The Two-Way Role of Jagged1 in Cancer: A Focus on CRC
Highlights
- Jagged1 is cleaved, releasing an intracellular domain (Jag1-ICD).
- Jag1-ICD acquires oncogenic properties in CRC.
- Jagged1 is a novel oncogenic driver that contributes to the multistep genetic model underlying the adenoma-to-carcinoma sequence in CRC.
- Understanding the two-way role of Jagged1 could lead to new strategies for addressing drug resistance in CRC.
Abstract
1. Introduction
2. Colorectal Cancer: An Overview
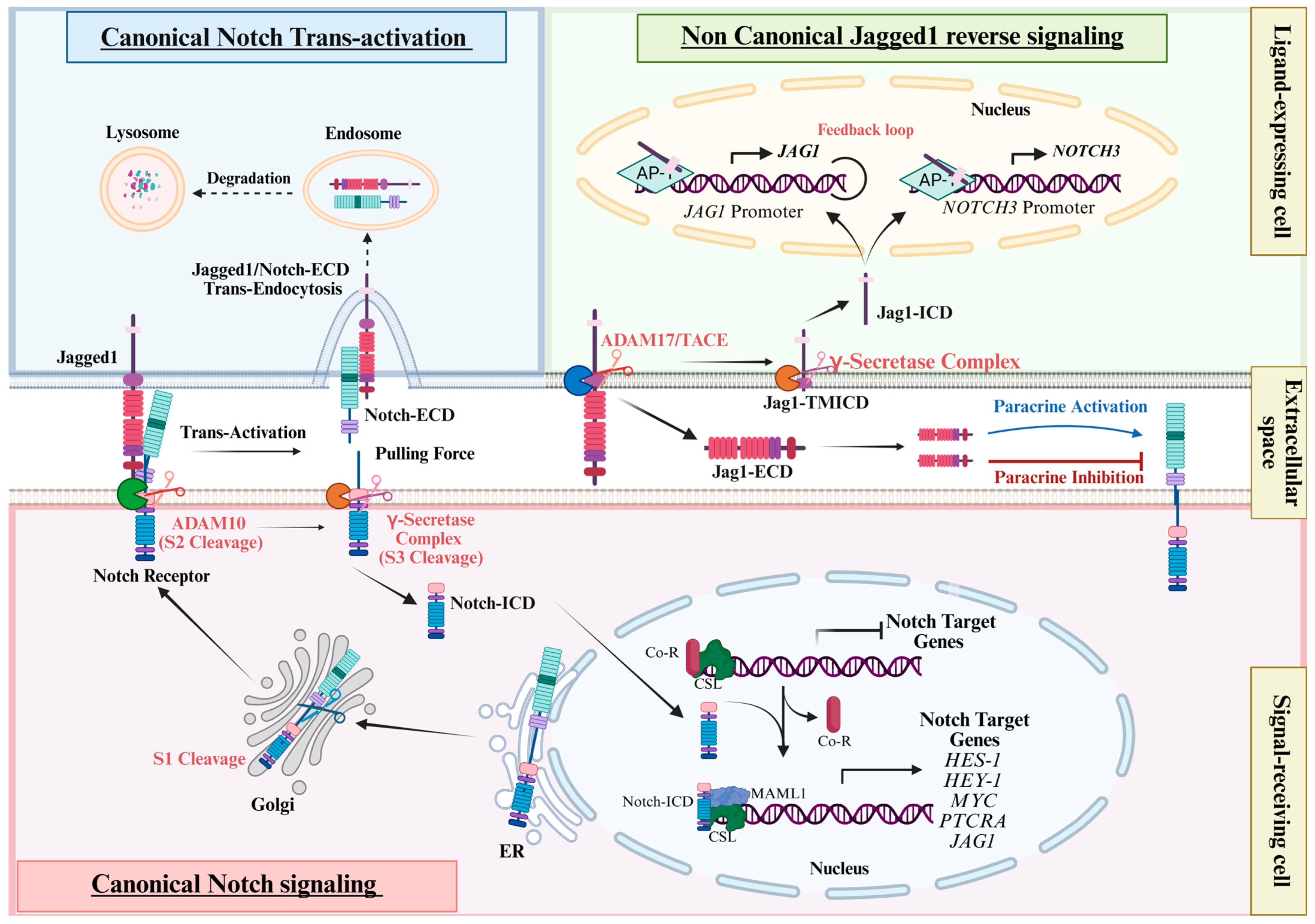
3. Jagged1 as a Component of the Canonical Notch Pathway
4. The Relevance of Jagged1 Intracellular Domain: From Development to Cancer
5. The Canonical and Non-Canonical Role of Jagged1 Ligand in CRC
5.1. The Canonical Notch Signaling in CRC
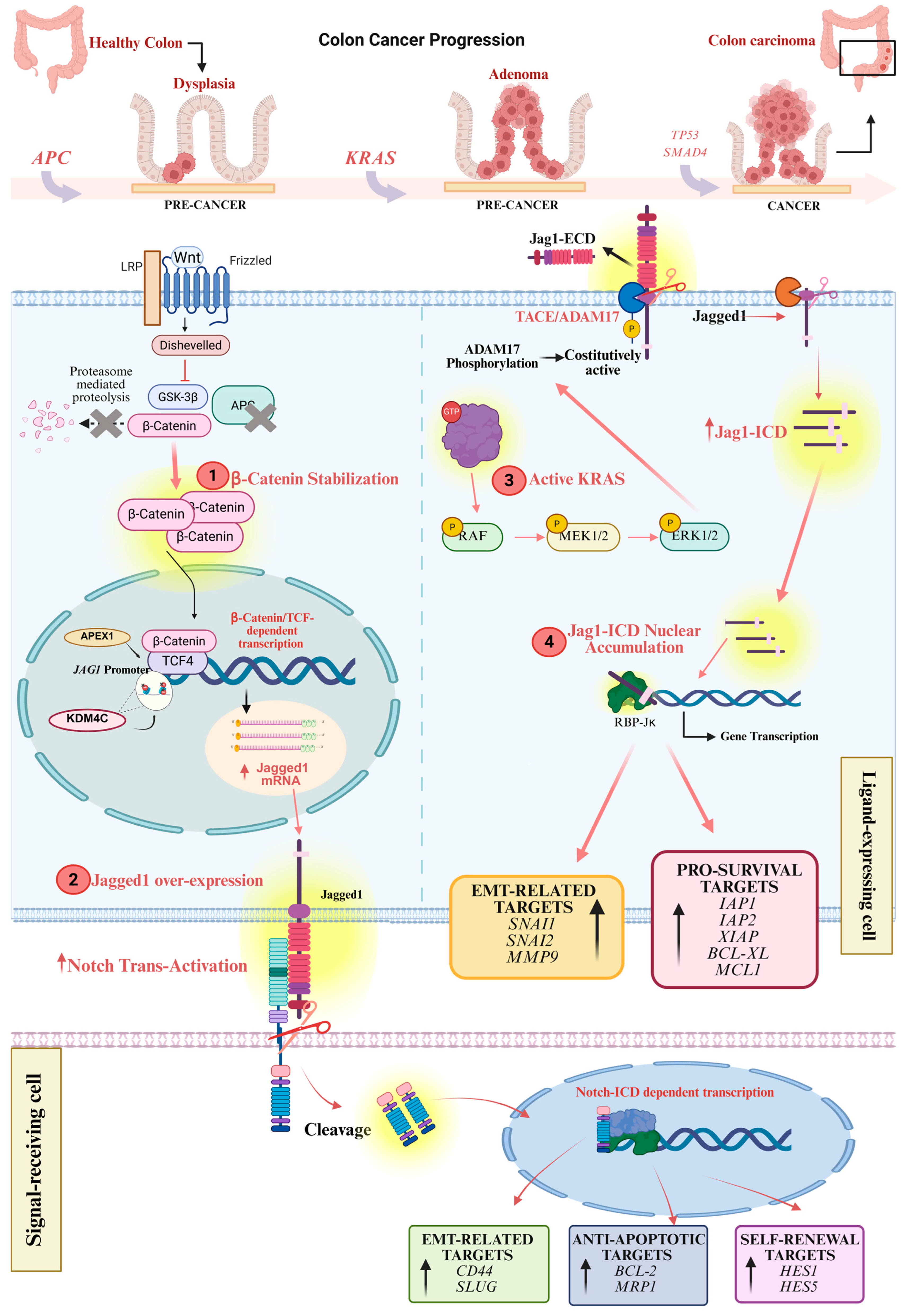
5.2. The Canonical Role of Jagged1 Ligand in CRC
5.3. The Non-Canonical Role of Jagged1 in CRC
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-fluorouracil |
| ANK | Ankyrin repeat domain |
| APC | Adenomatous polyposis coli |
| CIMP | CpG island methylator phenotype |
| CIN | Chromosomal instability |
| CLL | Chronic lymphocytic leukemia |
| CMS | Consensus molecular subtypes |
| CRC | Colorectal cancer |
| CRD | Cysteine-rich domain |
| CSC | Cancer stem cell |
| CSL | CBF1/RBP-Jκ/Su(H)/Lag-1 |
| DLL | Delta-like proteins |
| DSL | Delta/Serrate/Lag-2 |
| ECD | Extracellular domain |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial–mesenchymal transition |
| FAP | Familial Adenomatous Polyposis |
| GSI | γ-secretase inhibitor |
| ICD | Intracellular domain |
| LNR | Lin12-Notch repeats |
| MAPK | Mitogen-activated protein kinase |
| mCRC | Metastatic CRC |
| MDSC | Myeloid-derived suppressor cells |
| MFNG | Manic Fringe |
| MMR | Mismatch repair |
| MSI | Microsatellite instability |
| NRR | Negative regulatory region |
| OXA | Oxaliplatin |
| PI3K | Phosphatidylinositol 3-kinase |
| RAM | RBP-Jκ-associated module |
| T-ALL | T-cell acute lymphoblastic leukemia |
| TAM | Tumor-associated macrophages |
| TME | Tumor microenvironment |
| TMICD | Membrane-tethered intracellular fragment |
| TNM | Tumor, Node, Metastasis |
References
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Li, J.; Pan, J.; Wang, L.; Ji, G.; Dang, Y. Colorectal Cancer: Pathogenesis and Targeted Therapy. MedComm 2025, 6, e70127. [Google Scholar] [CrossRef]
- Fre, S.; Pallavi, S.K.; Huyghe, M.; Laé, M.; Janssen, K.-P.; Robine, S.; Artavanis-Tsakonas, S.; Louvard, D. Notch and Wnt Signals Cooperatively Control Cell Proliferation and Tumorigenesis in the Intestine. Proc. Natl. Acad. Sci. USA 2009, 106, 6309–6314. [Google Scholar] [CrossRef]
- Chu, D.; Wang, W.; Xie, H.; Li, Y.; Dong, G.; Xu, C.; Chen, D.; Zheng, J.; Li, M.; Lu, Z.; et al. Notch1 Expression in Colorectal Carcinoma Determines Tumor Differentiation Status. J. Gastrointest. Surg. 2009, 13, 253–260. [Google Scholar] [CrossRef]
- Zheng, C.-G.; Chen, R.; Xie, J.-B.; Liu, C.-B.; Jin, Z.; Jin, C. Immunohistochemical Expression of Notch1, Jagged1, NF-ΚB and MMP-9 in Colorectal Cancer Patients and the Relationship to Clinicopathological Parameters. Cancer Biomark. 2015, 15, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, M.; Odorcic, S.; Zhang, H.; Chetty, R.; Tennert, C.; Dickson, B.C.; Lockwood, G.; Gallinger, S.; Egan, S.E. Activation of Notch Signaling in Human Colon Adenocarcinoma. Int. J. Oncol. 2008, 33, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Jackstadt, R.; van Hooff, S.R.; Leach, J.D.; Cortes-Lavaud, X.; Lohuis, J.O.; Ridgway, R.A.; Wouters, V.M.; Roper, J.; Kendall, T.J.; Roxburgh, C.S.; et al. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell 2019, 36, 319–336.e7. [Google Scholar] [CrossRef] [PubMed]
- Gonulcu, S.C.; Unal, B.; Bassorgun, I.C.; Ozcan, M.; Coskun, H.S.; Elpek, G.O. Expression of Notch Pathway Components (Numb, Itch, and Siah-1) in Colorectal Tumors: A Clinicopathological Study. World J. Gastroenterol. 2020, 26, 3814–3833. [Google Scholar] [CrossRef]
- Fender, A.W.; Nutter, J.M.; Fitzgerald, T.L.; Bertrand, F.E.; Sigounas, G. Notch-1 Promotes Stemness and Epithelial to Mesenchymal Transition in Colorectal Cancer. J. Cell. Biochem. 2015, 116, 2517–2527. [Google Scholar] [CrossRef]
- Zhao, J.L.; Ye, Y.C.; Gao, C.C.; Wang, L.; Ren, K.X.; Jiang, R.; Hu, S.J.; Liang, S.Q.; Bai, J.; Liang, J.L.; et al. Notch-Mediated Lactate Metabolism Regulates MDSC Development through the Hes1/MCT2/c-Jun Axis. Cell Rep. 2022, 38, 110451. [Google Scholar] [CrossRef]
- Fre, S.; Huyghe, M.; Mourikis, P.; Robine, S.; Louvard, D.; Artavanis-Tsakonas, S. Notch Signals Control the Fate of Immature Progenitor Cells in the Intestine. Nature 2005, 435, 964–968. [Google Scholar] [CrossRef]
- Sikandar, S.S.; Pate, K.T.; Anderson, S.; Dizon, D.; Edwards, R.A.; Waterman, M.L.; Lipkin, S.M. NOTCH Signaling Is Required for Formation and Self-Renewal of Tumor-Initiating Cells and for Repression of Secretory Cell Differentiation in Colon Cancer. Cancer Res. 2010, 70, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Oki, E.; Nakaji, Y.; Tsutsumi, S.; Ono, N.; Nakanishi, R.; Sugiyama, M.; Nakashima, Y.; Sonoda, H.; Ohgaki, K.; et al. High Expression of the Notch Ligand Jagged-1 Is Associated with Poor Prognosis after Surgery for Colorectal Cancer. Cancer Sci. 2016, 107, 1705–1716. [Google Scholar] [CrossRef]
- Guilmeau, S.; Flandez, M.; Mariadason, J.M.; Augenlicht, L.H. Heterogeneity of Jagged1 Expression in Human and Mouse Intestinal Tumors: Implications for Targeting Notch Signaling. Oncogene 2010, 29, 992–1002. [Google Scholar] [CrossRef]
- Dai, Y.; Wilson, G.; Huang, B.; Peng, M.; Teng, G.; Zhang, D.; Zhang, R.; Ebert, M.P.A.; Chen, J.; Wong, B.C.Y.; et al. Silencing of Jagged1 Inhibits Cell Growth and Invasion in Colorectal Cancer. Cell Death Dis. 2014, 5, e1170. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Peng, J.; Wei, D.; Chen, P.; Zhao, Y. Effect of Jagged1 on the Proliferation and Migration of Colon Cancer Cells. Exp. Ther. Med. 2012, 4, 89–92. [Google Scholar] [CrossRef] [PubMed]
- De Falco, F.; Del Papa, B.; Baldoni, S.; Sabatini, R.; Falzetti, F.; Di Ianni, M.; Martelli, M.P.; Mezzasoma, F.; Pelullo, M.; Marconi, P.; et al. IL-4-Dependent Jagged1 Expression/Processing Is Associated with Survival of Chronic Lymphocytic Leukemia Cells but Not with Notch Activation. Cell Death Dis. 2018, 9, 1160. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, J.Y.; Kim, S.O.; Hong, N.; Choi, S.H.; Park, M.G.; Jang, J.; Ham, S.W.; Seo, S.; Lee, S.Y.; et al. The Oncogenic JAG1 Intracellular Domain Is a Transcriptional Cofactor That Acts in Concert with DDX17/SMAD3/TGIF2. Cell Rep. 2022, 41, 111626. [Google Scholar] [CrossRef]
- Kim, J.Y.; Hong, N.; Park, S.; Ham, S.W.; Kim, E.J.; Kim, S.O.; Jang, J.; Kim, Y.; Kim, J.K.; Kim, S.C.; et al. Jagged1 Intracellular Domain/SMAD3 Complex Transcriptionally Regulates TWIST1 to Drive Glioma Invasion. Cell Death Dis. 2023, 14, 822. [Google Scholar] [CrossRef]
- Pelullo, M.; Quaranta, R.; Talora, C.; Checquolo, S.; Cialfi, S.; Felli, M.P.; te Kronnie, G.; Borga, C.; Besharat, Z.M.; Palermo, R.; et al. Notch3/Jagged1 Circuitry Reinforces Notch Signaling and Sustains T-ALL. Neoplasia 2014, 16, 1007–1017. [Google Scholar] [CrossRef]
- Pelullo, M.; Nardozza, F.; Zema, S.; Quaranta, R.; Nicoletti, C.; Besharat, Z.M.Z.M.; Felli, M.P.M.P.; Cerbelli, B.; D’Amati, G.; Palermo, R.; et al. Kras/ADAM17-Dependent Jag1-ICD Reverse Signalling Sustains Colorectal Cancer Progression and Chemoresistance. Cancer Res. 2019, 79, 5575–5586. [Google Scholar] [CrossRef]
- Pelullo, M.; Zema, S.; De Carolis, M.; Cialfi, S.; Giuli, M.V.; Palermo, R.; Capalbo, C.; Giannini, G.; Screpanti, I.; Checquolo, S.; et al. 5FU/Oxaliplatin-Induced Jagged1 Cleavage Counteracts Apoptosis Induction in Colorectal Cancer: A Novel Mechanism of Intrinsic Drug Resistance. Front. Oncol. 2022, 12, 918763. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Karlitz, J.J.; Yen, T.; Lieu, C.H.; Boland, C.R. The Rising Tide of Early-Onset Colorectal Cancer: A Comprehensive Review of Epidemiology, Clinical Features, Biology, Risk Factors, Prevention, and Early Detection. Lancet Gastroenterol. Hepatol. 2022, 7, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Brisset, M.; Mehlen, P.; Meurette, O.; Hollande, F. Notch Receptor/Ligand Diversity: Contribution to Colorectal Cancer Stem Cell Heterogeneity. Front. Cell Dev. Biol. 2023, 11, 1231416. [Google Scholar] [CrossRef]
- Mundade, R.; Imperiale, T.F.; Prabhu, L.; Loehrer, P.J.; Lu, T. Genetic Pathways, Prevention, and Treatment of Sporadic Colorectal Cancer. Oncoscience 2014, 1, 400–406. [Google Scholar] [CrossRef]
- Weledji, E.P. The Etiology and Pathogenesis of Colorectal Cancer. Clin. Oncol. 2024, 9, 2046. [Google Scholar]
- Kastrinos, F.; Syngal, S. Inherited Colorectal Cancer Syndromes. Cancer J. 2011, 17, 405–415. [Google Scholar] [CrossRef]
- Yamane, L.; Scapulatempo-Neto, C.; Reis, R.M.; Guimarães, D.P. Serrated Pathway in Colorectal Carcinogenesis. World J. Gastroenterol. 2014, 20, 2634–2640. [Google Scholar] [CrossRef]
- Leslie, A.; Carey, F.A.; Pratt, N.R.; Steele, R.J.C. The Colorectal Adenoma-Carcinoma Sequence. Br. J. Surg. 2002, 89, 845–860. [Google Scholar] [CrossRef]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Smits, A.M.M.; Bos, J.L. Genetic Alterations during Colorectal-Tumor Development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef]
- Miyoshi, Y.; Nagase, H.; Horii, H.A.A.; Shigetoshi Ichii, S.N.; Aoki, T.; Miki, Y.; Mori, T.; Nakamura, Y. Somatic Mutations of the APC Gene in Colorectal Tumors: Mutation Cluster Region in the APC Gene. Hum. Mol. Genet. 1992, 1, 229–233. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Nilbert, M.C.; Su, L.K.; Vogelstein, B.; Bryan, T.M.; Levy, D.B.; Smith, K.J.; Preisinger, A.C.; Hedge, P.; McKechnie, D.; et al. Identification of FAP Locus Genes from Chromosome 5q21. Science 1991, 253, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Miyoshi, Y.; Horii, A.; Aoki, T.; Petersen, G.M.; Vogelstein, B.; Maher, E.; Ogawa, M.; Maruyama, M.; Utsunomiya, J.; et al. Screening for Germ-line Mutations in Familial Adenomatous Polyposis Patients: 61 New Patients and a Summary of 150 Unrelated Patients. Hum. Mutat. 1992, 1, 467–473. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Morin, P.J.; Van Wichen, D.; De Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC(−/−) Colon Carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Sancho, E.; Verweij, C.; De Lau, W.; Oving, I.; Hurlstone, A.; Van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The β-Catenin/TCF-4 Complex Imposes a Crypt Progenitor Phenotype on Colorectal Cancer Cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.-Q.; Luo, Q.; Wang, L.; Song, G.-B.; Sheng, J.-P.; Xu, B. Signaling Pathways Involved in Colorectal Cancer: Pathogenesis and Targeted Therapy. Signal Transduct. Target. Ther. 2024, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-Catenin-Tcf Signaling in Colon Cancer by Mutations in β-Catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef]
- Worthley, D.L.; Leggett, B.A. Colorectal Cancer: Molecular Features and Clinical Opportunities. Clin. Biochem. Rev. 2010, 31, 31–38. [Google Scholar] [PubMed]
- Pretlow, T.P.; Pretlow, T.G. Mutant KRAS in Aberrant Crypt Foci (ACF): Initiation of Colorectal Cancer? Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2005, 1756, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjöblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science 2007, 318, 1108–1113. [Google Scholar] [CrossRef] [PubMed]
- Frattini, M.; Balestra, D.; Suardi, S.; Oggionni, M.; Alberici, P.; Radice, P.; Costa, A.; Daidone, M.G.; Leo, E.; Pilotti, S.; et al. Different Genetic Features Associated with Colon and Rectal Carcinogenesis. Clin. Cancer Res. 2004, 10, 4015–4021. [Google Scholar] [CrossRef]
- Vasovcak, P.; Pavlikova, K.; Sedlacek, Z.; Skapa, P.; Kouda, M.; Hoch, J.; Krepelova, A. Molecular Genetic Analysis of 103 Sporadic Colorectal Tumours in Czech Patients. PLoS ONE 2011, 6, e24114. [Google Scholar] [CrossRef]
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef]
- Fabien, N.; Paulin, C.; Dubois, P.M.; Santoro, M.; Grieco, M.; Berger, N.; Fusco, A.; Galvain, D.; Barbier, Y. Detection of RET Oncogene Activation in Human Papillary Thyroid Carcinomas by in Situ Hybridisation. Br. J. Cancer 1992, 66, 1094–1098. [Google Scholar] [CrossRef]
- Van Schaeybroeck, S.; Kyula, J.N.; Fenton, A.; Fenning, C.S.; Sasazuki, T.; Shirasawa, S.; Longley, D.B.; Johnston, P.G. Oncogenic Kras Promotes Chemotherapy-Induced Growth Factor Shedding via ADAM17. Cancer Res. 2011, 71, 1071–1080. [Google Scholar] [CrossRef]
- Mologni, L.; Brussolo, S.; Ceccon, M.; Gambacorti-Passerini, C. Synergistic Effects of Combined Wnt/KRAS Inhibition in Colorectal Cancer Cells. PLoS ONE 2012, 7, e51449. [Google Scholar] [CrossRef]
- Lee, S.K.; Hwang, J.H.; Choi, K.Y. Interaction of the Wnt/β-Catenin and RAS-ERK Pathways Involving Co-Stabilization of Both β-Catenin and RAS Plays Important Roles in the Colorectal Tumorigenesis. Adv. Biol. Regul. 2018, 68, 46–54. [Google Scholar] [CrossRef]
- Saucier, C.; Rivard, N. Epithelial Cell Signalling in Colorectal Cancer Metastasis. In Metastasis of Colorectal Cancer; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Moon, B.-S.; Jeong, W.-J.; Park, J.; Kim, T.I.; Min, D.S.; Choi, K.-Y. Role of Oncogenic K-Ras in Cancer Stem Cell Activation by Aberrant Wnt/β-Catenin Signaling. J. Natl. Cancer Inst. 2014, 106, djt373. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, E.; Cagnol, S.; Beaudry, K.; Carrier, J.; Rivard, N. Oncogenic KRAS Signalling Promotes the Wnt/β-Catenin Pathway through LRP6 in Colorectal Cancer. Oncogene 2015, 34, 4914–4927. [Google Scholar] [CrossRef]
- D’Abaco, G.M.; Whitehead, R.H.; Burgess, A.W. Synergy between Apc Min and an Activated Ras Mutation Is Sufficient To Induce Colon Carcinomas. Mol. Cell. Biol. 1996, 16, 884–891. [Google Scholar] [CrossRef]
- Zeller, E.; Hammer, K.; Kirschnick, M.; Braeuning, A. Mechanisms of RAS/β-Catenin Interactions. Arch. Toxicol. 2013, 87, 611–632. [Google Scholar] [CrossRef]
- Kim, S.E.; Yoon, J.Y.; Jeong, W.J.; Jeon, S.H.; Park, Y.; Yoon, J.B.; Park, Y.N.; Kim, H.; Choi, K.Y. H-Ras Is Degraded by Wnt/β-Catenin Signaling via β-TrCP-Mediated Polyubiquitylation. J. Cell Sci. 2009, 122, 842–848. [Google Scholar] [CrossRef]
- Jeong, W.J.; Yoon, J.; Park, J.C.; Lee, S.H.; Lee, S.H.; Kaduwal, S.; Kim, H.; Yoon, J.B.; Choi, K.Y. Ras Stabilization through Aberrant Activation of Wnt/β-Catenin Signaling Promotes Intestinal Tumorigenesis. Sci. Signal 2012, 5, ra30. [Google Scholar] [CrossRef]
- Rubinfeld, B.; Albert, I.; Porfiri, E.; Fiol, C.; Munemitsu, S.; Polakis, P. Binding of GSK3β to the APC-β-Catenin Complex and Regulation of Complex Assembly. Science 1996, 272, 1023–1026. [Google Scholar] [CrossRef]
- Behrens, J.; Jerchow, B.A.; Würtele, M.; Grimm, J.; Asbrand, C.; Wirtz, R.; Kühl, M.; Wedlich, D.; Birchmeier, W. Functional Interaction of an Axin Homolog, Conductin, with β-Catenin, APC, and GSK3β. Science 1998, 280, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jeong, W.; Cho, Y.; Cha, P.; Yoon, J.; Ro, E.J.; Choi, S.; Oh, J.; Heo, Y.; Kim, H.; et al. Β-Catenin- RAS Interaction Serves as a Molecular Switch for RAS Degradation via GSK 3β. EMBO Rep. 2018, 19, e46060. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.J.; Preisinger, A.C.; Jessup, J.M.; Paraskeva, C.; Markowitz, S.; Willson, J.K.V.; Hamilton, S.; Vogelstein, B. P53 Gene Mutations Occur in Combination with 17p Allelic Deletions as Late Events in Colorectal Tumorigenesis. Cancer Res. 1990, 50, 7717–7722. [Google Scholar] [PubMed]
- Aiderus, A.; Barker, N.; Tergaonkar, V. Serrated Colorectal Cancer: Preclinical Models and Molecular Pathways. Trends Cancer 2024, 10, 76–91. [Google Scholar] [CrossRef]
- Rosenberg, D.W.; Yang, S.; Pleau, D.C.; Greenspan, E.J.; Stevens, R.G.; Rajan, T.V.; Heinen, C.D.; Levine, J.; Zhou, Y.; O’Brien, M.J. Mutations in BRAF and KRAS Differentially Distinguish Serrated versus Non-Serrated Hyperplastic Aberrant Crypt Foci in Humans. Cancer Res. 2007, 67, 3551–3554. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus Molecular Subtypes and the Evolution of Precision Medicine in Colorectal Cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Menter, D.G.; Davis, J.S.; Broom, B.M.; Overman, M.J.; Morris, J.; Kopetz, S. Back to the Colorectal Cancer Consensus Molecular Subtype Future. Curr. Gastroenterol. Rep. 2019, 21, 5. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, D.; Tang, Y.; Zhang, L.; Zhang, S.; Li, W.; Li, N.; Yan, X. Targeting KRAS in Colorectal Cancer (Review). Mol. Clin. Oncol. 2025, 23, 28. [Google Scholar] [CrossRef]
- Bteich, F.; Mohammadi, M.; Li, T.; Bhat, M.A.; Sofianidi, A.; Wei, N.; Kuang, C. Targeting KRAS in Colorectal Cancer: A Bench to Bedside Review. Int. J. Mol. Sci. 2023, 24, 12030. [Google Scholar] [CrossRef] [PubMed]
- Infante, J.R.; Fecher, L.A.; Falchook, G.S.; Nallapareddy, S.; Gordon, M.S.; Becerra, C.; DeMarini, D.J.; Cox, D.S.; Xu, Y.; Morris, S.R.; et al. Safety, Pharmacokinetic, Pharmacodynamic, and Efficacy Data for the Oral MEK Inhibitor Trametinib: A Phase 1 Dose-Escalation Trial. Lancet Oncol. 2012, 13, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.S.; LoRusso, P.; Ma, W.W.; Goldman, J.W.; Weise, A.; Colevas, A.D.; Adjei, A.; Yazji, S.; Shen, A.; Johnston, S.; et al. A First-in-Human Phase I Study to Evaluate the MEK1/2 Inhibitor, Cobimetinib, Administered Daily in Patients with Advanced Solid Tumors. Investig. New Drugs 2016, 34, 740–749. [Google Scholar] [CrossRef]
- Grilley-Olson, J.E.; Bedard, P.L.; Fasolo, A.; Cornfeld, M.; Cartee, L.; Razak, A.R.A.; Stayner, L.A.; Wu, Y.; Greenwood, R.; Singh, R.; et al. A Phase Ib Dose-Escalation Study of the MEK Inhibitor Trametinib in Combination with the PI3K/MTOR Inhibitor GSK2126458 in Patients with Advanced Solid Tumors. Investig. New Drugs 2016, 34, 740–749. [Google Scholar] [CrossRef]
- Deming, D.A.; Cavalcante, L.L.; Lubner, S.J.; Mulkerin, D.L.; Loconte, N.K.; Eickhoff, J.C.; Kolesar, J.M.; Fioravanti, S.; Greten, T.F.; Compton, K.; et al. A Phase i Study of Selumetinib (AZD6244/ARRY-142866), a MEK1/2 Inhibitor, in Combination with Cetuximab in Refractory Solid Tumors and KRAS Mutant Colorectal Cancer. Investig. New Drugs 2016, 34, 168–175. [Google Scholar] [CrossRef]
- Weisner, J.; Landel, I.; Reintjes, C.; Uhlenbrock, N.; Trajkovic-Arsic, M.; Dienstbier, N.; Hardick, J.; Ladigan, S.; Lindemann, M.; Smith, S.; et al. Preclinical Efficacy of Covalent-Allosteric AKT Inhibitor Borussertib in Combination with Trametinib in KRAS-Mutant Pancreatic and Colorectal Cancer. Cancer Res. 2019, 79, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Marie, P.K.; Bitner, L.; Syed, M.; Woods, M.; Manyam, G.; Kwong, L.N.; Johnson, B.; Morris, V.K.; Jones, P.; et al. Targeting RAS Mutant Colorectal Cancer with Dual Inhibition of MEK and CDK4/6. Cancer Res. 2022, 82, 3335–3344. [Google Scholar] [CrossRef]
- Sun, Q.; Burke, J.P.; Phan, J.; Burns, M.C.; Olejniczak, E.T.; Waterson, A.G.; Lee, T.; Rossanese, O.W.; Fesik, S.W. Discovery of Small Molecules That Bind to K-Ras and Inhibit Sos-Mediated Activation. Angew. Chem. Int. Ed. 2012, 51, 6140–6143. [Google Scholar] [CrossRef]
- Maurer, T.; Garrenton, L.S.; Oh, A.; Pitts, K.; Anderson, D.J.; Skelton, N.J.; Fauber, B.P.; Pan, B.; Malek, S.; Stokoe, D.; et al. Small-Molecule Ligands Bind to a Distinct Pocket in Ras and Inhibit SOS-Mediated Nucleotide Exchange Activity. Proc. Natl. Acad. Sci. USA 2012, 109, 5299–5304. [Google Scholar] [CrossRef]
- Cruz-Migoni, A.; Canning, P.; Quevedo, C.E.; Bataille, C.J.R.; Bery, N.; Miller, A.; Russell, A.J.; Phillips, S.E.V.; Carr, S.B.; Rabbitts, T.H. Structure-Based Development of New RAS-Effector Inhibitors from a Combination of Active and Inactive RAS-Binding Compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 2545–2550. [Google Scholar] [CrossRef]
- Quevedo, C.E.; Cruz-Migoni, A.; Bery, N.; Miller, A.; Tanaka, T.; Petch, D.; Bataille, C.J.R.; Lee, L.Y.W.; Fallon, P.S.; Tulmin, H.; et al. Small Molecule Inhibitors of RAS-Effector Protein Interactions Derived Using an Intracellular Antibody Fragment. Nat. Commun. 2018, 9, 3169. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) Inhibitors Allosterically Control GTP Affinity and Effector Interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e17. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an Undruggable Pocket on KRAS. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS G12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.I.; Jänne, P.A.; Leal, T.A.; Rybkin, I.I.; Sabari, J.K.; Barve, M.A.; Bazhenova, L.; Johnson, M.L.; Velastegui, K.L.; Cilliers, C.; et al. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients with Advanced KRAS G12CSolid Tumors (KRYSTAL-1). J. Clin. Oncol. 2022, 40, 2530–2538. [Google Scholar] [CrossRef]
- Amodio, V.; Yaeger, R.; Arcella, P.; Cancelliere, C.; Lamba, S.; Lorenzato, A.; Arena, S.; Montone, M.; Mussolin, B.; Bian, Y.; et al. Egfr Blockade Reverts Resistance to Krasg12c Inhibition in Colorectal Cancer. Cancer Discov. 2020, 10, 1129–1139. [Google Scholar] [CrossRef]
- Pellatt, A.J.; Bhamidipati, D.; Subbiah, V. Ready, Set, Go: Setting Off on the Mission to Target KRAS in Colorectal Cancer. JCO Oncol. Pr. 2024, 20, 1289–1292. [Google Scholar] [CrossRef]
- Kumar, A.; Gautam, V.; Sandhu, A.; Rawat, K.; Sharma, A.; Saha, L. Current and Emerging Therapeutic Approaches for Colorectal Cancer: A Comprehensive Review. World J. Gastrointest. Surg. 2023, 15, 495–519. [Google Scholar] [CrossRef]
- Oda, T.; Elkahloun, A.G.; Pike, B.L.; Okajima, K.; Krantz, I.D.; Genin, A.; Piccoli, D.A.; Meltzer, P.S.; Spinner, N.B.; Collins, F.S.; et al. Mutations in the Human Jagged1 Gene Are Responsible for Alagille Syndrome. Nat. Genet. 1997, 16, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Checquolo, S.; Campese, A.F.; Felli, M.P.; Gulino, A.; Screpanti, I. Notch3: From Subtle Structural Differences to Functional Diversity. Oncogene 2008, 27, 5092–5098. [Google Scholar] [CrossRef]
- Taylor, P.; Takeuchi, H.; Sheppard, D.; Chillakuri, C.; Lea, S.M.; Haltiwanger, R.S.; Handford, P.A. Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands. Proc. Natl. Acad. Sci. USA 2014, 111, 7290–7295. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Sanlidag, S.; Jensen, S.A.; Burnap, S.A.; Struwe, W.B.; Larsen, A.H.; Feng, X.; Mittal, S.; Sansom, M.S.P.; Sahlgren, C.; et al. An N-Glycan on the C2 Domain of JAGGED1 Is Important for Notch Activation. Sci. Signal 2022, 15, eabo3507. [Google Scholar] [CrossRef]
- Grochowski, C.M.; Loomes, K.M.; Spinner, N.B. Jagged1 (JAG1): Structure, Expression, and Disease Associations. Gene 2016, 576, 381–384. [Google Scholar] [CrossRef]
- Luca, V.C.; Kim, B.C.; Ge, C.; Kakuda, S.; Wu, D.; Roein-Peikar, M.; Haltiwanger, R.S.; Zhu, C.; Ha, T.; Garcia, K.C. Notch-Jagged Complex Structure Implicates a Catch Bond in Tuning Ligand Sensitivity. Science 2017, 355, 1320–1324. [Google Scholar] [CrossRef]
- Gordon, W.R.; Vardar-Ulu, D.; Histen, G.; Sanchez-Irizarry, C.; Aster, J.C.; Blacklow, S.C. Structural Basis for Autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007, 14, 295–300. [Google Scholar] [CrossRef]
- Zeronian, M.R.; Klykov, O.; Portell i de Montserrat, J.; Konijnenberg, M.J.; Gaur, A.; Scheltema, R.A.; Janssen, B.J.C. Notch–Jagged Signaling Complex Defined by an Interaction Mosaic. Proc. Natl. Acad. Sci. USA 2021, 118, e2102502118. [Google Scholar] [CrossRef] [PubMed]
- Lovendahl, K.N.; Blacklow, S.C.; Gordon, W.R. The Molecular Mechanism of Notch Activation. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; Volume 1066. [Google Scholar]
- Langridge, P.D.; Struhl, G. Epsin-Dependent Ligand Endocytosis Activates Notch by Force. Cell 2017, 171, 1383–1396. [Google Scholar] [CrossRef]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Palermo, R.; Checquolo, S.; Bellavia, D.; Screpanti, C.T. and I. The Molecular Basis of Notch Signaling Regulation: A Complex Simplicity. Curr. Mol. Med. 2014, 14, 34–44. [Google Scholar] [CrossRef]
- Shimizu, K.; Chiba, S.; Kumano, K.; Hosoya, N.; Takahashi, T.; Kanda, Y.; Hamada, Y.; Yazaki, Y.; Hirai, H. Mouse Jagged1 Physically Interacts with Notch2 and Other Notch Receptors. Assessment by Quantitative Methods. J. Biol. Chem. 1999, 274, 32961–32969. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Chiba, S.; Hosoya, N.; Kumano, K.; Saito, T.; Kurokawa, M.; Kanda, Y.; Hamada, Y.; Hirai, H. Binding of Delta1, Jagged1, and Jagged2 to Notch2 Rapidly Induces Cleavage, Nuclear Translocation, and Hyperphosphorylation of Notch2. Mol. Cell Biol. 2000, 20, 6913–6922. [Google Scholar] [CrossRef]
- Chapman, G.; Major, J.A.; Iyer, K.; James, A.C.; Pursglove, S.E.; Moreau, J.L.M.; Dunwoodie, S.L. Notch1 Endocytosis Is Induced by Ligand and Is Required for Signal Transduction. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 166–177. [Google Scholar] [CrossRef]
- Parks, A.L.; Klueg, K.M.; Stout, J.R.; Muskavitch, M.A.T. Ligand Endocytosis Drives Receptor Dissociation and Activation in the Notch Pathway. Development 2000, 127, 1373–1385. [Google Scholar] [CrossRef]
- Weinmaster, G. The Ins and Outs of Notch Signaling. Mol. Cell. Neurosci. 1997, 9, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Talora, C.; Campese, A.F.; Bellavia, D.; Pascucci, M.; Checquolo, S.; Groppioni, M.; Frati, L.; von Boehmer, H.; Gulino, A.; Screpanti, I. Pre-TCR-Triggered ERK Signalling-Dependent Downregulation of E2A Activity in Notch3-Induced T-Cell Lymphoma. EMBO Rep. 2003, 4, 1067–1071. [Google Scholar] [CrossRef]
- Bellavia, D.; Campese, A.F.; Checquolo, S.; Balestri, A.; Biondi, A.; Cazzaniga, G.; Lendahl, U.; Fehling, H.J.; Hayday, A.C.; Frati, L.; et al. Combined Expression of PTα and Notch3 in T Cell Leukemia Identifies the Requirement of PreTCR for Leukemogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 3788–3793. [Google Scholar] [CrossRef]
- Petrovic, J.; Zhou, Y.; Fasolino, M.; Goldman, N.; Schwartz, G.W.; Mumbach, M.R.; Nguyen, S.C.; Rome, K.S.; Sela, Y.; Zapataro, Z.; et al. Oncogenic Notch Promotes Long-Range Regulatory Interactions within Hyperconnected 3D Cliques. Mol. Cell 2019, 73, 1174–1190.e12. [Google Scholar] [CrossRef]
- Chen, X.; Stoeck, A.; Lee, S.J.; Shih, I.-M.; Wang, M.M.; Wang, T.-L. Jagged1 Expression Regulated by Notch3 and Wnt/β-Catenin Signaling Pathways in Ovarian Cancer. Oncotarget 2010, 1, 210–218. [Google Scholar] [CrossRef]
- Zhang, R.; Engler, A.; Taylor, V. Notch: An Interactive Player in Neurogenesis and Disease. Cell Tissue Res. 2018, 371, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Artavanis-Tsakonas, S. Notch Signalling in Vertebrate Neural Development. Nat. Rev. Neurosci. 2006, 7, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Akil, A.; Gutiérrez-García, A.K.; Guenter, R.; Rose, J.B.; Beck, A.W.; Chen, H.; Ren, B. Notch Signaling in Vascular Endothelial Cells, Angiogenesis, and Tumor Progression: An Update and Prospective. Front. Cell Dev. Biol. 2021, 9, 642352. [Google Scholar] [CrossRef]
- Pui, J.C.; Allman, D.; Xu, L.; DeRocco, S.; Karnell, F.G.; Bakkour, S.; Lee, J.Y.; Kadesch, T.; Hardy, R.R.; Aster, J.C.; et al. Notch1 Expression in Early Lymphopoiesis Influences B versus T Lineage Determination. Immunity 1999, 11, 299–308. [Google Scholar] [CrossRef]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; Van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T Cell Fate Specification in Mice with an Induced Inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Felli, M.P.; Maroder, M.; Mitsiadis, T.A.; Campese, A.F.; Bellavia, D.; Vacca, A.; Mann, R.S.; Frati, L.; Lendahl, U.; Gulino, A.; et al. Expression Pattern of Notch1, 2 and 3 and Jagged1 and 2 in Lymphoid and Stromal Thymus Components: Distinct Ligand-Receptor Interactions in Intrathymic T Cell Development. Int. Immunol. 1999, 11, 1017–1025. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch Signaling Pathway: Architecture, Disease, and Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Brai, E.; Alina Raio, N.; Alberi, L. Notch1 Hallmarks Fibrillary Depositions in Sporadic Alzheimer’s Disease. Acta Neuropathol. Commun. 2016, 4, 64. [Google Scholar] [CrossRef]
- Mašek, J.; Andersson, E.R. The Developmental Biology of Genetic Notch Disorders. Development 2017, 144, 1743–1763. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.M.; Artavanis-Tsakonas, S.; Louvi, A. The Notch Pathway in CNS Homeostasis and Neurodegeneration. Wiley Interdiscip. Rev. Dev. Biol. 2020, 9, e358. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch Signaling Pathway in Cancer: From Mechanistic Insights to Targeted Therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef]
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.L.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the Human Homolog of the Drosophila Notch Gene, Is Broken by Chromosomal Translocations in T Lymphoblastic Neoplasms. Cell 1991, 66, 649–661. [Google Scholar] [CrossRef]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris, J.P.; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef]
- Pear, W.S.; Aster, J.C.; Scott, M.L.; Hasserjian, R.P.; Soffer, B.; Sklar, J.; Baltimore, D. Exclusive Development of T Cell Neoplasms in Mice Transplanted with Bone Marrow Expressing Activated Notch Alleles. J. Exp. Med. 1996, 183, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi-Elias, P.; Hu, T.; Jenkins, D.; Firestone, B.; Gans, S.; Kurth, E.; Capodieci, P.; Deplazes-Lauber, J.; Petropoulos, K.; Thiel, P.; et al. Characterization of Activating Mutations of NOTCH3 in T-Cell Acute Lymphoblastic Leukemia and Anti-Leukemic Activity of NOTCH3 Inhibitory Antibodies. Oncogene 2016, 35, 6077–6086. [Google Scholar] [CrossRef]
- Bellavia, D.; Campese, A.F.; Alesse, E.; Vacca, A.; Felli, M.P.; Balestri, A.; Stoppacciaro, A.; Tiveron, C.; Tatangelo, L.; Giovarelli, M.; et al. Constitutive Activation of NF-KappaB and T-Cell Leukemia/Lymphoma in Notch3 Transgenic Mice. EMBO J. 2000, 19, 3337–3348. [Google Scholar] [CrossRef]
- Giuli, M.V.; Diluvio, G.; Giuliani, E.; Franciosa, G.; Di Magno, L.; Pignataro, M.G.; Tottone, L.; Nicoletti, C.; Besharat, Z.M.; Peruzzi, G.; et al. Notch3 Contributes to T-Cell Leukemia Growth via Regulation of the Unfolded Protein Response. Oncogenesis 2020, 9, 93. [Google Scholar] [CrossRef]
- Bellavia, D.; Campese, A.F.; Vacca, A.; Gulino, A.; Screpanti, I. Notch3, Another Notch in T Cell Development. Semin. Immunol. 2003, 15, 107–112. [Google Scholar] [CrossRef]
- Campese, A.F.; Grazioli, P.; Colantoni, S.; Anastasi, E.; Mecarozzi, M.; Checquolo, S.; De Luca, G.; Bellavia, D.; Frati, L.; Gulino, A.; et al. Notch3 and PTα/Pre-TCR Sustain the in Vivo Function of Naturally Occurring Regulatory T Cells. Int. Immunol. 2009, 21, 727–743. [Google Scholar] [CrossRef]
- Tardivon, D.; Antoszewski, M.; Zangger, N.; Nkosi, M.; Sordet-Dessimoz, J.; Hendriks, R.; Koch, U.; Radtke, F. Notch Signaling Promotes Disease Initiation and Progression in Murine Chronic Lymphocytic Leukemia. Blood 2021, 137, 3079–3092. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Palermo, R.; Felli, M.P.; Screpanti, I.; Checquolo, S. Notch Signaling as a Therapeutic Target for Acute Lymphoblastic Leukemia. Expert Opin. Ther. Targets 2018, 22, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Pelullo, M.; Zema, S.; Nardozza, F.; Checquolo, S.; Screpanti, I.; Bellavia, D. Wnt, Notch, and TGF-β Pathways Impinge on Hedgehog Signaling Complexity: An Open Window on Cancer. Front. Genet. 2019, 10, 711. [Google Scholar] [CrossRef]
- Zema, S.; Pelullo, M.; Nardozza, F.; Felli, M.P.; Screpanti, I.; Bellavia, D. A Dynamic Role of Mastermind-Like 1: A Journey Through the Main (Path)Ways Between Development and Cancer. Front. Cell Dev. Biol. 2020, 8, 1615. [Google Scholar] [CrossRef]
- Gragnani, L.; Lorini, S.; Marri, S.; Zignego, A.L. Role of Notch Receptors in Hematologic Malignancies. Cells 2021, 10, 16. [Google Scholar] [CrossRef]
- Sorrentino, C.; Cuneo, A.; Roti, G. Therapeutic Targeting of Notch Signaling Pathway in Hematological Malignancies. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019037. [Google Scholar] [CrossRef]
- Nicolas, M.; Wolfer, A.; Raj, K.; Kummer, J.A.; Mill, P.; van Noort, M.; Hui, C.-C.; Clevers, H.; Dotto, G.P.; Radtke, F. Notch1 Functions as a Tumor Suppressor in Mouse Skin. Nat. Genet. 2003, 33, 416–421. [Google Scholar] [CrossRef]
- Wang, N.J.; Sanborn, Z.; Arnett, K.L.; Bayston, L.J.; Liao, W.; Proby, C.M.; Leigh, I.M.; Collisson, E.A.; Gordon, P.B.; Jakkula, L.; et al. Loss-of-Function Mutations in Notch Receptors in Cutaneous and Lung Squamous Cell Carcinoma. Proc. Natl. Acad. Sci. USA 2011, 108, 17761–17766. [Google Scholar] [CrossRef] [PubMed]
- Lefort, K.; Mandinova, A.; Ostano, P.; Kolev, V.; Calpini, V.; Kolfschoten, I.; Devgan, V.; Lieb, J.; Raffoul, W.; Hohl, D.; et al. Notch1 Is a P53 Target Gene Involved in Human Keratinocyte Tumor Suppression through Negative Regulation of ROCK1/2 and MRCKα Kinases. Genes. Dev. 2007, 21, 562–577. [Google Scholar] [CrossRef]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J.; et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational Landscape and Clonal Architecture in Grade II and III Gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretia, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive Genomic Profiles of Small Cell Lung Cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Maraver, A.; Fernandez-Marcos, P.J.; Cash, T.P.; Mendez-Pertuz, M.; Dueñas, M.; Maietta, P.; Martinelli, P.; Muñoz-Martin, M.; Martínez-Fernández, M.; Cañamero, M.; et al. NOTCH Pathway Inactivation Promotes Bladder Cancer Progression. J. Clin. Investig. 2015, 125, 824–830. [Google Scholar] [CrossRef]
- Rampias, T.; Vgenopoulou, P.; Avgeris, M.; Polyzos, A.; Stravodimos, K.; Valavanis, C.; Scorilas, A.; Klinakis, A. A New Tumor Suppressor Role for the Notch Pathway in Bladder Cancer. Nat. Med. 2014, 20, 1199–1205. [Google Scholar] [CrossRef]
- Rekhtman, N.; Pietanza, M.C.; Hellmann, M.D.; Naidoo, J.; Arora, A.; Won, H.; Halpenny, D.F.; Wang, H.; Tian, S.K.; Litvak, A.M.; et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin. Cancer Res. 2016, 22, 3618–3629. [Google Scholar] [CrossRef]
- Pecora, G.; Mancini, C.; Mazzilli, R.; Zamponi, V.; Telese, S.; Scalera, S.; Maugeri-Saccà, M.; Ciuffreda, L.; De Nicola, F.; Fanciulli, M.; et al. Genetic Insight into Lung Neuroendocrine Tumors: Notch and Wnt Signaling Pathways as Potential Targets. J. Transl. Med. 2025, 23, 538. [Google Scholar] [CrossRef]
- Kunnimalaiyaan, M.; Vaccaro, A.M.; Ndiaye, M.A.; Chen, H. Overexpression of the NOTCH1 Intracellular Domain Inhibits Cell Proliferation and Alters the Neuroendocrine Phenotype of Medullary Thyroid Cancer Cells. J. Biol. Chem. 2006, 281, 39819–39830. [Google Scholar] [CrossRef]
- Nowell, C.S.; Radtke, F. Notch as a Tumour Suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Rodilla, V.; Villanueva, A.; Obrador-Hevia, A.; Robert-Moreno, A.; Fernández-Majada, V.; Grilli, A.; López-Bigas, N.; Bellora, N.; Albà, M.M.; Torres, F.; et al. Jagged1 Is the Pathological Link between Wnt and Notch Pathways in Colorectal Cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Storci, G.; Tavolari, S.; Guarnieri, T.; Giovannini, C.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Paterini, P.; Marcu, K.B.; et al. IL-6 Triggers Malignant Features in Mammospheres from Human Ductal Breast Carcinoma and Normal Mammary Gland. J. Clin. Investig. 2007, 17, 3988–4002. [Google Scholar] [CrossRef]
- Zavadil, J.; Cermak, L.; Soto-Nieves, N.; Böttinger, E.P. Integration of TGF-Beta/Smad and Jagged1/Notch Signalling in Epithelial-to-Mesenchymal Transition. EMBO J. 2004, 23, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Taguchi, Y.; Ito-Kureha, T.; Semba, K.; Yamaguchi, N.; Inoue, J. NF-ΚB Non-Cell-Autonomously Regulates Cancer Stem Cell Populations in the Basal-like Breast Cancer Subtype. Nat. Commun. 2013, 4, 2299. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, F.; Weng, J.; Zheng, Y.; Lin, J.; Qi, T.; Wei, Y.; Wang, D.; Zeng, H. SOX12 Promotes Stem Cell-Like Phenotypes and Osteosarcoma Tumor Growth by Upregulating JAGGED1. Stem Cells Int. 2021, 2021, 9941733. [Google Scholar] [CrossRef]
- Santagata, S.; Demichelis, F.; Riva, A.; Varambally, S.; Hofer, M.D.; Kutok, J.L.; Kim, R.; Tang, J.; Montie, J.E.; Chinnaiyan, A.M.; et al. JAGGED1 Expression Is Associated with Prostate Cancer Metastasis and Recurrence. Cancer Res. 2004, 64, 6854–6857. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, H.; Liang, Y.; Liang, L.; Zheng, G.; Huang, H.; Wu, J.; Liao, G. Suppression of Tongue Squamous Cell Carcinoma Growth by Inhibition of Jagged1 in Vitro and in Vivo. J. Oral. Pathol. Med. 2013, 42, 322–331. [Google Scholar] [CrossRef]
- Wu, K.; Xu, L.; Zhang, L.; Lin, Z.; Hou, J. High Jagged1 Expression Predicts Poor Outcome in Clear Cell Renal Cell Carcinoma. Jpn. J. Clin. Oncol. 2010, 41, 411–416. [Google Scholar] [CrossRef]
- Reedijk, M.; Pinnaduwage, D.; Dickson, B.C.; Mulligan, A.M.; Zhang, H.; Bull, S.B.; O’Malley, F.P.; Egan, S.E.; Andrulis, I.L. JAG1 Expression Is Associated with a Basal Phenotype and Recurrence in Lymph Node-Negative Breast Cancer. Breast Cancer Res. Treat. 2008, 111, 439–448. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.; Kim, J.H. Association of Jagged1 Expression with Malignancy and Prognosis in Human Pancreatic Cancer. Cell. Oncol. 2020, 43, 821–834. [Google Scholar] [CrossRef]
- Platonova, N.; Lazzari, E.; Colombo, M.; Falleni, M.; Tosi, D.; Giannandrea, D.; Citro, V.; Casati, L.; Ronchetti, D.; Bolli, N.; et al. The Potential of JAG Ligands as Therapeutic Targets and Predictive Biomarkers in Multiple Myeloma. Int. J. Mol. Sci. 2023, 24, 14558. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Garavelli, S.; Mazzola, M.; Platonova, N.; Giannandrea, D.; Colella, R.; Apicella, L.; Lancellotti, M.; Lesma, E.; Ancona, S.; et al. Multiple Myeloma Exploits Jagged1 and Jagged2 to Promote Intrinsic and Bone Marrow-Dependent Drug Resistance. Haematologica 2020, 105, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-H.; Park, J.T.; Davidson, B.; Morin, P.J.; Shih, I.-M.; Wang, T.-L. Jagged-1 and Notch3 Juxtacrine Loop Regulates Ovarian Tumor Growth and Adhesion. Cancer Res. 2008, 68, 5716–5723. [Google Scholar] [CrossRef] [PubMed]
- Xiu, M.-X.; Liu, Y.-M.; Kuang, B.-H. The Oncogenic Role of Jagged1/Notch Signaling in Cancer. Biomed. Pharmacother. 2020, 129, 110416. [Google Scholar] [CrossRef]
- Qi, H.; Rand, M.D.; Wu, X.; Sestan, N.; Wang, W.; Rakic, P.; Xu, T.; Artavanis-Tsakonas, S. Processing of the Notch Ligand Delta by the Metalloprotease Kuzbanian. Science 1999, 283, 91–94. [Google Scholar] [CrossRef]
- LaVoie, M.J.; Selkoe, D.J. The Notch Ligands, Jagged and Delta, Are Sequentially Processed by α-Secretase and Presenilin/γ-Secretase and Release Signaling Fragments. J. Biol. Chem. 2003, 278, 34427–34437. [Google Scholar] [CrossRef]
- Ascano, J.M.; Beverly, L.J.; Capobianco, A.J. The C-Terminal PDZ-Ligand of JAGGED1 Is Essential for Cellular Transformation. J. Biol. Chem. 2003, 278, 8771–8779. [Google Scholar] [CrossRef]
- Caolo, V.; Schulten, H.M.; Zhuang, Z.W.; Murakami, M.; Wagenaar, A.; Verbruggen, S.; Molin, D.G.M.; Post, M.J. Soluble Jagged-1 Inhibits Neointima Formation by Attenuating Notch-Herp2 Signaling. Arter. Thromb. Vasc. Biol. 2011, 31, 1059–1065. [Google Scholar] [CrossRef]
- Xiao, Y.; Gong, D.; Wang, W. Soluble Jagged1 Inhibits Pulmonary Hypertension by Attenuating Notch Signaling. Arter. Thromb. Vasc. Biol. 2013, 33, 2733–2739. [Google Scholar] [CrossRef] [PubMed]
- Urs, S.; Roudabush, A.; O’Neill, C.F.; Pinz, I.; Prudovsky, I.; Kacer, D.; Tang, Y.; Liaw, L.; Small, D.J. Soluble Forms of the Notch Ligands Delta1 and Jagged1 Promote in Vivo Tumorigenicity in NIH3T3 Fibroblasts with Distinct Phenotypes. Am. J. Pathol. 2008, 173, 865–878. [Google Scholar] [CrossRef]
- Sun, J.; Luo, Z.; Wang, G.; Wang, Y.; Wang, Y.; Olmedo, M.; Morandi, M.M.; Barton, S.; Kevil, C.G.; Shu, B.; et al. Notch Ligand Jagged1 Promotes Mesenchymal Stromal Cell-Based Cartilage Repair. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Aho, S. Soluble Form of Jagged1: Unique Product of Epithelial Keratinocytes and a Regulator of Keratinocyte Differentiation. J. Cell Biochem. 2004, 92, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Karanu, F.N.; Murdoch, B.; Gallacher, L.; Wu, D.M.; Koremoto, M.; Sakano, S.; Bhatia, M. The Notch Ligand Jagged-1 Represents a Novel Growth Factor of Human Hematopoietic Stem Cells. J. Exp. Med. 2000, 192, 1365–1372. [Google Scholar] [CrossRef]
- Masuya, M.; Katayama, N.; Hoshino, N.; Nishikawa, H.; Sakano, S.; Araki, H.; Mitani, H.; Suzuki, H.; Miyashita, H.; Kobayashi, K.; et al. The Soluble Notch Ligand, Jagged-1, Inhibits Proliferation of CD34+ Macrophage Progenitors. Int. J. Hematol. 2002, 75, 269–276. [Google Scholar] [CrossRef]
- Campese, A.F.; Grazioli, P.; de Cesaris, P.; Riccioli, A.; Bellavia, D.; Pelullo, M.; Padula, F.; Noce, C.; Verkhovskaia, S.; Filippini, A.; et al. Mouse Sertoli Cells Sustain De Novo Generation of Regulatory T Cells by Triggering the Notch Pathway Through Soluble JAGGED1. Biol. Reprod. 2014, 90, 53. [Google Scholar] [CrossRef]
- Metrich, M.; Bezdek Pomey, A.; Berthonneche, C.; Sarre, A.; Nemir, M.; Pedrazzini, T. Jagged1 Intracellular Domain-Mediated Inhibition of Notch1 Signalling Regulates Cardiac Homeostasis in the Postnatal Heart. Cardiovasc. Res. 2015, 108, 74–86. [Google Scholar] [CrossRef]
- Azimi, M.; Brown, N.L. Jagged1 Protein Processing in the Developing Mammalian Lens. Biol. Open 2019, 8, bio041095. [Google Scholar] [CrossRef]
- Kumar, S.; Park, H.S.; Lee, K. Jagged1 Intracellular Domain Modulates Steroidogenesis in Testicular Leydig Cells. PLoS ONE 2020, 15, e0244553. [Google Scholar] [CrossRef]
- Hock, B.; Böhme, B.; Karn, T.; Yamamoto, T.; Kaibuchi, K.; Holtrich, U.; Holland, S.; Pawson, T.; Rübsamen-Waigmann, H.; Strebhardt, K. PDZ-Domain-Mediated Interaction of the Eph-Related Receptor Tyrosine Kinase EphB3 and the Ras-Binding Protein AF6 Depends on the Kinase Activity of the Receptor. Proc. Natl. Acad. Sci. USA 1998, 95, 9779–9784. [Google Scholar] [CrossRef]
- Popovic, M.; Bella, J.; Zlatev, V.; Hodnik, V.; Anderluh, G.; Barlow, P.N.; Pintar, A.; Pongor, S. The Interaction of Jagged-1 Cytoplasmic Tail with Afadin PDZ Domain Is Local, Folding-Independent, and Tuned by Phosphorylation. J. Mol. Recognit. 2011, 24, 245–253. [Google Scholar] [CrossRef]
- Carmena, A.; Speicher, S.; Baylies, M. The PDZ Protein Canoe/AF-6 Links Ras-MAPK, Notch and Wingless/Wnt Signaling Pathways by Directly Interacting with Ras, Notch and Dishevelled. PLoS ONE 2006, 1, e66. [Google Scholar] [CrossRef]
- Liu, H.; Kennard, S.; Lilly, B. NOTCH3 Expression Is Induced in Mural Cells through an Autoregulatory Loop That Requires Endothelial-Expressed JAGGED1. Circ. Res. 2009, 104, 466–475. [Google Scholar] [CrossRef]
- Tran, T.T.; Lee, K. JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells. Cancers 2022, 14, 5714. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Tang, J.; Li, W.; Li, Y.; Mei, Y.; He, L.; Zhong, K.; Xu, R. Mutual Regulation of JAG2 and PRAF2 Promotes Migration and Invasion of Colorectal Cancer Cells Uncoupled from Epithelial-Mesenchymal Transition. Cancer Cell Int. 2019, 19, 160. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhou, Z.; Zhou, H.; Zhao, L.; Chen, D.; Chen, H.; Zou, H.; Qi, Y.; Jia, W.; Pang, L. The Expression Profile and Clinicopathological Significance of Notch1 in Patients with Colorectal Cancer: A Meta-Analysis. Future Oncol. 2017, 13, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Zheng, J.; Wang, W.; Zhao, Q.; Li, Y.; Li, J.; Xie, H.; Zhang, H.; Dong, G.; Xu, C.; et al. Notch2 Expression Is Decreased in Colorectal Cancer and Related to Tumor Differentiation Status. Ann. Surg. Oncol. 2009, 16, 3259–3266. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Z.; Zhou, Y.; Wang, W.; Li, Y.; Zhang, H.; Dong, G.; Zhao, Q.; Ji, G. Notch1 and Notch2 Have Opposite Prognostic Effects on Patients with Colorectal Cancer. Ann. Oncol. 2011, 22, 2440–2447. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Kazama, S.; Akiyoshi, T.; Murono, K.; Yoneyama, S.; Tanaka, T.; Tanaka, J.; Kiyomatsu, T.; Kawai, K.; Nozawa, H.; et al. Nuclear Notch3 Expression Is Associated with Tumor Recurrence in Patients with Stage II and III Colorectal Cancer. Ann. Surg. Oncol. 2014, 21, 2650–2658. [Google Scholar] [CrossRef] [PubMed]
- Serafin, V.; Persano, L.; Moserle, L.; Esposito, G.; Ghisi, M.; Curtarello, M.; Bonanno, L.; Masiero, M.; Ribatti, D.; Stürzl, M.; et al. Notch3 Signalling Promotes Tumour Growth in Colorectal Cancer. J. Pathol. 2011, 224, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Luo, W.; Fang, J.; Yu, C.; Liu, G.; Yuan, X.; Liu, Y.; Wu, W. Notch3 Signaling Promotes Colorectal Tumor Growth by Enhancing Immunosuppressive Cells Infiltration in the Microenvironment. BMC Cancer 2023, 23, 55. [Google Scholar] [CrossRef]
- George, D.C.; Bertrand, F.E.; Sigounas, G. Notch-3 Affects Chemoresistance in Colorectal Cancer via DNA Base Excision Repair Enzymes. Adv. Biol. Regul. 2024, 91, 101013. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nimisha; Apurva; Kumar, A.; Ali, A.; Singh Saluja, S.; Prasad, B. Elevated Expression of Notch 2 & Notch 3 Is Associated with Disease Progression in Colorectal Cancer. Int. J. Appl. Biol. Pharm. 2022, 13, 33–50. [Google Scholar] [CrossRef]
- Wang, F.; Long, J.; Li, L.; Wu, Z.X.; Da, T.T.; Wang, X.Q.; Huang, C.; Jiang, Y.H.; Yao, X.Q.; Ma, H.Q.; et al. Single-Cell and Spatial Transcriptome Analysis Reveals the Cellular Heterogeneity of Liver Metastatic Colorectal Cancer. Sci. Adv. 2023, 9, eadf5464. [Google Scholar] [CrossRef]
- Gowrikumar, S.; Ahmad, R.; Uppada, S.B.; Washington, M.K.; Shi, C.; Singh, A.B.; Dhawan, P. Upregulated Claudin-1 Expression Promotes Colitis-Associated Cancer by Promoting β-Catenin Phosphorylation and Activation in Notch/p-AKT-Dependent Manner. Oncogene 2019, 38, 5321–5337. [Google Scholar] [CrossRef]
- Meng, R.D.; Shelton, C.C.; Li, Y.M.; Qin, L.X.; Notterman, D.; Paty, P.B.; Schwartz, G.K. γ-Secretase Inhibitors Abrogate Oxaliplatin-Induced Activation of the Notch-1 Signaling Pathway in Colon Cancer Cells Resulting in Enhanced Chemosensitivity. Cancer Res. 2009, 69, 573–582. [Google Scholar] [CrossRef]
- Liu, H.; Yin, Y.; Hu, Y.; Feng, Y.; Bian, Z.; Yao, S.; Li, M.; You, Q.; Huang, Z. MiR-139-5p Sensitizes Colorectal Cancer Cells to 5-Fluorouracil by Targeting NOTCH-1. Pathol. Res. Pr. 2016, 212, 643–649. [Google Scholar] [CrossRef]
- Riccio, O.; van Gijn, M.E.; Bezdek, A.C.; Pellegrinet, L.; van Es, J.H.; Zimber-Strobl, U.; Strobl, L.J.; Honjo, T.; Clevers, H.; Radtke, F. Loss of Intestinal Crypt Progenitor Cells Owing to Inactivation of Both Notch1 and Notch2 Is Accompanied by Derepression of CDK Inhibitors P27Kip1 and P57Kip2. EMBO Rep. 2008, 9, 377–383. [Google Scholar] [CrossRef]
- Van Es, J.H.; Van Gijn, M.E.; Riccio, O.; Van Den Born, M.; Vooijs, M.; Begthel, H.; Cozijnsen, M.; Robine, S.; Winton, D.J.; Radtke, F.; et al. Notch/γ-Secretase Inhibition Turns Proliferative Cells in Intestinal Crypts and Adenomas into Goblet Cells. Nature 2005, 435, 959–963. [Google Scholar] [CrossRef]
- Wang, R.; Ye, X.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Ellis, L.M. A Disintegrin and Metalloproteinase Domain 17 Regulates Colorectal Cancer Stem Cells and Chemosensitivity Via Notch1 Signaling. Stem Cells Transl. Med. 2016, 5, 331–338. [Google Scholar] [CrossRef]
- Li, D.D.; Zhao, C.H.; Ding, H.W.; Wu, Q.; Ren, T.S.; Wang, J.; Chen, C.Q.; Zhao, Q.C. A Novel Inhibitor of ADAM17 Sensitizes Colorectal Cancer Cells to 5-Fluorouracil by Reversing Notch and Epithelial-Mesenchymal Transition in Vitro and in Vivo. Cell Prolif. 2018, 51, e12480. [Google Scholar] [CrossRef]
- Dosch, J.; Ziemke, E.; Wan, S.; Luker, K.; Welling, T.; Hardiman, K.; Fearon, E.; Thomas, S.; Flynn, M.; Rios-Doria, J.; et al. Targeting ADAM17 Inhibits Human Colorectal Adenocarcinoma Progression and Tumor-Initiating Cell Frequency. Oncotarget 2017, 8, 65090–65099. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Luna, C.; González-Flores, E.; Ortiz, R.; Martínez-González, L.J.; Antúnez-Rodríguez, A.; Expósito-Ruiz, M.; Melguizo, C.; Caba, O.; Prados, J. Circulating Ptgs2, Jag1, Gucy2c and Pgf Mrna in Peripheral Blood and Serum as Potential Biomarkers for Patients with Metastatic Colon Cancer. J. Clin. Med. 2021, 10, 2248. [Google Scholar] [CrossRef]
- Nakata, T.; Shimizu, H.; Nagata, S.; Ito, G.; Fujii, S.; Suzuki, K.; Kawamoto, A.; Ishibashi, F.; Kuno, R.; Anzai, S.; et al. Indispensable Role of Notch Ligand-Dependent Signaling in the Proliferation and Stem Cell Niche Maintenance of APC-Deficient Intestinal Tumors. Biochem. Biophys. Res. Commun. 2017, 482, 1296–1303. [Google Scholar] [CrossRef]
- Yamamoto, S.; Tateishi, K.; Kudo, Y.; Yamamoto, K.; Isagawa, T.; Nagae, G.; Nakatsuka, T.; Asaoka, Y.; Ijichi, H.; Hirata, Y.; et al. Histone Demethylase KDM4C Regulates Sphere Formation by Mediating the Cross Talk between Wnt and Notch Pathways in Colonic Cancer Cells. Carcinogenesis 2013, 34, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Pannequin, J.; Bonnans, C.; Delaunay, N.; Ryan, J.; Bourgaux, J.-F.; Joubert, D.; Hollande, F. The Wnt Target Jagged-1 Mediates the Activation of Notch Signaling by Progastrin in Human Colorectal Cancer Cells. Cancer Res. 2009, 69, 6065–6073. [Google Scholar] [CrossRef] [PubMed]
- López-Arribillaga, E.; Rodilla, V.; Colomer, C.; Vert, A.; Shelton, A.; Cheng, J.H.; Yan, B.; Gonzalez-Perez, A.; Junttila, M.R.; Iglesias, M.; et al. Manic Fringe Deficiency Imposes Jagged1 Addiction to Intestinal Tumor Cells. Nat. Commun. 2018, 9, 2992. [Google Scholar] [CrossRef]
- Kuintzle, R.; Santat, L.A.; Elowitz, M.B. Diversity in Notch Ligand-Receptor Signaling Interactions. Elife 2025, 12, RP91422. [Google Scholar] [CrossRef]
- Stanley, P.; Okajima, T. Roles of Glycosylation in Notch Signaling. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 92. [Google Scholar]
- Haltiwanger, R.S.; Stanley, P. Modulation of Receptor Signaling by Glycosylation: Fringe Is an O-Fucose-Β1,3-N-Acetylglucosaminyltransferase. Biochim. Biophys. Acta Gen. Subj. 2002, 1573, 328–335. [Google Scholar] [CrossRef]
- Tan, J.B.; Xu, K.; Cretegny, K.; Visan, I.; Yuan, J.S.; Egan, S.E.; Guidos, C.J. Lunatic and Manic Fringe Cooperatively Enhance Marginal Zone B Cell Precursor Competition for Delta-like 1 in Splenic Endothelial Niches. Immunity 2009, 30, 254–263. [Google Scholar] [CrossRef]
- Kim, M.-H.; Kim, H.-B.; Yoon, S.P.; Lim, S.-C.; Cha, M.J.; Jeon, Y.J.; Park, S.G.; Chang, I.-Y.; You, H.J. Colon Cancer Progression Is Driven by APEX1-Mediated Upregulation of Jagged. J. Clin. Investig. 2013, 123, 3211–3230. [Google Scholar] [CrossRef]
- Kim, H.B.; Lim, H.J.; Lee, H.J.; Park, J.H.; Park, S.G. Evaluation and Clinical Significance of Jagged-1-Activated Notch Signaling by APEX1 in Colorectal Cancer. Anticancer. Res. 2019, 39, 6097–6105. [Google Scholar] [CrossRef] [PubMed]
- Van Schaeybroeck, S.; Kalimutho, M.; Dunne, P.D.; Carson, R.; Allen, W.; Jithesh, P.V.; Redmond, K.L.; Sasazuki, T.; Shirasawa, S.; Blayney, J.; et al. ADAM17-Dependent c-MET-STAT3 Signaling Mediates Resistance to MEK Inhibitors in KRAS Mutant Colorectal Cancer. Cell Rep. 2014, 7, 1940–1955. [Google Scholar] [CrossRef]
- Saad, M.I.; Alhayyani, S.; McLeod, L.; Yu, L.; Alanazi, M.; Deswaerte, V.; Tang, K.; Jarde, T.; Smith, J.A.; Prodanovic, Z.; et al. ADAM 17 Selectively Activates the IL -6 Trans-signaling/ERK MAPK Axis in KRAS -addicted Lung Cancer. EMBO Mol. Med. 2019, 11, e9976. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rodríguez, E.; Montero, J.C.; Esparís-Ogando, A.; Yuste, L.; Pandiella, A. Extracellular Signal-Regulated Kinase Phosphorylates Tumor Necrosis Factor Alpha-Converting Enzyme at Threonine 735: A Potential Role in Regulated Shedding. Mol. Biol. Cell 2002, 13, 2031–2044. [Google Scholar] [CrossRef]
- Lu, J.; Ye, X.; Fan, F.; Xia, L.; Bhattacharya, R.; Bellister, S.; Tozzi, F.; Sceusi, E.; Zhou, Y.; Tachibana, I.; et al. Endothelial Cells Promote the Colorectal Cancer Stem Cell Phenotype through a Soluble Form of Jagged-1. Cancer Cell 2013, 23, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Martínez-Cardus, A.; Gines, A.; Ruiz De Porras, V.; Moutinho, C.; Layos, L.; Manzano, J.L.; Buges, C.; Bystrup, S.; Esteller, M.; et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol. Cancer Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef]
- Briffa, R.; Langdon, S.P.; Grech, G.; Harrison, D.J. Acquired and Intrinsic Resistance to Colorectal Cancer Treatment. In Colorectal Cancer—Diagnosis, Screening and Management; InTechOpen: London, UK, 2018. [Google Scholar]
- Curry, C.L.; Reed, L.L.; Golde, T.E.; Miele, L.; Nickoloff, B.J.; Foreman, K.E. Gamma Secretase Inhibitor Blocks Notch Activation and Induces Apoptosis in Kaposi’s Sarcoma Tumor Cells. Oncogene 2005, 24, 6333–6344. [Google Scholar] [CrossRef]
- Farnie, G.; Clarke, R.B.; Spence, K.; Pinnock, N.; Brennan, K.; Anderson, N.G.; Bundred, N.J. Novel Cell Culture Technique for Primary Ductal Carcinoma in Situ: Role of Notch and Epidermal Growth Factor Receptor Signaling Pathways. J. Natl. Cancer Inst. 2007, 99, 616–627. [Google Scholar] [CrossRef]
- Gounder, M.; Ratan, R.; Alcindor, T.; Schöffski, P.; van der Graaf, W.T.; Wilky, B.A.; Riedel, R.F.; Lim, A.; Smith, L.M.; Moody, S.; et al. Nirogacestat, a γ-Secretase Inhibitor for Desmoid Tumors. N. Engl. J. Med. 2023, 388, 898–912. [Google Scholar] [CrossRef]
- Sanchez-Martin, M.; Ambesi-Impiombato, A.; Qin, Y.; Herranz, D.; Bansal, M.; Girardi, T.; Paietta, E.; Tallman, M.S.; Rowe, J.M.; De Keersmaecker, K.; et al. Synergistic Antileukemic Therapies in NOTCH1-Induced T-ALL. Proc. Natl. Acad. Sci. USA 2017, 114, 2006–2011. [Google Scholar] [CrossRef]
- Govaerts, I.; Prieto, C.; Vandersmissen, C.; Gielen, O.; Jacobs, K.; Provost, S.; Nittner, D.; Maertens, J.; Boeckx, N.; De Keersmaecker, K.; et al. PSEN1-Selective Gamma-Secretase Inhibition in Combination with Kinase or XPO-1 Inhibitors Effectively Targets T Cell Acute Lymphoblastic Leukemia. J. Hematol. Oncol. 2021, 14, 97. [Google Scholar] [CrossRef]
- Kukcinaviciute, E.; Jonusiene, V.; Sasnauskiene, A.; Dabkeviciene, D.; Eidenaite, E.; Laurinavicius, A. Significance of Notch and Wnt Signaling for Chemoresistance of Colorectal Cancer Cells HCT116. J. Cell Biochem. 2018, 119, 5913–5920. [Google Scholar] [CrossRef]
- Aleksic, T.; Feller, S.M. Gamma-Secretase Inhibition Combined with Platinum Compounds Enhances Cell Death in a Large Subset of Colorectal Cancer Cells. Cell Commun. Signal. 2008, 6, 8. [Google Scholar] [CrossRef]
- Timme, C.R.; Gruidl, M.; Yeatman, T.J. Gamma-Secretase Inhibition Attenuates Oxaliplatin-Induced Apoptosis through Increased Mcl-1 and/or Bcl-XL in Human Colon Cancer Cells. Apoptosis 2013, 18, 1163–1174. [Google Scholar] [CrossRef]
- Strosberg, J.R.; Yeatman, T.; Weber, J.; Coppola, D.; Schell, M.J.; Han, G.; Almhanna, K.; Kim, R.; Valone, T.; Jump, H.; et al. A Phase II Study of RO4929097 in Metastatic Colorectal Cancer. Eur. J. Cancer 2012, 48, 997–1003. [Google Scholar] [CrossRef]

| CMS Subtype | Subtype Name | Molecular and Genomic Features | Pathway Activation/Biological Characteristics | Clinical/Prognostic Features |
|---|---|---|---|---|
| CMS1 | MSI Immune | Hypermutated DNA MSI CIMP-High Frequent BRAF mutations | Strong immune activation and infiltration | Poor overall survival and relapse outcomes |
| CMS2 | Canonical | Chromosomal instability (CIN) High somatic copy number alterations (SCNA) | Activation of Wnt and Myc pathways | Typical “classical” subtype with intermediate prognosis |
| CMS3 | Metabolic | Mixed MSI status Low SCNA and low CIMP KRAS mutations enriched | Epithelial features with metabolic dysregulation | Variable prognosis; often linked to altered metabolism |
| CMS4 | Mesenchymal | High SCNA Stromal infiltration and angiogenesis | Activation of TGFβ pathway | Worst prognosis, associated with poor relapse-free and overall survival |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zema, S.; Di Fazio, F.; Palermo, R.; Talora, C.; Bellavia, D. The Two-Way Role of Jagged1 in Cancer: A Focus on CRC. Cells 2025, 14, 1815. https://doi.org/10.3390/cells14221815
Zema S, Di Fazio F, Palermo R, Talora C, Bellavia D. The Two-Way Role of Jagged1 in Cancer: A Focus on CRC. Cells. 2025; 14(22):1815. https://doi.org/10.3390/cells14221815
Chicago/Turabian StyleZema, Sabrina, Francesca Di Fazio, Rocco Palermo, Claudio Talora, and Diana Bellavia. 2025. "The Two-Way Role of Jagged1 in Cancer: A Focus on CRC" Cells 14, no. 22: 1815. https://doi.org/10.3390/cells14221815
APA StyleZema, S., Di Fazio, F., Palermo, R., Talora, C., & Bellavia, D. (2025). The Two-Way Role of Jagged1 in Cancer: A Focus on CRC. Cells, 14(22), 1815. https://doi.org/10.3390/cells14221815







