst. 1993, 85, 875–884. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Hoffman, R.M. Is DNA methylation the new guardian of the genome? Mol. Cytogenet. 2017, 10, 11. [Google Scholar] [CrossRef]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef]
- Lien, E.C.; Ghisolfi, L.; Geck, R.C.; Asara, J.M.; Toker, A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Lui, V.W.Y.; Hedberg, M.L.; Li, H.; Vangara, B.S.; Pendleton, K.; Zeng, Y.; Lu, Y.; Zhang, Q.; Du, Y.; Gilbert, B.R.; et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013, 3, 761–769. [Google Scholar] [CrossRef]
- Yonezawa, K.; Nishiumi, S.; Kitamoto-Matsuda, J.; Fujita, T.; Morimoto, K.; Yamashita, D.; Saito, M.; Otsuki, N.; Irino, Y.; Shinohara, M.; et al. Serum and tissue metabolomics of head and neck cancer. Cancer Genom. Proteom. 2013, 10, 233–238. [Google Scholar]
- Munasinghe, L.L.; Robinson, J.L.; Harding, S.V.; Brunton, J.A.; Bertolo, R.F. Protein Synthesis in Mucin-Producing Tissues Is Conserved When Dietary Threonine Is Limiting in Piglets. J. Nutr. 2017, 147, 202–210. [Google Scholar] [CrossRef]
- Faure, M.; Moënnoz, D.; Montigon, F.; Mettraux, C.; Breuillé, D.; Ballèvre, O. Dietary Threonine Restriction Specifically Reduces Intestinal Mucin Synthesis in Rats. J. Nutr. 2005, 135, 486–491. [Google Scholar] [CrossRef]
- Holmes, A.J.; Chew, Y.V.; Colakoglu, F.; Cliff, J.B.; Klaassens, E.; Read, M.N.; Solon-Biet, S.M.; McMahon, A.C.; Cogger, V.C.; Ruohonen, K.; et al. Diet-Microbiome Interactions in Health Are Controlled by Intestinal Nitrogen Source Constraints. Cell Metab. 2017, 25, 140–151. [Google Scholar] [CrossRef]
- Engen, P.A.; Green, S.J.; Voigt, R.M.; Forsyth, C.B.; Keshavarzian, A. The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol Res. 2015, 37, 223–236. [Google Scholar]
- Touchefeu, Y.; Montassier, E.; Nieman, K.; Gastinne, T.; Potel, G.; Bruley des Varannes, S.; Le Vacon, F.; de La Cochetiere, M.F. Systematic review: The role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis - current evidence and potential clinical applications. Aliment. Pharm. Ther. 2014, 40, 409–421. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vétizou, M.; Daillère, R.; Merad, M.; Kroemer, G. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015, 7, 271ps1. [Google Scholar] [CrossRef]
- Fukutake, N.; Ueno, M.; Hiraoka, N.; Shimada, K.; Shiraishi, K.; Saruki, N.; Ito, T.; Yamakado, M.; Ono, N.; Imaizumi, A.; et al. A Novel Multivariate Index for Pancreatic Cancer Detection Based On the Plasma Free Amino Acid Profile. PLoS ONE 2015, 10, e0132223. [Google Scholar] [CrossRef]
- Mattaini, K.R.; Sullivan, M.R.; Vander Heiden, M.G. The importance of serine metabolism in cancer. J. Cell Biol. 2016, 214, 249–257. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef]
- de Arruda Leme, I.; Portari, G.V.; Padovan, G.J.; Rosa, F.T.; de Mello-Filho, F.V.; Marchini, J.S. Amino acids in squamous cell carcinomas and adjacent normal tissues from patients with larynx and oral cavity lesions. Clinics 2012, 67, 1225–1227. [Google Scholar] [CrossRef]
- Gao, X.; Lee, K.; Reid, M.A.; Sanderson, S.M.; Qiu, C.; Li, S.; Liu, J.; Locasale, J.W. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018, 22, 3507–3520. [Google Scholar] [CrossRef]
- LeBleu, V.S.; O’Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; de Carvalho, F.M.; Damascena, A.; Domingos Chinen, L.T.; Rocha, R.M.; et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003. [Google Scholar] [CrossRef]
- Viale, A.; Pettazzoni, P.; Lyssiotis, C.A.; Ying, H.; Sanchez, N.; Marchesini, M.; Carugo, A.; Green, T.; Seth, S.; Giuliani, V.; et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514, 628–632. [Google Scholar] [CrossRef]
- Poschke, I.; Mao, Y.; Kiessling, R.; de Boniface, J. Tumor-dependent increase of serum amino acid levels in breast cancer patients has diagnostic potential and correlates with molecular tumor subtypes. J. Transl. Med. 2013, 11, 290. [Google Scholar] [CrossRef]
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS ONE 2011, 6, e24143. [Google Scholar] [CrossRef]
- Bi, X.; Henry, C.J. Plasma-free amino acid profiles are predictors of cancer and diabetes development. Nutr. Diabetes 2017, 7, e249. [Google Scholar] [CrossRef]
| Parameter | Level of Parameter | n |
|---|---|---|
| Sex | ||
| Male | 133 | |
| Female | 7 | |
| Median Age (Range) | 67 (48–93) | |
| Tumor Location | ||
| Oral Cavity | 20 | |
| Oropharynx | 54 | |
| Hypopharynx | 23 | |
| Larynx | 43 | |
| Differentiation | ||
| Low Grade | 8 | |
| Intermediate Grade | 101 | |
| High Grade | 31 | |
| T stage | ||
| T1 | 17 | |
| T2 | 44 | |
| T3 | 29 | |
| T4 | 50 | |
| N stage | ||
| N0 | 52 | |
| N1 | 19 | |
| N2 | 60 | |
| N3 | 9 | |
| M stage | ||
| M0 | 135 | |
| M1 | 5 | |
| Treatment | ||
| Surgery | 18 | |
| Surgery with adjuvant RT/CT | 44 | |
| Primary RT/CT | 63 | |
| Palliative | 15 |
| Parameter | Univariate | Multivariate | |
|---|---|---|---|
| p | HR (95% CI) | p | |
| OS | |||
| Hypopharynx | >0.001 | 1.3596 (0.7103–2.6025) | 0.3538 |
| Oropharynx | 0.0314 | 0.7232 (0.3910–1.3376) | 0.3017 |
| T 3,4 vs. T 1,2 | 0.0486 | 1.5954 (0.9483–2.6843) | 0.0784 |
| N 2,3 vs. N 0,1 | 0.00776 | 1.7314 (0.9869–3.0374) | 0.0556 |
| Palliative Treatment | >0.001 | 3.6380 (1.2499–10.5885) | 0.0178 |
| Primary RT/CT | 0.0258 | 1.1562 (0.4655–2.8715) | 0.7545 |
| Surgery with adjuvant RT/CT | >0.001 | 0.3771 (0.1375–1.0338) | 0.0580 |
| Alanine | 0.00482 | 0.7182 (0.4138–1.2467) | 0.2395 |
| Cystine | 0.0331 | 1.0765 (0.6204–1.8679) | 0.7932 |
| Methionine | >0.001 | 0.5248 (0.3056–0.9013) | 0.0195 |
| RFS | |||
| Oral Cavity | 0.0223 | 4.6898 (1.49124–14.7489) | 0.00820 |
| Oropharynx | 0.00292 | 0.2998 (0.09951–0.9034) | 0.03232 |
| Primary RT/CT | 0.0181 | 5.1464 (1.89193–13.9992) | 0.00133 |
| Threonine | 0.033 | 0.2893 (0.06630–1.2623) | 0.09892 |
| Methionine | 0.0136 | 0.4373 (0.19240–0.9941) | 0.04836 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vsiansky, V.; Svobodova, M.; Gumulec, J.; Cernei, N.; Sterbova, D.; Zitka, O.; Kostrica, R.; Smilek, P.; Plzak, J.; Betka, J.; et al. Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers. Cells 2019, 8, 428. https://doi.org/10.3390/cells8050428
Vsiansky V, Svobodova M, Gumulec J, Cernei N, Sterbova D, Zitka O, Kostrica R, Smilek P, Plzak J, Betka J, et al. Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers. Cells. 2019; 8(5):428. https://doi.org/10.3390/cells8050428
Chicago/Turabian StyleVsiansky, Vit, Marketa Svobodova, Jaromir Gumulec, Natalia Cernei, Dagmar Sterbova, Ondrej Zitka, Rom Kostrica, Pavel Smilek, Jan Plzak, Jan Betka, and et al. 2019. "Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers" Cells 8, no. 5: 428. https://doi.org/10.3390/cells8050428
APA StyleVsiansky, V., Svobodova, M., Gumulec, J., Cernei, N., Sterbova, D., Zitka, O., Kostrica, R., Smilek, P., Plzak, J., Betka, J., Kalfert, D., Masarik, M., & Raudenska, M. (2019). Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers. Cells, 8(5), 428. https://doi.org/10.3390/cells8050428