Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron–Glia Cultures Involves Histone Serotonylation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Rat Primary Cortical Neuron Cultures
2.3. Drugs
2.4. Cell Proliferation Assay
2.5. RNA Extraction and Sequencing
2.6. RNA Sequence Alignment and Analysis
2.7. Visualization and Functional Annotation of DEGs
2.8. qPCR Analysis
2.9. Immunocytochemistry (ICC)
2.10. Microscopy and Analysis
3. Results
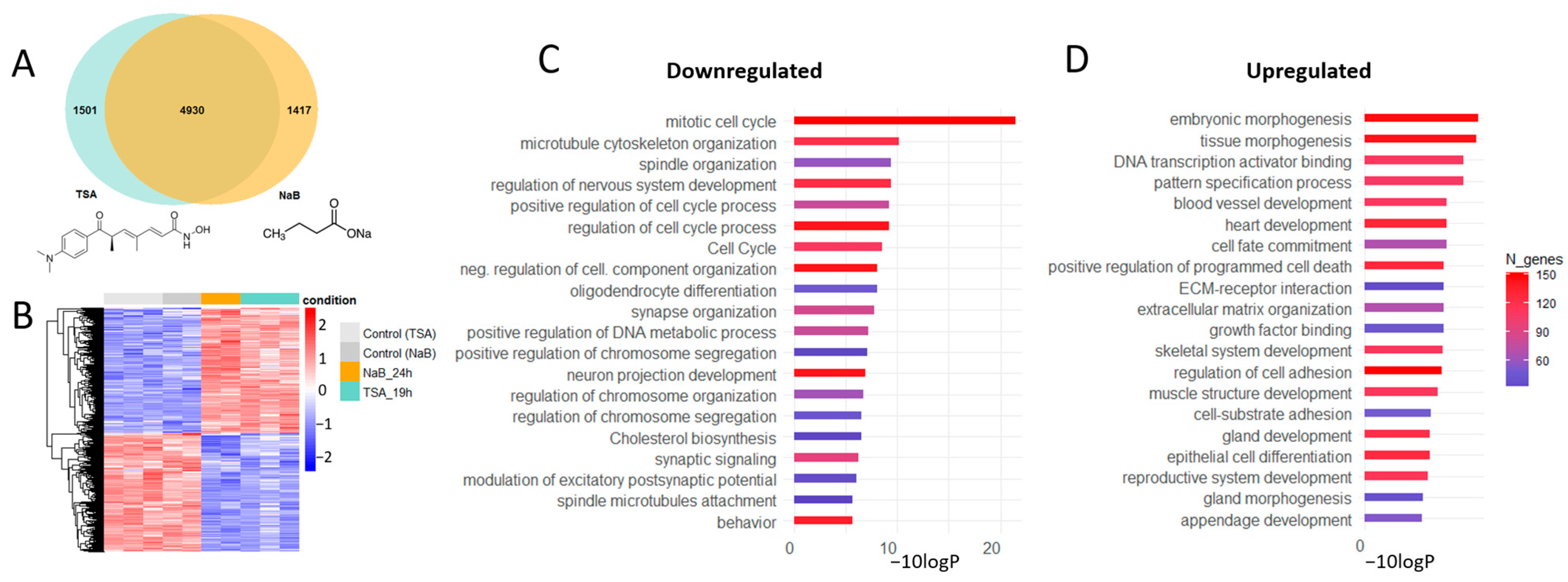
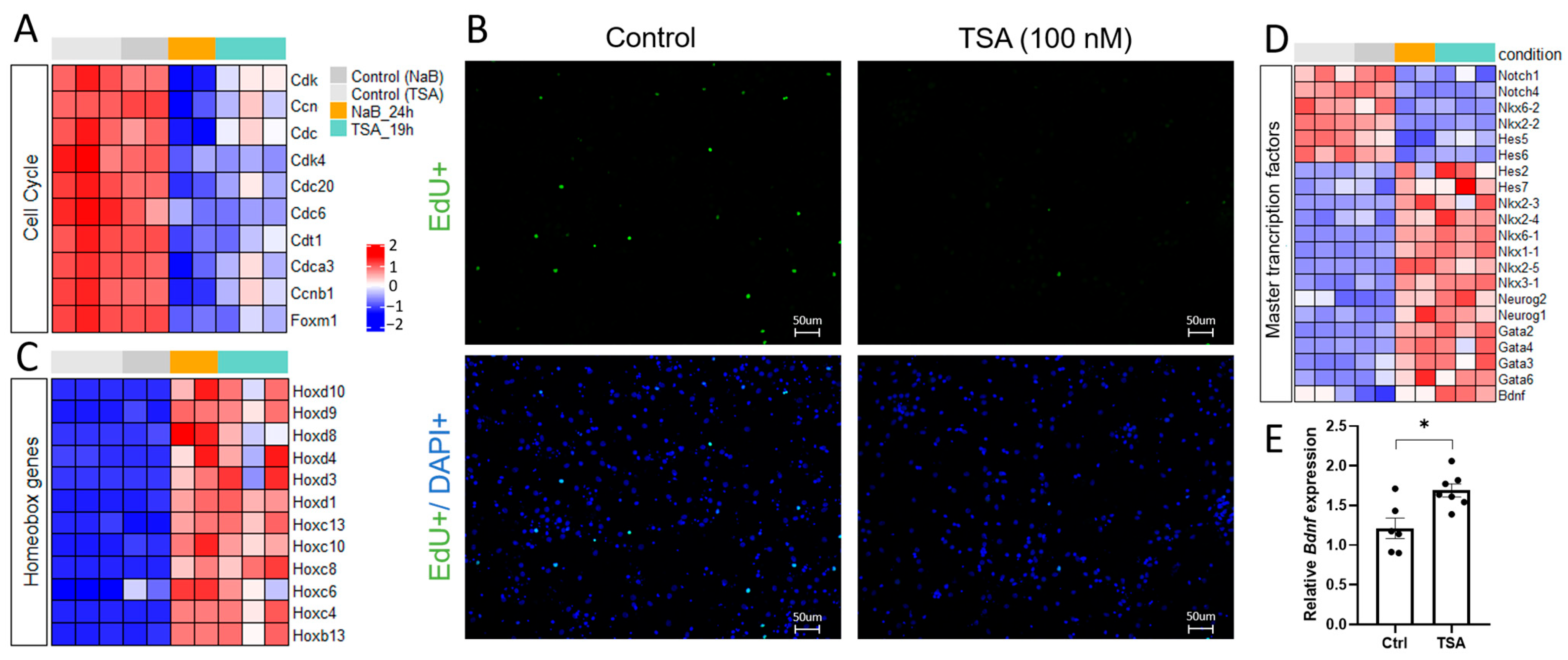
3.1. HDAC Inhibitors Shift the Gene Expression Profiles in Primary Neuron Cultures from Proliferation to Differentiation and Affect the Expression of Master Transcription Factors
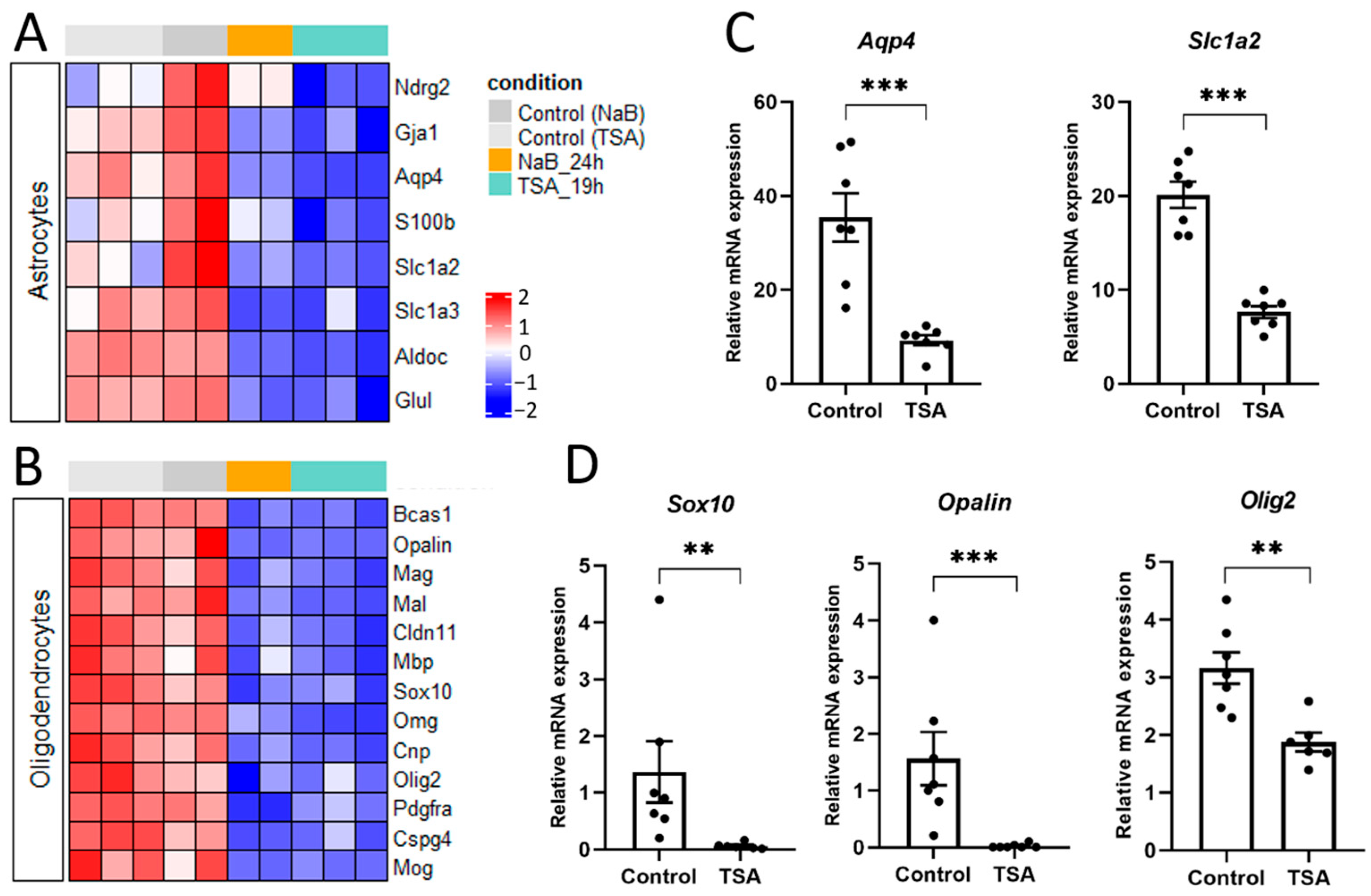
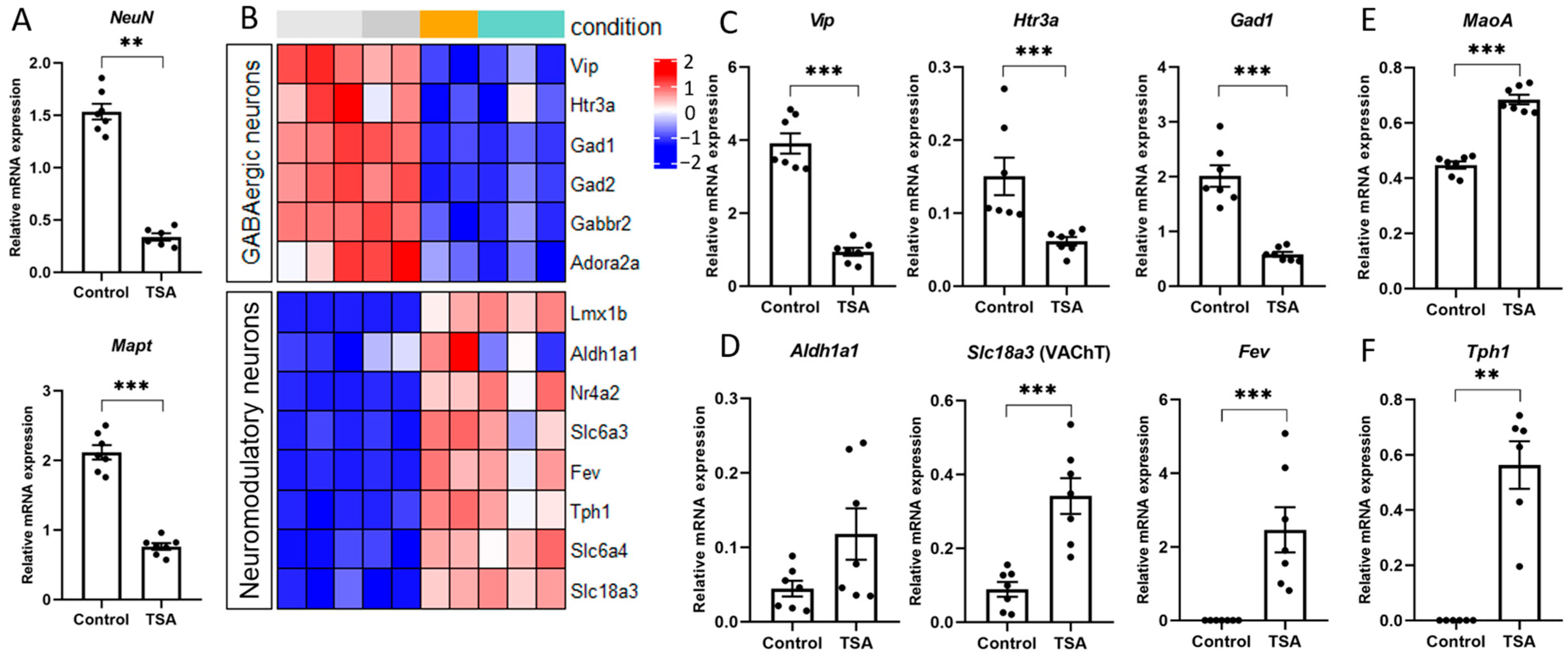
3.2. HDAC Inhibitors Regulate the Expression of Specific Markers of Various Brain Cells
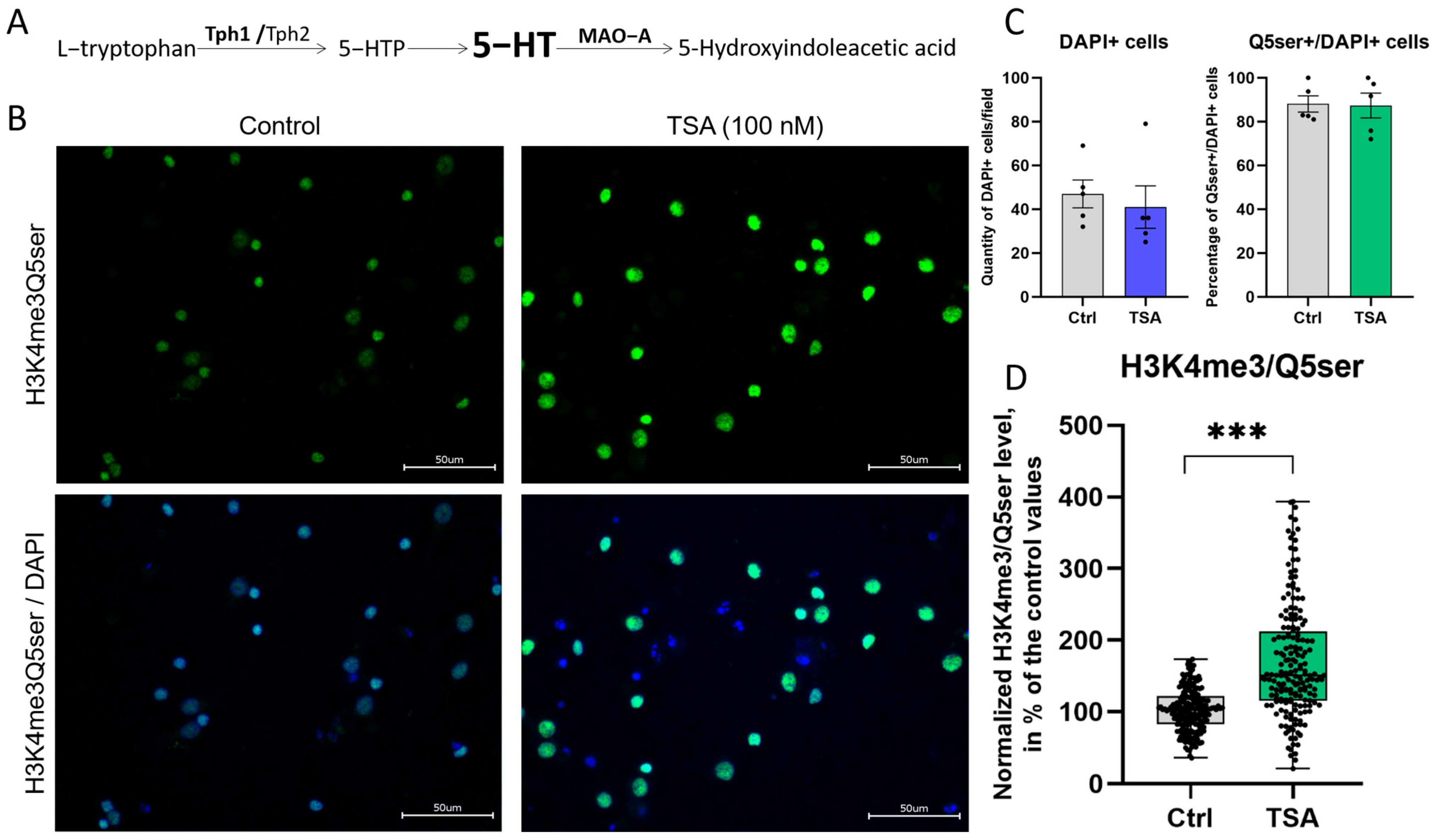
3.3. HDAC Inhibitor Trichostatin A Elevates the Expression of Genes Associated with Serotonergic Secretory Phenotype and Stimulates Histone Serotonylation
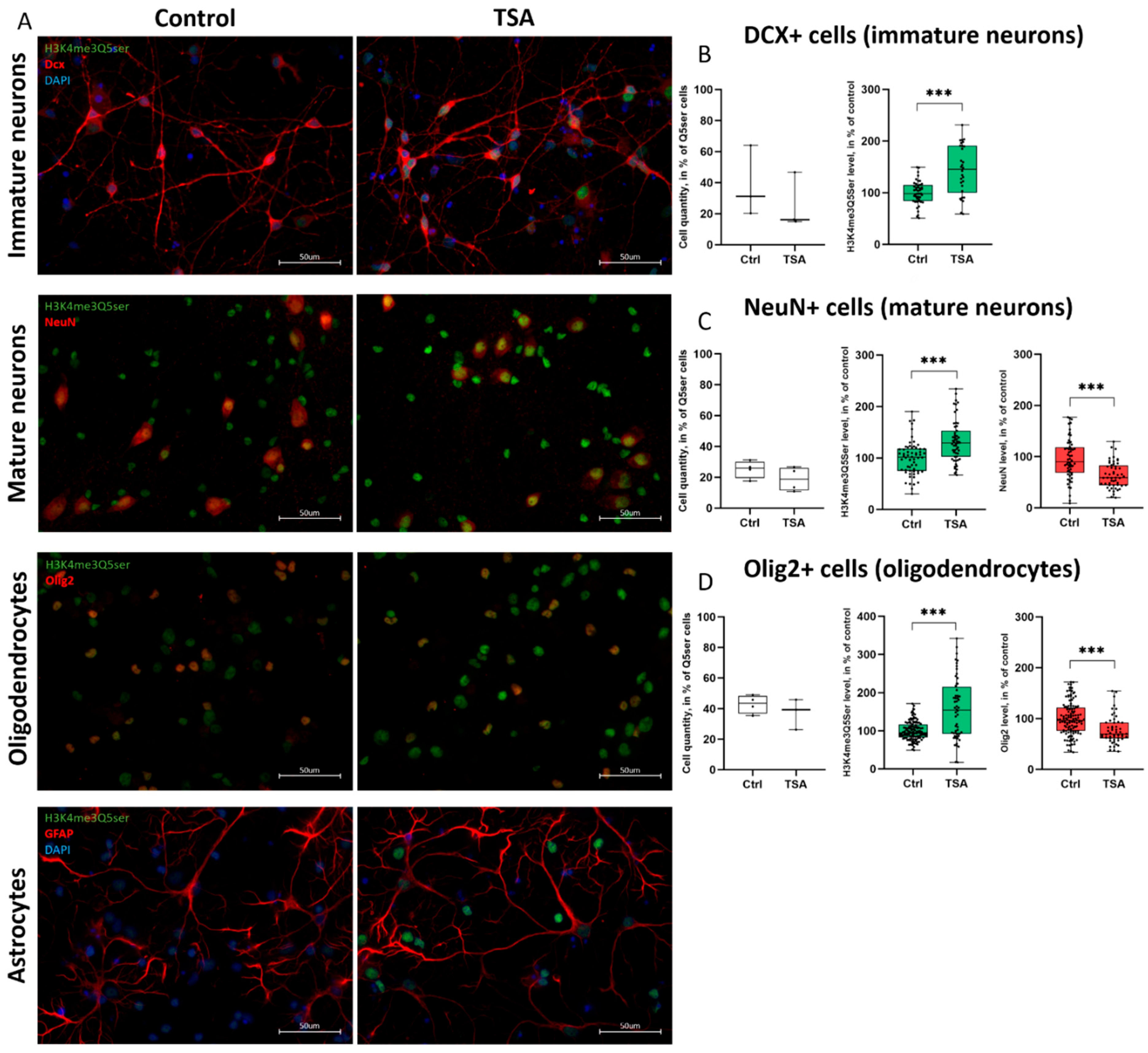
3.4. Histone Serotonylation Marks Are Widely Distributed Across Different Populations of Both Neurons and Glia
4. Discussion
4.1. HDAC Inhibition Suppresses Pro-Glial Transcriptional Programs and Promotes Neuronal Differentiation
4.2. HDAC Inhibition Mediates Neuron Specialization
4.3. HDAC off, Serotonin on: Serotonylation as a Novel Epigenetic Mechanism
4.4. Limitations of the Study
- Bulk RNA sequencing results should be taken with caution because primary neuronal cultures consist of highly heterogeneous populations of various types of brain cells, likely at different stages of development. More precise assessment of gene expression programs in individual brain cells using scRNA-seq would be preferable.
- The incubation times for TSA and NaB were different. As a continuation of previous research [37], TSA was applied for 19 h. The sodium butyrate data with the appropriate time-matched controls were taken from a separate project where conditions were the same except for the longer incubation time (24 h), which was chosen as a more generally accepted value. These two datasets were taken to strengthen our conclusions and to avoid possible side effects caused by each drug. We believe that these differences in the incubation times for TSA and NaB were not relevant since the initial goal of long-term use of HDAC inhibitors was to assess which late-response genes were affected following chromatin rearrangements. We understand that some genes may be underestimated with this approach, but the overall gene expression pattern appears to be the same.
- Most of our conclusions regarding changes in transcriptional programs were based on RNA sequencing. Only a few transcriptional factors were confirmed by the ICC staining. We can only speculate that transcriptional changes may lead to changes in the proteome, but this requires additional verification.
- We speculate that induced histone modifications may trigger processes associated with cell identity regulation and cellular reprogramming. However, actual complete reprogramming of one cell into another cannot be confirmed in this experimental paradigm, as it requires a longer time, possibly several days/weeks.
4.5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juliandi, B.; Abematsu, M.; Nakashima, K. Chromatin Remodeling in Neural Stem Cell Differentiation. Curr. Opin. Neurobiol. 2010, 20, 408–415. [Google Scholar] [CrossRef]
- Hsieh, J.; Nakashima, K.; Kuwabara, T.; Mejia, E.; Gage, F.H. Histone Deacetylase Inhibition-Mediated Neuronal Differentiation of Multipotent Adult Neural Progenitor Cells. Proc. Natl. Acad. Sci. USA 2004, 101, 16659–16664. [Google Scholar] [CrossRef]
- Huang, Y.; Myers, S.J.; Dingledine, R. Transcriptional Repression by REST: Recruitment of Sin3A and Histone Deacetylase to Neuronal Genes. Nat. Neurosci. 1999, 2, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, G.W.; Wang, Y.H.; Hirai, T.; Padmanabhan, R.; Panchision, D.M.; Newell, L.F.; McKay, R.D.; Howard, B.H. Complementary Roles for Histone Deacetylases, 1, 2, and 3 in Differentiation of Pluripotent Stem Cells. Differentiation 2008, 76, 348–356. [Google Scholar] [CrossRef]
- Ye, F.; Chen, Y.; Hoang, T.; Montgomery, R.L.; Zhao, X.H.; Bu, H.; Hu, T.; Taketo, M.M.; van Es, J.H.; Clevers, H.; et al. HDAC1 and HDAC2 Regulate Oligodendrocyte Differentiation by Disrupting the β-Catenin–TCF Interaction. Nat. Neurosci. 2009, 12, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Conway, G.D.; O’Bara, M.A.; Vedia, B.H.; Pol, S.U.; Sim, F.J. Histone Deacetylase Activity Is Required for Human Oligodendrocyte Progenitor Differentiation. Glia 2012, 60, 1944–1953. [Google Scholar] [CrossRef]
- Castelo-Branco, G.; Lilja, T.; Wallenborg, K.; Falcão, A.M.; Marques, S.C.; Gracias, A.; Solum, D.; Paap, R.; Walfridsson, J.; Teixeira, A.I.; et al. Neural Stem Cell Differentiation Is Dictated by Distinct Actions of Nuclear Receptor Corepressors and Histone Deacetylases. Stem Cell Rep. 2014, 3, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, X.; Liu, L.; Jiang, M.; Zhao, C.; Wang, H.; He, D.; Zheng, T.; Zhou, X.; Hassan, A.; et al. Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev. Cell 2016, 36, 316–330. [Google Scholar] [CrossRef]
- Harrison, M.R.; Georgiou, A.S.; Spaink, H.P.; Cunliffe, V.T. The epigenetic regulator Histone Deacetylase 1 promotes transcription of a core neurogenic programme in zebrafish embryos. BMC Genom. 2011, 12, 24. [Google Scholar] [CrossRef]
- Nieto-Estevez, V.; Changarathil, G.; Adeyeye, A.O.; Coppin, M.O.; Kassim, R.S.; Zhu, J.; Hsieh, J. HDAC1 Regulates Neuronal Differentiation. Front. Mol. Neurosci. 2022, 14, 815808. [Google Scholar] [CrossRef]
- Montgomery, R.L.; Hsieh, J.; Barbosa, A.C.; Richardson, J.A.; Olson, E.N. Histone Deacetylases 1 and 2 Control the Progression of Neural Precursors to Neurons during Brain Development. Proc. Natl. Acad. Sci. USA 2009, 106, 7876–7881. [Google Scholar] [CrossRef] [PubMed]
- Lyssiotis, C.A.; Walker, J.; Wu, C.; Kondo, T.; Schultz, P.G.; Wu, X. Inhibition of Histone Deacetylase Activity Induces Developmental Plasticity in Oligodendrocyte Precursor Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 14982–14987. [Google Scholar] [CrossRef] [PubMed]
- Siebzehnrubl, F.A.; Buslei, R.; Eyupoglu, I.Y.; Seufert, S.; Hahnen, E.; Blumcke, I. Histone Deacetylase Inhibitors Increase Neuronal Differentiation in Adult Forebrain Precursor Cells. Exp. Brain Res. 2007, 176, 672–678. [Google Scholar] [CrossRef]
- Yu, I.T.; Park, J.Y.; Kim, S.H.; Lee, J.S.; Kim, Y.S.; Son, H. Valproic Acid Promotes Neuronal Differentiation by Induction of Proneural Factors in Association with H4 Acetylation. Neuropharmacology 2009, 56, 473–480. [Google Scholar] [CrossRef]
- Fukuchi, M.; Nii, T.; Ishimaru, N.; Minamino, A.; Hara, D.; Takasaki, I.; Tabuchi, A.; Tsuda, M. Valproic Acid Induces Up- or down-Regulation of Gene Expression Responsible for the Neuronal Excitation and Inhibition in Rat Cortical Neurons through Its Epigenetic Actions. Neurosci. Res. 2009, 65, 35–43. [Google Scholar] [CrossRef]
- Asaoka, N.; Nagayasu, K.; Nishitani, N.; Yamashiro, M.; Shirakawa, H.; Nakagawa, T.; Kaneko, S. Inhibition of Histone Deacetylases Enhances the Function of Serotoninergic Neurons in Organotypic Raphe Slice Cultures. Neurosci. Lett. 2015, 593, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, L.A.; Thompson, R.E.; Zhao, S.; Lepack, A.E.; Lyu, Y.; Bhanu, N.V.; Zhang, B.; Loh, Y.E.; Ramakrishnan, A.; Vadodaria, K.C.; et al. Histone Serotonylation Is a Permissive Modification That Enhances TFIID Binding to H3K4me3. Nature 2019, 567, 535–539. [Google Scholar] [CrossRef]
- Sardar, D.; Cheng, Y.T.; Woo, J.; Choi, D.J.; Lee, Z.F.; Kwon, W.; Chen, H.C.; Lozzi, B.; Cervantes, A.; Rajendran, K.; et al. Induction of Astrocytic Slc22a3 Regulates Sensory Processing through Histone Serotonylation. Science 2023, 380, eade0027. [Google Scholar] [CrossRef]
- Al-Kachak, A.; Di Salvo, G.; Fulton, S.L.; Chan, J.C.; Farrelly, L.A.; Lepack, A.E.; Bastle, R.M.; Kong, L.; Cathomas, F.; Newman, E.L.; et al. Histone Serotonylation in Dorsal Raphe Nucleus Contributes to Stress- and Antidepressant-Mediated Gene Expression and Behavior. Nat. Commun. 2024, 15, 5042. [Google Scholar] [CrossRef]
- Chan, J.C.; Alenina, N.; Cunningham, A.M.; Ramakrishnan, A.; Shen, L.; Bader, M.; Maze, I. Serotonin Transporter-dependent Histone Serotonylation in Placenta Contributes to the Neurodevelopmental Transcriptome. J. Mol. Biol. 2024, 436, 168454. [Google Scholar] [CrossRef]
- Ehrensberger, A.H.; Svejstrup, J.Q. Reprogramming Chromatin. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 464–482. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.L.; Liu, C.W.; Shen, S.W.; Yu, Z.; Mo, J.L.; Chen, X.H.; Sun, F.Y. Striatal Astrocytes Transdifferentiate into Functional Mature Neurons Following Ischemic Brain Injury. Glia 2015, 63, 1660–1670. [Google Scholar] [CrossRef]
- Tai, W.; Wu, W.; Wang, L.L.; Ni, H.; Chen, C.; Yang, J.; Zang, T.; Zou, Y.; Xu, X.M.; Zhang, C.L. In Vivo Reprogramming of NG2 Glia Enables Adult Neurogenesis and Functional Recovery Following Spinal Cord Injury. Cell Stem Cell 2021, 28, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Zang, T.; Zou, Y.; Fang, S.; Smith, D.K.; Bachoo, R.; Zhang, C.L. In Vivo Reprogramming of Astrocytes to Neuroblasts in the Adult Brain. Nat. Cell Biol. 2013, 15, 1164–1175. [Google Scholar] [CrossRef]
- Boshans, L.L.; Factor, D.C.; Singh, V.; Liu, J.; Zhao, C.; Mandoiu, I.; Lu, Q.R.; Casaccia, P.; Tesar, P.J.; Nishiyama, A. The Chromatin Environment Around Interneuron Genes in Oligodendrocyte Precursor Cells and Their Potential for Interneuron Reprograming. Front. Neurosci. 2019, 13, 829. [Google Scholar] [CrossRef]
- Pavlou, M.A.S.; Grandbarbe, L.; Buckley, N.J.; Niclou, S.P.; Michelucci, A. Transcriptional and Epigenetic Mechanisms Underlying Astrocyte Identity. Prog. Neurobiol. 2019, 174, 36–52. [Google Scholar] [CrossRef]
- Griffiths, B.B.; Bhutani, A.; Stary, C.M. Adult Neurogenesis from Reprogrammed Astrocytes. Neural Regen. Res. 2020, 15, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef]
- Lentini, C.; d’Orange, M.; Marichal, N.; Trottmann, M.M.; Vignoles, R.; Foucault, L.; Verrier, C.; Massera, C.; Raineteau, O.; Conzelmann, K.K.; et al. Reprogramming Reactive Glia into Interneurons Reduces Chronic Seizure Activity in a Mouse Model of Mesial Temporal Lobe Epilepsy. Cell Stem Cell 2021, 28, 2104–2121. [Google Scholar] [CrossRef]
- Weinberg, M.S.; Criswell, H.E.; Powell, S.K.; Bhatt, A.P.; McCown, T.J. Viral Vector Reprogramming of Adult Resident Striatal Oligodendrocytes into Functional Neurons. Mol. Ther. 2017, 25, 928–934. [Google Scholar] [CrossRef]
- Mo, J.L.; Liu, Q.; Kou, Z.W.; Wu, K.W.; Yang, P.; Chen, X.H.; Sun, F.Y. MicroRNA-365 Modulates Astrocyte Conversion into Neuron in Adult Rat Brain after Stroke by Targeting Pax6. Glia 2018, 66, 1346–1362. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Zhao, A.; Fu, X. Small molecules for reprogramming and transdifferentiation. Cell Mol. Life Sci. 2017, 74, 3553–3575. [Google Scholar] [CrossRef]
- Ma, Y.; Xie, H.; Du, X.; Wang, L.; Jin, X.; Zhang, Q.; Han, Y.; Sun, S.; Wang, L.; Li, X.; et al. In Vivo Chemical Reprogramming of Astrocytes into Neurons. Cell Discov. 2021, 7, 12. [Google Scholar] [CrossRef]
- Pavlova, G.; Kolesnikova, V.; Samoylenkova, N.; Drozd, S.; Revishchin, A.; Shamadykova, D.; Usachev, D.Y.; Kopylov, A. A Combined Effect of G-Quadruplex and Neuro-Inducers as an Alternative Approach to Human Glioblastoma Therapy. Front. Oncol. 2022, 12, 880740. [Google Scholar] [CrossRef]
- Yavarpour-Bali, H.; Ghasemi-Kasman, M.; Shojaei, A. Direct Reprogramming of Terminally Differentiated Cells into Neurons: A Novel and Promising Strategy for Alzheimer’s Disease Treatment. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 98, 109820. [Google Scholar] [CrossRef] [PubMed]
- Giehrl-Schwab, J.; Giesert, F.; Rauser, B.; Lao, C.L.; Hembach, S.; Lefort, S.; Ibarra, I.L.; Koupourtidou, C.; Luecken, M.D.; Truong, D.J.; et al. Parkinson’s disease motor symptoms rescue by CRISPRa-reprogramming astrocytes into GABAergic neurons. EMBO Mol. Med. 2022, 14, e14797. [Google Scholar] [CrossRef] [PubMed]
- Borodinova, A.A.; Kuznetsova, M.A.; Alekseeva, V.S.; Balaban, P.M. Histone Acetylation Determines Transcription of Atypical Protein Kinases in Rat Neurons. Sci. Rep. 2019, 9, 4332. [Google Scholar] [CrossRef]
- Yoshida, M.; Kijima, M.; Akita, M.; Beppu, T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990, 265, 17174–17179. [Google Scholar] [CrossRef]
- Park, I.H.; Kang, J.H.; Shin, J.M.; Lee, H.M. Trichostatin A Inhibits Epithelial Mesenchymal Transition Induced by TGF-β1 in Airway Epithelium. PLoS ONE 2016, 11, e0162058. [Google Scholar] [CrossRef]
- Cousens, L.S.; Gallwitz, D.; Alberts, B.M. Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 1979, 254, 1716–1723. [Google Scholar] [CrossRef]
- Parker, M.I.; de Haan, J.B.; Gevers, W. DNA hypermethylation in sodium butyrate-treated WI-38 fibroblasts. J. Biol. Chem. 1986, 261, 2786–2790. [Google Scholar] [CrossRef]
- Chen, X.; Xiao, W.; Chen, W.; Luo, L.; Ye, S.; Liu, Y. The epigenetic modifier trichostatin A, a histone deacetylase inhibitor, sup-presses proliferation and epithelial-mesenchymal transition of lens epithelial cells. Cell Death Dis. 2013, 4, e884. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Sock, E.; Wegner, M. Using the Lineage Determinants Olig2 and Sox10 to Explore Transcriptional Regulation of Oligodendrocyte Development. Dev. Neurobiol. 2021, 81, 892–901. [Google Scholar] [CrossRef]
- Rumajogee, P.; Madeira, A.; Vergé, D.; Hamon, M.; Miquel, M.C. Up-Regulation of the Neuronal Serotoninergic Phenotype in Vitro: BDNF and CAMP Share Trk B-Dependent Mechanisms. J. Neurochem. 2002, 83, 1525–1528. [Google Scholar] [CrossRef]
- Craven, S.E.; Lim, K.C.; Ye, W.; Engel, J.D.; de Sauvage, F.; Rosenthal, A. Gata2 Specifies Serotonergic Neurons Downstream of Sonic Hedgehog. Development 2004, 131, 1165–1173. [Google Scholar] [CrossRef]
- Dixit, R.; Wilkinson, G.; Cancino, G.I.; Shaker, T.; Adnani, L.; Li, S.; Dennis, D.; Kurrasch, D.; Chan, J.A.; Olson, E.C.; et al. Neurog1 and Neurog2 Control Two Waves of Neuronal Differentiation in the Piriform Cortex. J. Neurosci. 2014, 34, 539–553. [Google Scholar] [CrossRef]
- Seifert, A.; Werheid, D.F.; Knapp, S.M.; Tobiasch, E. Role of Hox genes in stem cell differentiation. World J. Stem Cells 2015, 7, 583–595. [Google Scholar] [CrossRef]
- Haugas, M.; Tikker, L.; Achim, K.; Salminen, M.; Partanen, J. Gata2 and Gata3 Regulate the Differentiation of Serotonergic and Glutamatergic Neuron Subtypes of the Dorsal Raphe. Development 2016, 143, 4495–4508. [Google Scholar] [CrossRef]
- Shimojo, H.; Masaki, T.; Kageyama, R. The Neurog2-Tbr2 Axis Forms a Continuous Transition to the Neurogenic Gene Expression State in Neural Stem Cells. Dev. Cell 2024, 59, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Jurga, A.M.; Paleczna, M.; Kadluczka, J.; Kuter, K.Z. Beyond the GFAP-Astrocyte Protein Markers in the Brain. Biomolecules 2021, 11, 1361. [Google Scholar] [CrossRef]
- Teng, X.Y.; Hu, P.; Zhang, C.M.; Zhang, Q.X.; Yang, G.; Zang, Y.Y.; Liu, Z.X.; Chen, G.; Shi, Y.S. OPALIN is an LGI1 receptor promoting oligodendrocyte differentiation. Proc. Natl. Acad. Sci. USA. 2024, 121, e2403652121. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, W.; Yan, Q.; Ou, Y.; Long, Q.; Zhang, P. Biomarkers of mature neuronal differentiation and related diseases. Future Sci. OA 2024, 10, 2410146. [Google Scholar] [CrossRef]
- Rudy, B.; Fishell, G.; Lee, S.; Hjerling-Leffler, J. Three Groups of Interneurons Account for Nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 2011, 71, 45–61. [Google Scholar] [CrossRef]
- Bence, M.; Koller, J.; Sasvari-Szekely, M.; Keszler, G. Transcriptional modulation of monoaminergic neurotransmission genes by the histone deacetylase inhibitor trichostatin A in neuroblastoma cells. J. Neural Transm. 2012, 119, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yu, J.; Ding, J.; Xie, C.; Sun, L.; Rudenko, I.; Zheng, W.; Sastry, N.; Luo, J.; Rudow, G.; et al. Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. J. Clin. Investig. 2014, 124, 3032–3046. [Google Scholar] [CrossRef]
- Hendricks, T.; Francis, N.; Fyodorov, D.; Deneris, E.S. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 1999, 19, 10348–10356. [Google Scholar] [CrossRef]
- Liu, C.; Maejima, T.; Wyler, S.C.; Casadesus, G.; Herlitze, S.; Deneris, E.S. Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 2010, 13, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.S.; Peng, G.S.; Li, G.; Yang, S.; Wu, X.; Wang, C.C.; Wilson, B.; Lu, R.B.; Gean, P.W.; Chuang, D.M.; et al. Valproate Protects Dopaminergic Neurons in Midbrain Neuron/Glia Cultures by Stimulating the Release of Neurotrophic Factors from Astrocytes. Mol. Psychiatry 2006, 11, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Phi van, D.K.; Mühlbauer, E.; Phi-van, L. Histone Deacetylase HDAC1 Downregulates Transcription of the Serotonin Transporter (5-HTT) Gene in Tumor Cells. Biochim. Biophys. Acta 2015, 1849, 909–918. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Zhang, L.; Zhou, F.; Zhu, K.; Zhu, Q.; Liu, Q.; Liu, Y.; Jiang, L.; Ning, G.; et al. Protein Acetylation Derepresses Serotonin Synthesis to Potentiate Pancreatic Beta-Cell Function through HDAC1-PKA-Tph1 Signaling. Theranostics 2020, 10, 7351–7368. [Google Scholar] [CrossRef] [PubMed]
- Lavdas, A.A.; Blue, M.E.; Lincoln, J.; Parnavelas, J.G. Serotonin Promotes the Differentiation of Glutamate Neurons in Organotypic Slice Cultures of the Developing Cerebral Cortex. J. Neurosci. 1997, 17, 7872–7880. [Google Scholar] [CrossRef]
- Jiang, Y.; Hsieh, J. HDAC3 Controls Gap 2/Mitosis Progression in Adult Neural Stem/Progenitor Cells by Regulating CDK1 Levels. Proc. Natl. Acad. Sci. USA 2014, 111, 13541–13546. [Google Scholar] [CrossRef]
- Tang, T.; Zhang, Y.; Wang, Y.; Cai, Z.; Lu, Z.; Li, L.; Huang, R.; Hagelkruys, A.; Matthias, P.; Zhang, H.; et al. HDAC1 and HDAC2 Regulate Intermediate Progenitor Positioning to Safeguard Neocortical Development. Neuron 2019, 101, 1117–1133. [Google Scholar] [CrossRef]
- Douvaras, P.; Rusielewicz, T.; Kim, K.H.; Haines, J.D.; Casaccia, P.; Fossati, V. Epigenetic Modulation of Human Induced Pluripotent Stem Cell Differentiation to Oligodendrocytes. Int. J. Mol. Sci. 2016, 17, 614. [Google Scholar] [CrossRef]
- Borodinova, A.A.; Balaban, P.M. Epigenetic Regulation as a Basis for Long-Term Changes in the Nervous System: In Search of Specificity Mechanisms. Biochemistry 2020, 85, 994–1010. [Google Scholar] [CrossRef]
- Balasubramaniyan, V.; Boddeke, E.; Bakels, R.; Küst, B.; Kooistra, S.; Veneman, A.; Copray, S. Effects of Histone Deacetylation Inhibition on Neuronal Differentiation of Embryonic Mouse Neural Stem Cells. Neuroscience 2006, 143, 939–951. [Google Scholar] [CrossRef]
- Darlington, P.J.; Goldman, J.S.; Cui, Q.L.; Antel, J.P.; Kennedy, T.E. Widespread immunoreactivity for neuronal nuclei in cultured human and rodent astrocytes. J. Neurochem. 2008, 104, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.R.; Greenamyre, J.T. NeuN is not a reliable marker of dopamine neurons in rat substantia nigra. Neurosci. Lett. 2009, 464, 14–17. [Google Scholar] [CrossRef]
- Seiberlich, V.; Bauer, N.G.; Schwarz, L.; Ffrench-Constant, C.; Goldbaum, O.; Richter-Landsberg, C. Downregulation of the microtubule associated protein tau impairs process outgrowth and myelin basic protein mRNA transport in oligodendrocytes. Glia 2015, 63, 1621–1635. [Google Scholar] [CrossRef]
- Krueger, K.C.; Deneris, E.S. Serotonergic Transcription of Human FEV Reveals Direct GATA Factor Interactions and Fate of Pet-1-Deficient Serotonin Neuron Precursors. J. Neurosci. 2008, 28, 12748–12758. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, E.J.; Mesman, S.; de Munnik, W.A.; Smidt, M.P. LMX1B Is Part of a Transcriptional Complex with PSPC1 and PSF. PLoS ONE 2013, 8, e53122. [Google Scholar] [CrossRef] [PubMed]
- Rossin, F.; Ciccosanti, F.; D’Eletto, M.; Occhigrossi, L.; Fimia, G.M.; Piacentini, M. Type 2 Transglutaminase in the Nucleus: The New Epigenetic Face of a Cytoplasmic Enzyme. Cell Mol. Life Sci. 2023, 80, 52. [Google Scholar] [CrossRef]
- Ballestar, E.; Abad, C.; Franco, L. Core histones are glutaminyl substrates for tissue transglutaminase. J. Biol. Chem. 1996, 271, 18817–18824. [Google Scholar] [CrossRef]
- Bader, M. Serotonylation: Serotonin Signaling and Epigenetics. Front. Mol. Neurosci. 2019, 12, 288. [Google Scholar] [CrossRef]
- Zhao, S.; Chuh, K.N.; Zhang, B.; Dul, B.E.; Thompson, R.E.; Farrelly, L.A.; Liu, X.; Xu, N.; Xue, Y.; Roeder, R.G.; et al. Histone H3Q5 Serotonylation Stabilizes H3K4 Methylation and Potentiates Its Readout. Proc. Natl. Acad. Sci. USA 2021, 118, e2016742118. [Google Scholar] [CrossRef]
- Pereira, M.; Birtele, M.; Rylander Ottosson, D. Direct Reprogramming into Interneurons: Potential for Brain Repair. Cell Mol. Life Sci. 2019, 76, 3953–3967. [Google Scholar] [CrossRef]
- Raina, A.; Mahajani, S.; Bähr, M.; Kügler, S. Neuronal Trans-Differentiation by Transcription Factors Ascl1 and Nurr1: Induction of a Dopaminergic Neurotransmitter Phenotype in Cortical GABAergic Neurons. Mol. Neurobiol. 2020, 57, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Rivetti di Val Cervo, P.; Romanov, R.A.; Spigolon, G.; Masini, D.; Martín-Montañez, E.; Toledo, E.M.; La Manno, G.; Feyder, M.; Pifl, C.; Ng, Y.H.; et al. Induction of Functional Dopamine Neurons from Human Astrocytes in Vitro and Mouse Astrocytes in a Parkinson’s Disease Model. Nat. Biotechnol. 2017, 35, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Lukasak, B.J.; Mitchener, M.M.; Kong, L.; Dul, B.E.; Lazarus, C.D.; Ramakrishnan, A.; Ni, J.; Shen, L.; Maze, I.; Muir, T.W. TGM2-Mediated Histone Transglutamination Is Dictated by Steric Accessibility. Proc. Natl. Acad. Sci. USA 2022, 119, e2208672119. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Tsai, L.H. The potential of HDAC inhibitors as cognitive enhancers. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 311–330. [Google Scholar] [CrossRef]
- D’Mello, S.R. Histone deacetylase-3: Friend and foe of the brain. Exp. Biol. Med. 2020, 245, 1130–1141. [Google Scholar] [CrossRef]
- Kim, H.J.; Leeds, P.; Chuang, D.M. The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. J. Neurochem. 2009, 110, 1226–1240. [Google Scholar] [CrossRef]
- Kiyooka, M.; Shimizu, Y.; Ohshima, T. Histone deacetylase inhibition promotes regenerative neurogenesis after stab wound injury in the adult zebrafish optic tectum. Biochem. Biophys. Res. Commun. 2020, 529, 366–371. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borodinova, A.A.; Leontovich, Y.A.; Beletskiy, A.P.; Revishchin, A.V.; Pavlova, G.V.; Balaban, P.M. Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron–Glia Cultures Involves Histone Serotonylation. Cells 2025, 14, 905. https://doi.org/10.3390/cells14120905
Borodinova AA, Leontovich YA, Beletskiy AP, Revishchin AV, Pavlova GV, Balaban PM. Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron–Glia Cultures Involves Histone Serotonylation. Cells. 2025; 14(12):905. https://doi.org/10.3390/cells14120905
Chicago/Turabian StyleBorodinova, Anastasia A., Yulia A. Leontovich, Alexander P. Beletskiy, Alexander V. Revishchin, Galina V. Pavlova, and Pavel M. Balaban. 2025. "Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron–Glia Cultures Involves Histone Serotonylation" Cells 14, no. 12: 905. https://doi.org/10.3390/cells14120905
APA StyleBorodinova, A. A., Leontovich, Y. A., Beletskiy, A. P., Revishchin, A. V., Pavlova, G. V., & Balaban, P. M. (2025). Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron–Glia Cultures Involves Histone Serotonylation. Cells, 14(12), 905. https://doi.org/10.3390/cells14120905





