Nuclear Mechanics in the Fission Yeast
Abstract
1. Introduction
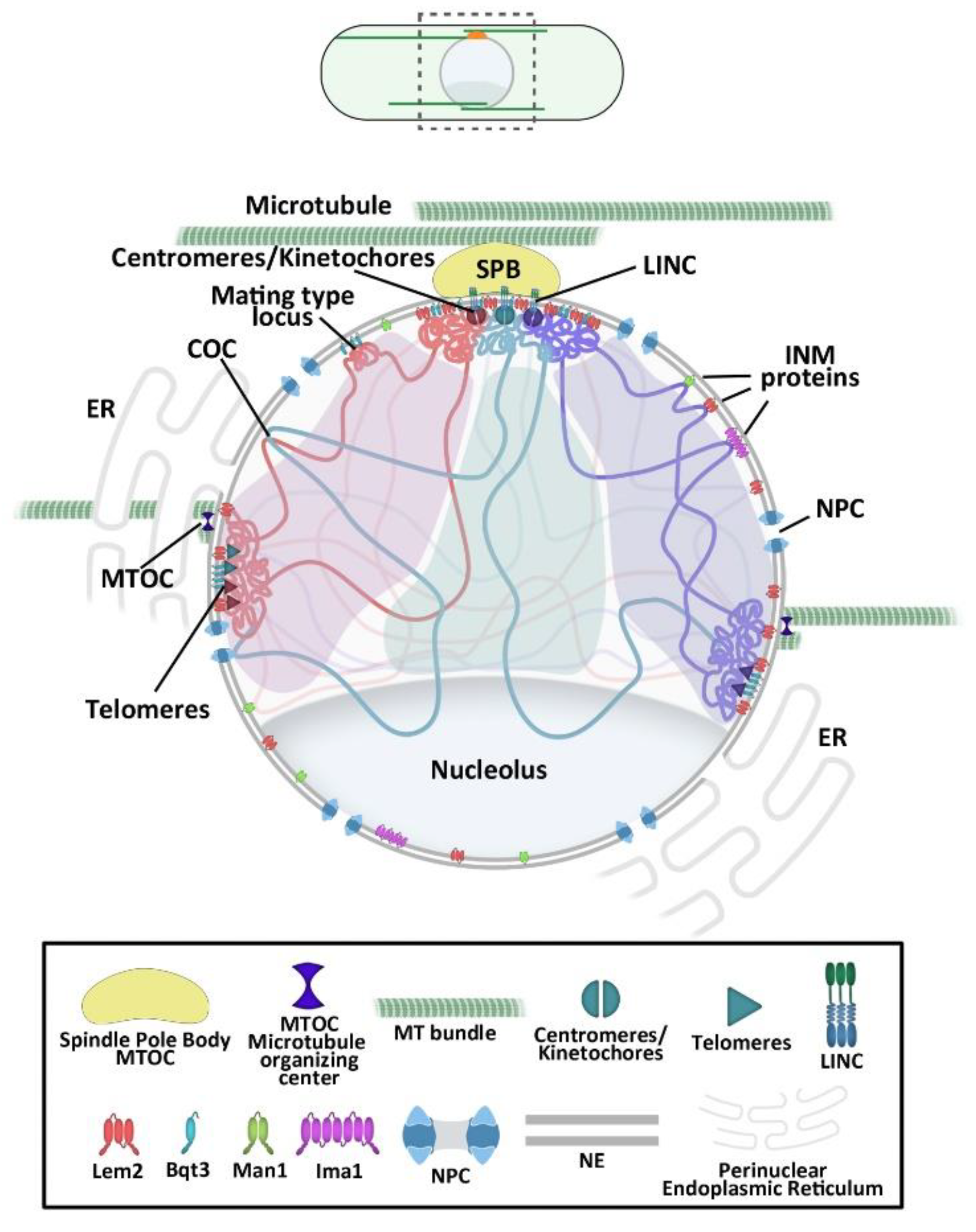
2. Overall Chromosomal Organization in the Fission Yeast
3. The NE as Genome 3D Organizer
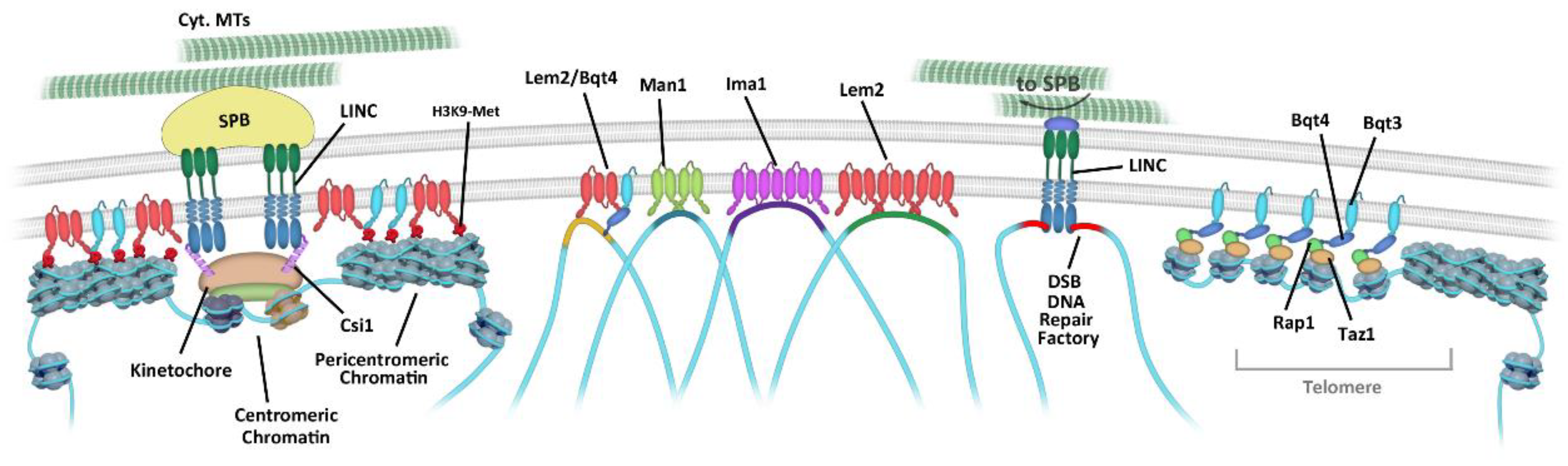
3.1. Centromere Attachment to the NE
3.2. Telomere Tethering to the NE
3.3. INM Microdomains
3.4. Transcription Boundaries
4. Nuclear Organization is Necessary to Support Nuclear Mechanics
Chromatin Tethers to the NE Influence the Mechanical Response of the Nucleus
5. Effect of MT Cytoskeletal Forces on Chromatin Dynamics
5.1. MTs and Dynein Modulate the Extent of Chromatin Contacts During Meiotic Prophase
5.2. Interphase MT Movements Promote the Repair of Persistent DSBs
5.3. Interphase Chromosomal Movements Affect Distribution of Cohesin Into Chromosomes and the Efficiency of DNA Repair
6. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mekhail, K.; Moazed, D. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 2010, 11, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T.; Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009, 10, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Fabre, E.; Zimmer, C. From dynamic chromatin architecture to DNA damage repair and back. Nucleus 2018, 9, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Grosschedl, R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007, 21, 3027–3043. [Google Scholar] [CrossRef]
- Gong, K.; Tjong, H.; Zhou, X.J.; Alber, F. Comparative 3D genome structure analysis of the fission and the budding yeast. PLoS ONE 2015, 10, e0119672. [Google Scholar] [CrossRef]
- Matsuda, A.; Asakawa, H.; Haraguchi, T.; Hiraoka, Y. Spatial organization of the Schizosaccharomyces pombe genome within the nucleus. Yeast 2017, 34, 55–66. [Google Scholar] [CrossRef]
- Misteli, T. Beyond the sequence: Cellular organization of genome function. Cell 2007, 128, 787–800. [Google Scholar] [CrossRef]
- Mizuguchi, T.; Barrowman, J.; Grewal, S.I. Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS Lett. 2015, 589, 2975–2986. [Google Scholar] [CrossRef]
- Noma, K.I. The yeast genomes in three dimensions: Mechanisms and functions. Annu. Rev. Genet. 2017, 51, 23–44. [Google Scholar] [CrossRef]
- Romero-Bueno, R.; Ruiz, P.D.; Artal-Sanz, M.; Askjaer, P.; Dobrzynska, A. Nuclear organization in stress and aging. Cells 2019, 8, 664. [Google Scholar] [CrossRef]
- Li, F.; An, Z.; Zhang, Z. The dynamic 3D genome in gametogenesis and early embryonic development. Cells 2019, 8, 788. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.L.; Soshnev, A.A.; Geyer, P.K. Networking in the nucleus: A spotlight on LEM-domain proteins. Curr. Opin. Cell Biol. 2015, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Towbin, D.B.; Gonzalez-Sandoval, A.; Gasser, S.M. Mechanisms of heterochromatin subnuclear localization. Trends Biochem. Sci. 2013, 38, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Cabianca, S.D.; Gasser, S.M. Spatial segregation of heterochromatin: Uncovering functionality in a multicellular organism. Nucleus 2016, 7, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Buchwalter, A.; Kaneshiro, J.M.; Hetzer, M.W. Coaching from the sidelines: The nuclear periphery in genome regulation. Nat. Rev. Genet. 2019, 20, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Politz, C.J.; Scalzo, D.; Groudine, M. Something silent this way forms: The functional organization of the repressive nuclear compartment. Annu. Rev. Cell Dev. Biol. 2013, 29, 241–270. [Google Scholar] [CrossRef]
- Agrawal, A.; Lele, T.P. Mechanics of nuclear membranes. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Kirby, J.T.; Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef]
- Maurer, M.; Lammerding, J. The driving force: Nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef]
- Fedorchak, R.G.; Kaminski, A.; Lammerding, J. Cellular mechanosensing: Getting to the nucleus of it all. Prog. Biophys. Mol. Biol. 2014, 115, 76–92. [Google Scholar] [CrossRef]
- Hieda, M. Signal transduction across the nuclear envelope: Role of the LINC complex in bidirectional signaling. Cells 2019, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Allshire, R.C.; Ekwall, K. Epigenetic regulation of chromatin states in schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 2015, 7, a018770. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, H.; Iwasaki, O.; Tanaka, A.; Capizzi, J.R.; Wickramasinghe, P.; Lee, M.; Fu, Z.; Noma, K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucl. Acids Res. 2010, 38, 8164–8177. [Google Scholar] [CrossRef] [PubMed]
- Kniola, B.; O’Toole, E.; McIntosh, J.R.; Mellone, B.; Allshire, R.; Mengarelli, S.; Hultenby, K.; Ekwall, K. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 2001, 12, 2767–2775. [Google Scholar] [CrossRef]
- Gonzalez, Y.; Saito, A.; Sazer, S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 2012, 3, 60–76. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Maekawa, H.; Asakawa, H.; Chikashige, Y.; Kojidani, T.; Osakada, H.; Matsuda, A.; Haraguchi, T. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 2011, 16, 1000–1011. [Google Scholar] [CrossRef]
- Steglich, B.; Filion, G.J.; van Steensel, B.; Ekwall, K. The inner nuclear membrane proteins Man1 and Ima1 link to two different types of chromatin at the nuclear periphery in S. pombe. Nucleus 2012, 3, 77–87. [Google Scholar] [CrossRef]
- Sawin, E.K.; Tran, P.T. Cytoplasmic microtubule organization in fission yeast. Yeast 2006, 23, 1001–1014. [Google Scholar] [CrossRef]
- Tran, T.P.; Marsh, L.; Doye, V.; Inoue, S.; Chang, F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001, 153, 397–411. [Google Scholar] [CrossRef]
- Bao, X.X.; Spanos, C.; Kojidani, T.; Lynch, E.M.; Rappsilber, J.; Hiraoka, Y.; Haraguchi, T.; Sawin, K.E. Exportin Crm1 is repurposed as a docking protein to generate microtubule organizing centers at the nuclear pore. Elife 2018, 7, e33465. [Google Scholar] [CrossRef]
- Daga, R.R.; Chang, F. Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA 2005, 102, 8228–8232. [Google Scholar] [CrossRef] [PubMed]
- Chacon, R.M.; Delivani, P.; Tolic, I.M. Meiotic nuclear oscillations are necessary to avoid excessive chromosome associations. Cell Rep. 2016, 17, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Tanizawa, H.; Iwasaki, O.; Corcoran, C.J.; Capizzi, J.R.; Hayden, J.E.; Noma, K. Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast. J. Cell Sci. 2013, 126, 5271–5283. [Google Scholar] [CrossRef] [PubMed]
- Swartz, K.R.; Rodriguez, E.C.; King, M.C. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol. Biol. Cell 2014, 25, 2461–2471. [Google Scholar] [CrossRef]
- Zhurinsky, J.; Salas-Pino, S.; Iglesias-Romero, A.B.; Torres-Mendez, A.; Knapp, B.; Flor-Parra, I.; Wang, J.; Bao, K.; Jia, S.; Chang, F.; et al. Effects of the microtubule nucleator Mto1 on chromosome movement, DNA repair and sister chromatid cohesion in fission yeast. Mol. Biol. Cell 2019. [Google Scholar] [CrossRef]
- Daga, R.R.; Yonetani, A.; Chang, F. Asymmetric microtubule pushing forces in nuclear centering. Curr. Biol. 2006, 16, 1544–1550. [Google Scholar] [CrossRef]
- Funabiki, H.; Hagan, I.; Uzawa, S.; Yanagida, M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993, 121, 961–976. [Google Scholar] [CrossRef]
- Ekwall, K.; Javerzat, J.P.; Lorentz, A.; Schmidt, H.; Cranston, G.; Allshire, R. The chromodomain protein Swi6: A key component at fission yeast centromeres. Science 1995, 269, 1429–1431. [Google Scholar] [CrossRef]
- Noma, K.; Allis, C.D.; Grewal, S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 2001, 293, 1150–1155. [Google Scholar] [CrossRef]
- Uzawa, S.; Yanagida, M. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 1992, 101, 267–275. [Google Scholar]
- Alfredsson-Timmins, J.; Henningson, F.; Bjerling, P. The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J. Cell Sci. 2007, 120, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, H.; Masuda, H.; Jain, D.; Cooper, J.P. Distinct ‘safe zones’ at the nuclear envelope ensure robust replication of heterochromatic chromosome regions. Elife 2018, 7, e32911. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, O.; Tanaka, A.; Tanizawa, H.; Grewal, S.I.; Noma, K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell 2010, 21, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Noma, K.; Cam, H.P.; Maraia, R.J.; Grewal, S.I. A role for TFIIIC transcription factor complex in genome organization. Cell 2006, 125, 859–872. [Google Scholar] [CrossRef]
- Woolcock, J.K.; Stunnenberg, R.; Gaidatzis, D.; Hotz, H.R.; Emmerth, S.; Barraud, P.; Buhler, M. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 2012, 26, 683–692. [Google Scholar] [CrossRef][Green Version]
- Harr, C.J.; Gonzalez-Sandoval, A.; Gasser, S.M. Histones and histone modifications in perinuclear chromatin anchoring: From yeast to man. EMBO Rep. 2016, 17, 139–155. [Google Scholar] [CrossRef]
- Mizuguchi, T.; Fudenberg, G.; Mehta, S.; Belton, J.M.; Taneja, N.; Folco, H.D.; FitzGerald, P.; Dekker, J.; Mirny, L.; Barrowman, J.; et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 2014, 516, 432–435. [Google Scholar] [CrossRef]
- Hou, H.; Zhou, Z.; Wang, Y.; Wang, J.; Kallgren, S.P.; Kurchuk, T.; Miller, E.A.; Chang, F.; Jia, S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J. Cell Biol. 2012, 199, 735–744. [Google Scholar] [CrossRef]
- Fernandez-Alvarez, A.; Bez, C.; O’Toole, E.T.; Morphew, M.; Cooper, J.P. Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev. Cell 2016, 39, 544–559. [Google Scholar] [CrossRef]
- Goto, B.; Okazaki, K.; Niwa, O. Cytoplasmic microtubular system implicated in de novo formation of a Rabl-like orientation of chromosomes in fission yeast. J. Cell Sci. 2001, 114, 2427–2435. [Google Scholar]
- Miki, F.; Kurabayashi, A.; Tange, Y.; Okazaki, K.; Shimanuki, M.; Niwa, O. Two-Hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genom. 2004, 270, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.; Yanagida, M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995, 129, 1033–1047. [Google Scholar] [CrossRef] [PubMed]
- Barrales, R.R.; Forn, M.; Georgescu, P.R.; Sarkadi, Z.; Braun, S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 2016, 30, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Tange, Y.; Chikashige, Y.; Takahata, S.; Kawakami, K.; Higashi, M.; Mori, C.; Kojidani, T.; Hirano, Y.; Asakawa, H.; Murakami, Y.; et al. Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells 2016, 21, 812–832. [Google Scholar] [CrossRef] [PubMed]
- Banday, S.; Farooq, Z.; Rashid, R.; Abdullah, E.; Altaf, M. Role of inner nuclear membrane protein complex Lem2-Nur1 in heterochromatic gene silencing. J. Biol. Chem. 2016, 291, 20021–20029. [Google Scholar] [CrossRef]
- Barrales, R.R.; Braun, S. Chromatin binding and silencing: Two roles of the same protein Lem2. Microbiol. Cell 2016, 3, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.; Barrales, R.R. Beyond tethering and the LEM domain: MSCellaneous functions of the inner nuclear membrane Lem2. Nucleus 2016, 7, 523–531. [Google Scholar] [CrossRef]
- Steglich, B.; Stralfors, A.; Khorosjutina, O.; Persson, J.; Smialowska, A.; Javerzat, J.P.; Ekwall, K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015, 11, e1005101. [Google Scholar] [CrossRef]
- Chikashige, Y.; Yamane, M.; Okamasa, K.; Tsutsumi, C.; Kojidani, T.; Sato, M.; Haraguchi, T.; Hiraoka, Y. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009, 187, 413–427. [Google Scholar] [CrossRef]
- Fujita, I.; Nishihara, Y.; Tanaka, M.; Tsujii, H.; Chikashige, Y.; Watanabe, Y.; Saito, M.; Ishikawa, F.; Hiraoka, Y.; Kanoh, J. Telomere-nuclear envelope dissociation promoted by Rap1 phosphorylation ensures faithful chromosome segregation. Curr. Biol. 2012, 22, 1932–1937. [Google Scholar] [CrossRef]
- Hirano, Y.; Kinugasa, Y.; Asakawa, H.; Chikashige, Y.; Obuse, C.; Haraguchi, T.; Hiraoka, Y. Lem2 is retained at the nuclear envelope through its interaction with Bqt4 in fission yeast. Genes Cells 2018, 23, 122–135. [Google Scholar] [CrossRef] [PubMed]
- Chikashige, Y.; Haraguchi, T.; Hiraoka, Y. Nuclear envelope attachment is not necessary for telomere function in fission yeast. Nucleus 2010, 1, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lawry, S.T.; Cohen, A.L.; Jia, S. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol. Life Sci. 2014, 71, 4841–4852. [Google Scholar] [CrossRef] [PubMed]
- Cam, P.H.; Sugiyama, T.; Chen, E.S.; Chen, X.; FitzGerald, P.C.; Grewal, S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005, 37, 809–819. [Google Scholar] [CrossRef]
- Stralfors, A.; Walfridsson, J.; Bhuiyan, H.; Ekwall, K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011, 7, e1001334. [Google Scholar] [CrossRef]
- Iwasaki, O.; Noma, K. Global genome organization mediated by RNA polymerase III-transcribed genes in fission yeast. Gene 2012, 493, 195–200. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanizawa, H.; Sriswasdi, S.; Iwasaki, O.; Chatterjee, A.G.; Speicher, D.W.; Levin, H.L.; Noguchi, E.; Noma, K. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell 2012, 48, 532–546. [Google Scholar] [CrossRef]
- Kim, D.K.; Tanizawa, H.; Iwasaki, O.; Noma, K. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat. Genet. 2016, 48, 1242–1252. [Google Scholar] [CrossRef]
- Hiraga, S.; Botsios, S.; Donze, D.; Donaldson, A.D. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol. Biol. Cell 2012, 23, 2741–2754. [Google Scholar] [CrossRef]
- Dahl, N.K.; Engler, A.J.; Pajerowski, J.D.; Discher, D.E. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 2005, 89, 2855–2864. [Google Scholar] [CrossRef]
- Furusawa, T.; Rochman, M.; Taher, L.; Dimitriadis, E.K.; Nagashima, K.; Anderson, S.; Bustin, M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 2015, 6, 6138. [Google Scholar] [CrossRef] [PubMed]
- Gerlitz, G.; Bustin, M. The role of chromatin structure in cell migration. Trends Cell Biol. 2011, 21, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Stephens, D.A.; Banigan, E.J.; Marko, J.F. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 2019, 58, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Dreger, M.; Madrazo, E.; Hurlstone, A.; Redondo-Munoz, J. Novel contribution of epigenetic changes to nuclear dynamics. Nucleus 2019, 10, 42–47. [Google Scholar] [CrossRef]
- Shimamoto, Y.; Tamura, S.; Masumoto, H.; Maeshima, K. Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol. Biol. Cell 2017, 28, 1580–1589. [Google Scholar] [CrossRef]
- Stephens, D.A.; Banigan, E.J.; Marko, J.F. Separate roles for chromatin and lamins in nuclear mechanics. Nucleus 2018, 9, 119–124. [Google Scholar] [CrossRef]
- Schreiner, M.S.; Koo, P.K.; Zhao, Y.; Mochrie, S.G.; King, M.C. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 2015, 6, 7159. [Google Scholar] [CrossRef]
- Kume, K.; Cantwell, H.; Burrell, A.; Nurse, P. Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat. Commun. 2019, 10, 1871. [Google Scholar] [CrossRef]
- Kinugasa, Y.; Hirano, Y.; Sawai, M.; Ohno, Y.; Shindo, T.; Asakawa, H.; Chikashige, Y.; Shibata, S.; Kihara, A.; Haraguchi, T.; et al. The very-long-chain fatty acid elongase Elo2 rescues lethal defects associated with loss of the nuclear barrier function in fission yeast cells. J. Cell Sci. 2019, 132, jcs229021. [Google Scholar] [CrossRef]
- Raucher, D.; Sheetz, M.P. Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 1999, 77, 1992–2002. [Google Scholar] [CrossRef]
- Enyedi, B.; Niethammer, P. Nuclear membrane stretch and its role in mechanotransduction. Nucleus 2017, 8, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Wolf, K.; Lammerding, J. Bursting the bubble—Nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 2017, 27, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Denais, M.C.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; Lindert, M.T.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Vargas, D.J.; Hatch, E.M.; Anderson, D.J.; Hetzer, M.W. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 2012, 3, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Isermann, P.; Lammerding, J. Consequences of a tight squeeze: Nuclear envelope rupture and repair. Nucleus 2017, 8, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Foethke, D.; Makushok, T.; Brunner, D.; Nedelec, F. Force- and length-dependent catastrophe activities explain interphase microtubule organization in fission yeast. Mol. Syst. Biol. 2009, 5, 241. [Google Scholar] [CrossRef] [PubMed]
- Gluncic, M.; Maghelli, N.; Krull, A.; Krstic, V.; Ramunno-Johnson, D.; Pavin, N.; Tolic, I.M. Kinesin-8 motors improve nuclear centering by promoting microtubule catastrophe. Phys. Rev. Lett. 2015, 114, 078103. [Google Scholar] [CrossRef]
- Dogterom, M.; Yurke, B. Measurement of the force-velocity relation for growing microtubules. Science 1997, 278, 856–860. [Google Scholar] [CrossRef]
- Ananthanarayanan, V.; Schattat, M.; Vogel, S.K.; Krull, A.; Pavin, N.; Tolic-Norrelykke, I.M. Dynein motion switches from diffusive to directed upon cortical anchoring. Cell 2013, 153, 1526–1536. [Google Scholar] [CrossRef]
- Vogel, K.S.; Pavin, N.; Maghelli, N.; Julicher, F.; Tolic-Norrelykke, I.M. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009, 7, e1000087. [Google Scholar] [CrossRef]
- Yamamoto, A.; West, R.R.; McIntosh, J.R.; Hiraoka, Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 1999, 145, 1233–1249. [Google Scholar] [CrossRef] [PubMed]
- Chikashige, Y.; Haraguchi, T.; Hiraoka, Y. Another way to move chromosomes. Chromosoma 2007, 116, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, Y.; Dernburg, A.F. The SUN rises on meiotic chromosome dynamics. Dev. Cell 2009, 17, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Chikashige, Y.; Ding, D.Q.; Funabiki, H.; Haraguchi, T.; Mashiko, S.; Yanagida, M.; Hiraoka, Y. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994, 264, 270–273. [Google Scholar] [CrossRef]
- Ding, Q.D.; Chikashige, Y.; Haraguchi, T.; Hiraoka, Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 1998, 111, 701–712. [Google Scholar]
- Ding, Q.D.; Yamamoto, A.; Haraguchi, T.; Hiraoka, Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 2004, 6, 329–341. [Google Scholar] [CrossRef]
- Christophorou, N.; Rubin, T.; Bonnet, I.; Piolot, T.; Arnaud, M.; Huynh, J.R. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 2015, 17, 1388–1400. [Google Scholar] [CrossRef]
- Caridi, C.P.; Delabaere, L.; Zapotoczny, G.; Chiolo, I. And yet, it moves: Nuclear and chromatin dynamics of a heterochromatic double-strand break. Philos. Trans. R Soc. Lond B Biol. Sci. 2017, 372, 20160291. [Google Scholar] [CrossRef]
- Ryu, T.; Spatola, B.; Delabaere, L.; Bowlin, K.; Hopp, H.; Kunitake, R.; Karpen, G.H.; Chiolo, I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015, 17, 1401–1411. [Google Scholar] [CrossRef]
- Oza, P.; Jaspersen, S.L.; Miele, A.; Dekker, J.; Peterson, C.L. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009, 23, 912–927. [Google Scholar] [CrossRef]
- Horigome, C.; Bustard, D.E.; Marcomini, I.; Delgoshaie, N.; Tsai-Pflugfelder, M.; Cobb, J.A.; Gasser, S.M. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 2016, 30, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Kalocsay, M.; Hiller, N.J.; Jentsch, S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 2009, 33, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Samejima, I.; Miller, V.J.; Groocock, L.M.; Sawin, K.E. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J. Cell Sci. 2008, 121, 3971–3980. [Google Scholar] [CrossRef] [PubMed]
- Samejima, I.; Miller, V.J.; Rincon, S.A.; Sawin, K.E. Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr. Biol. 2010, 20, 1959–1965. [Google Scholar] [CrossRef]
- Sawin, E.K.; Lourenco, P.C.; Snaith, H.A. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 2004, 14, 763–775. [Google Scholar] [CrossRef]
- Venkatram, S.; Tasto, J.J.; Feoktistova, A.; Jennings, J.L.; Link, A.J.; Gould, L.K. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 2004, 15, 2287–2301. [Google Scholar] [CrossRef]
- Zimmerman, S.; Chang, F. Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell 2005, 16, 2719–2733. [Google Scholar] [CrossRef]
- Lawrimore, J.; Barry, T.M.; Barry, R.M.; York, A.C.; Friedman, B.; Cook, D.M.; Akialis, K.; Tyler, J.; Vasquez, P.; Yeh, E.; et al. Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol. Biol. Cell 2017, 28, 1701–1711. [Google Scholar] [CrossRef]
- Mine-Hattab, J.; Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Bernard, P.; Maure, J.F.; Partridge, J.F.; Genier, S.; Javerzat, J.P.; Allshire, R.C. Requirement of heterochromatin for cohesion at centromeres. Science 2001, 294, 2539–2542. [Google Scholar] [CrossRef]
- Bhardwaj, S.; Schlackow, M.; Rabajdova, M.; Gullerova, M. Transcription facilitates sister chromatid cohesion on chromosomal arms. Nucl. Acids Res. 2016, 44, 6676–6692. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomonaga, T.; Nagao, K.; Kawasaki, Y.; Furuya, K.; Murakami, A.; Morishita, J.; Yuasa, T.; Sutani, T.; Kearsey, S.E.; Uhlmann, F.; et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000, 14, 2757–2770. [Google Scholar] [CrossRef] [PubMed]
- Sidhwani, P.; Yelon, D. Fluid forces shape the embryonic heart: Insights from zebrafish. Curr. Top Dev. Biol. 2019, 132, 395–416. [Google Scholar] [PubMed]
- Boselli, F.; Steed, E.; Freund, J.B.; Vermot, J. Anisotropic shear stress patterns predict the orientation of convergent tissue movements in the embryonic heart. Development 2017, 144, 4322–4327. [Google Scholar] [CrossRef] [PubMed]
- Huse, M. Mechanical forces in the immune system. Nat. Rev. Immunol. 2017, 17, 679–690. [Google Scholar] [CrossRef]
- Barzilai, S.; Yadav, S.K.; Morrell, S.; Roncato, F.; Klein, E.; Stoler-Barak, L.; Golani, O.; Feigelson, S.W.; Zemel, A.; Nourshargh, S.; et al. Leukocytes breach endothelial barriers by insertion of nuclear lobes and disassembly of endothelial actin filaments. Cell Rep. 2017, 18, 685–699. [Google Scholar] [CrossRef]
- Wilson, H.M.; Holzbaur, E.L. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 2015, 142, 218–228. [Google Scholar] [CrossRef]
- Wu, K.Y.; Umeshima, H.; Kurisu, J.; Kengaku, M. Nesprins and opposing microtubule motors generate a point force that drives directional nuclear motion in migrating neurons. Development 2018, 145, dev158782. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallardo, P.; Barrales, R.R.; Daga, R.R.; Salas-Pino, S. Nuclear Mechanics in the Fission Yeast. Cells 2019, 8, 1285. https://doi.org/10.3390/cells8101285
Gallardo P, Barrales RR, Daga RR, Salas-Pino S. Nuclear Mechanics in the Fission Yeast. Cells. 2019; 8(10):1285. https://doi.org/10.3390/cells8101285
Chicago/Turabian StyleGallardo, Paola, Ramón R. Barrales, Rafael R. Daga, and Silvia Salas-Pino. 2019. "Nuclear Mechanics in the Fission Yeast" Cells 8, no. 10: 1285. https://doi.org/10.3390/cells8101285
APA StyleGallardo, P., Barrales, R. R., Daga, R. R., & Salas-Pino, S. (2019). Nuclear Mechanics in the Fission Yeast. Cells, 8(10), 1285. https://doi.org/10.3390/cells8101285



