Non-Redundant Essential Roles of Proteasomal Ubiquitin Receptors Rpn10 and Rpn13 in Germ Cell Formation and Fertility
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Collection and Immunofluorescence or Immunohistochemical Staining
2.3. Immunoblotting
2.4. Hematoxylin and Eosin (H & E) Staining
2.5. Proteasome Activity Assay, Spermatocyte Spread and Immunolabeling
2.6. TUNEL Assay
2.7. Quantification and Statistical Analysis
3. Results
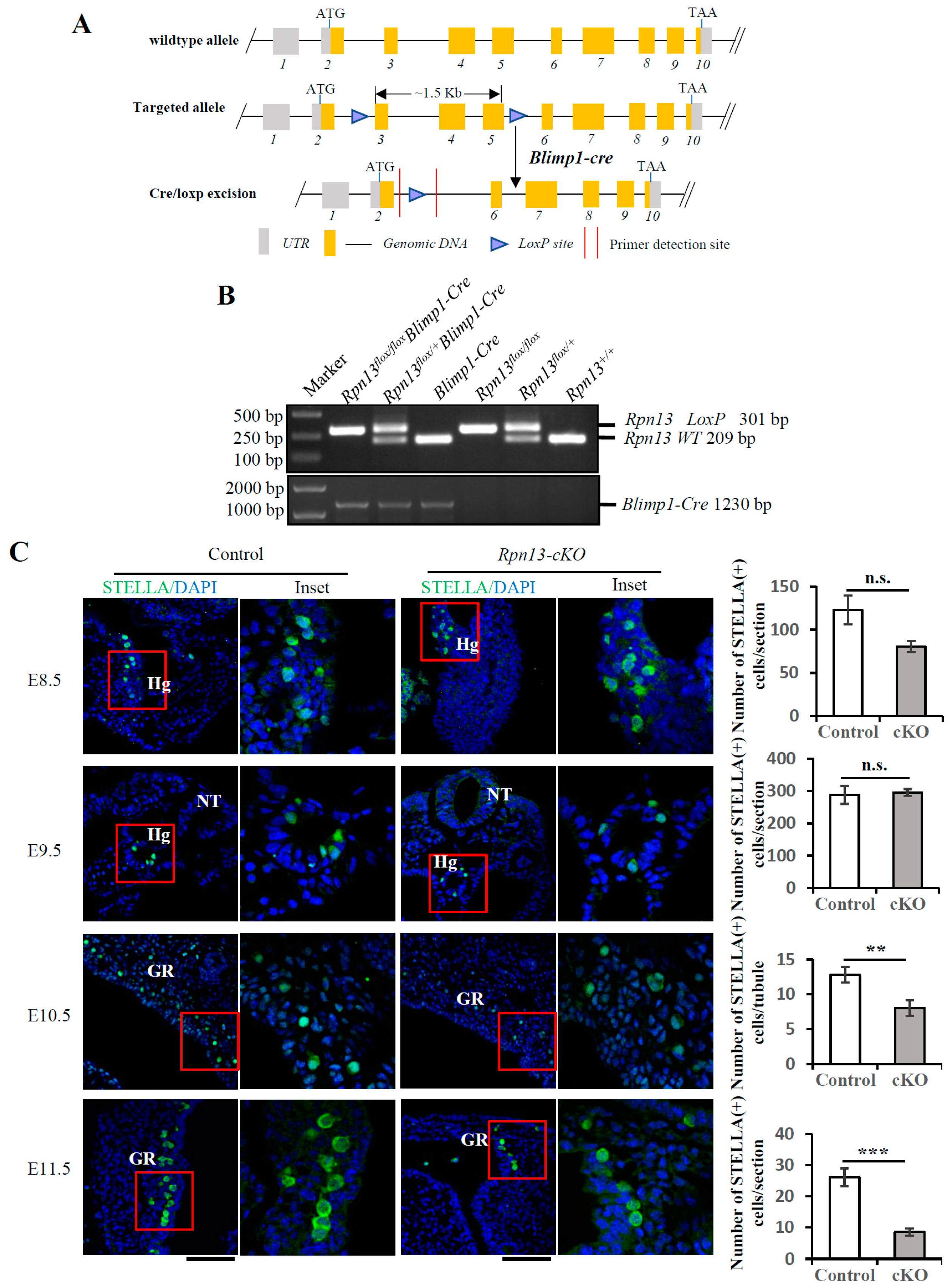
3.1. Conditional Deletion of Rpn13 in PGCs Reduces the PGC Number at E10.5
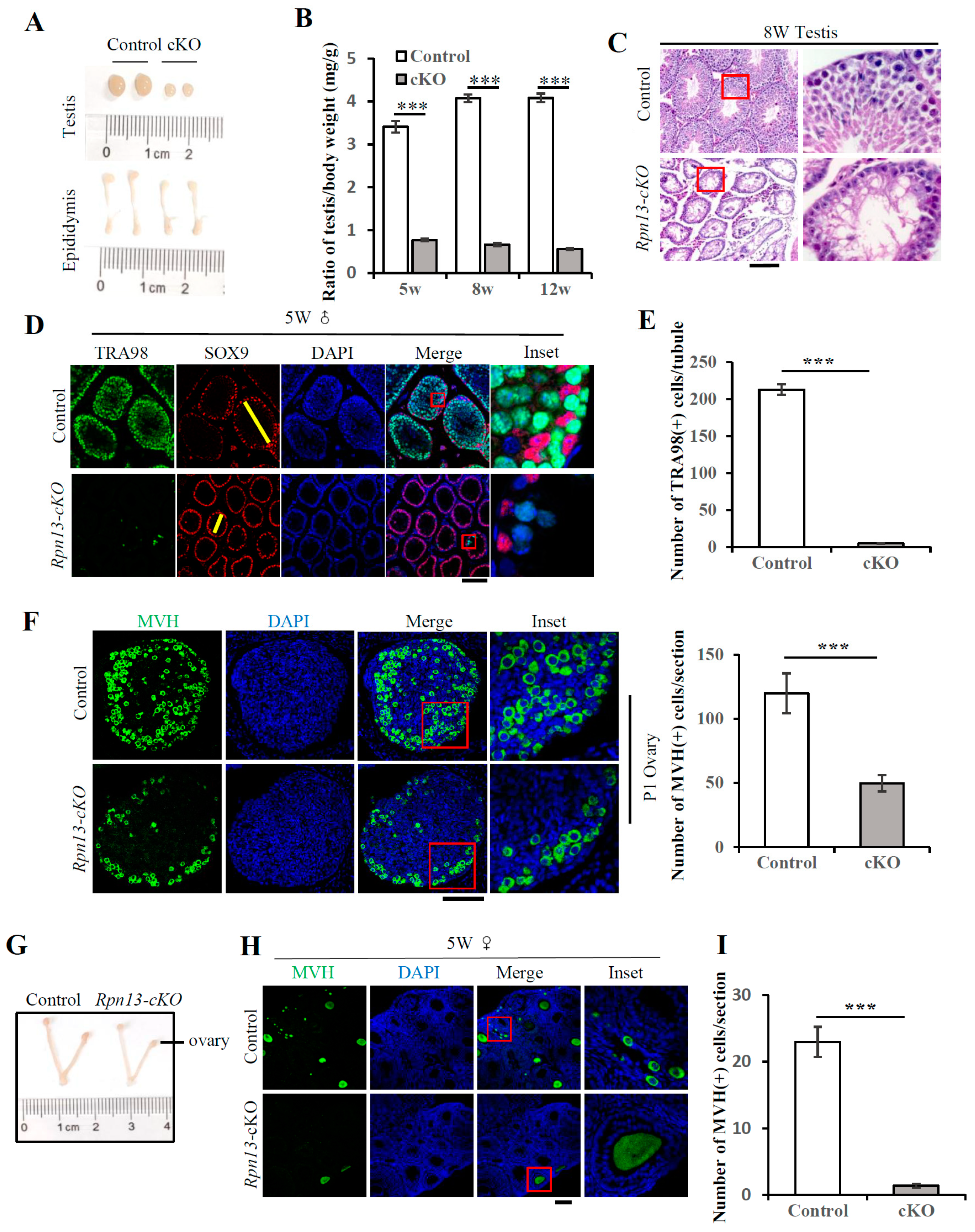
3.2. Conditional Deletion of Rpn13 Disrupts Germ Cell Development and Leads to Infertility in Both Males and Females
3.3. Deletion of Rpn13 Blocks Meiosis of Spermatocytes at Zygotene Stage During Prophase I
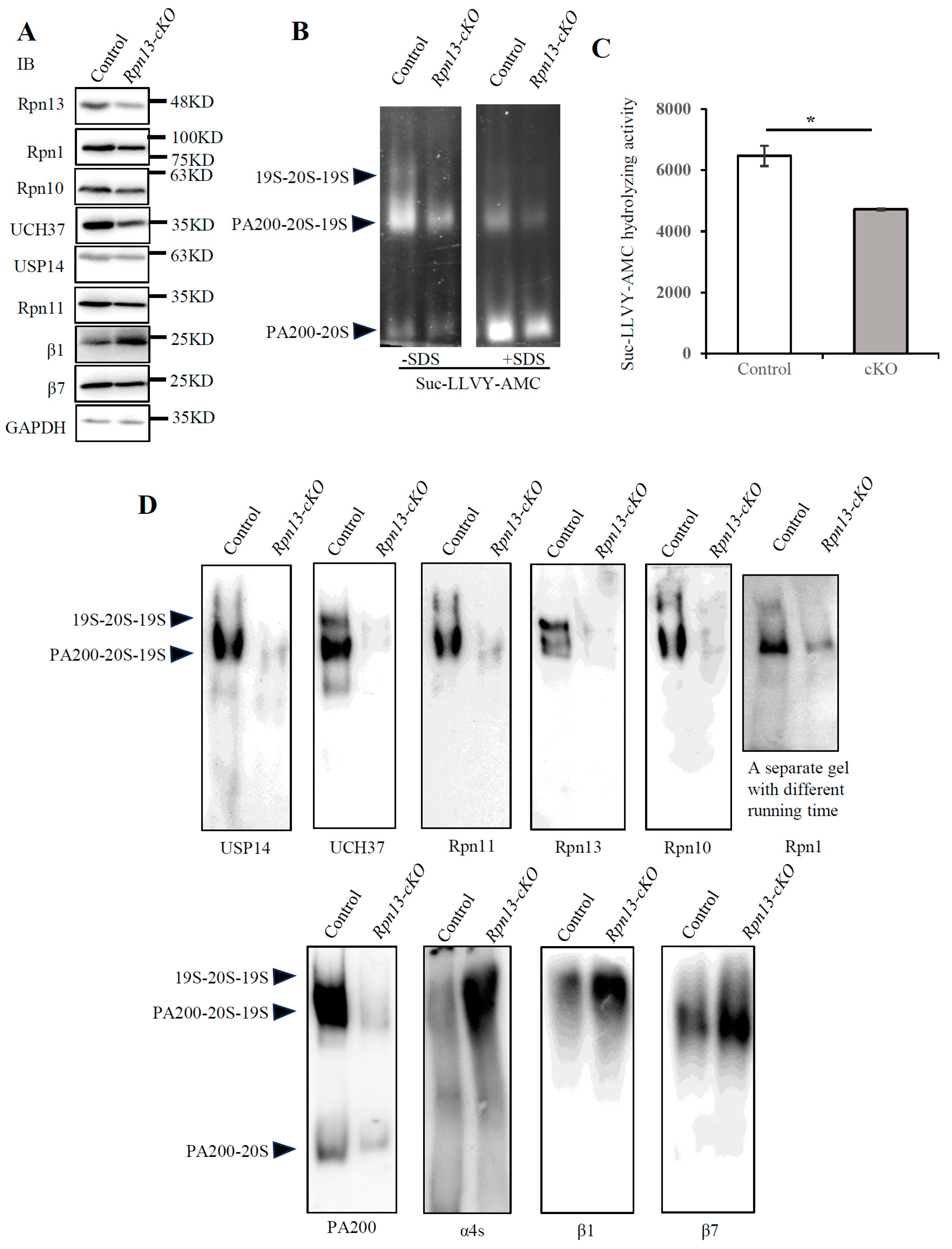
3.4. Conditional Deletion of Rpn13 Markedly Reduces Activity of the 26S Proteasome
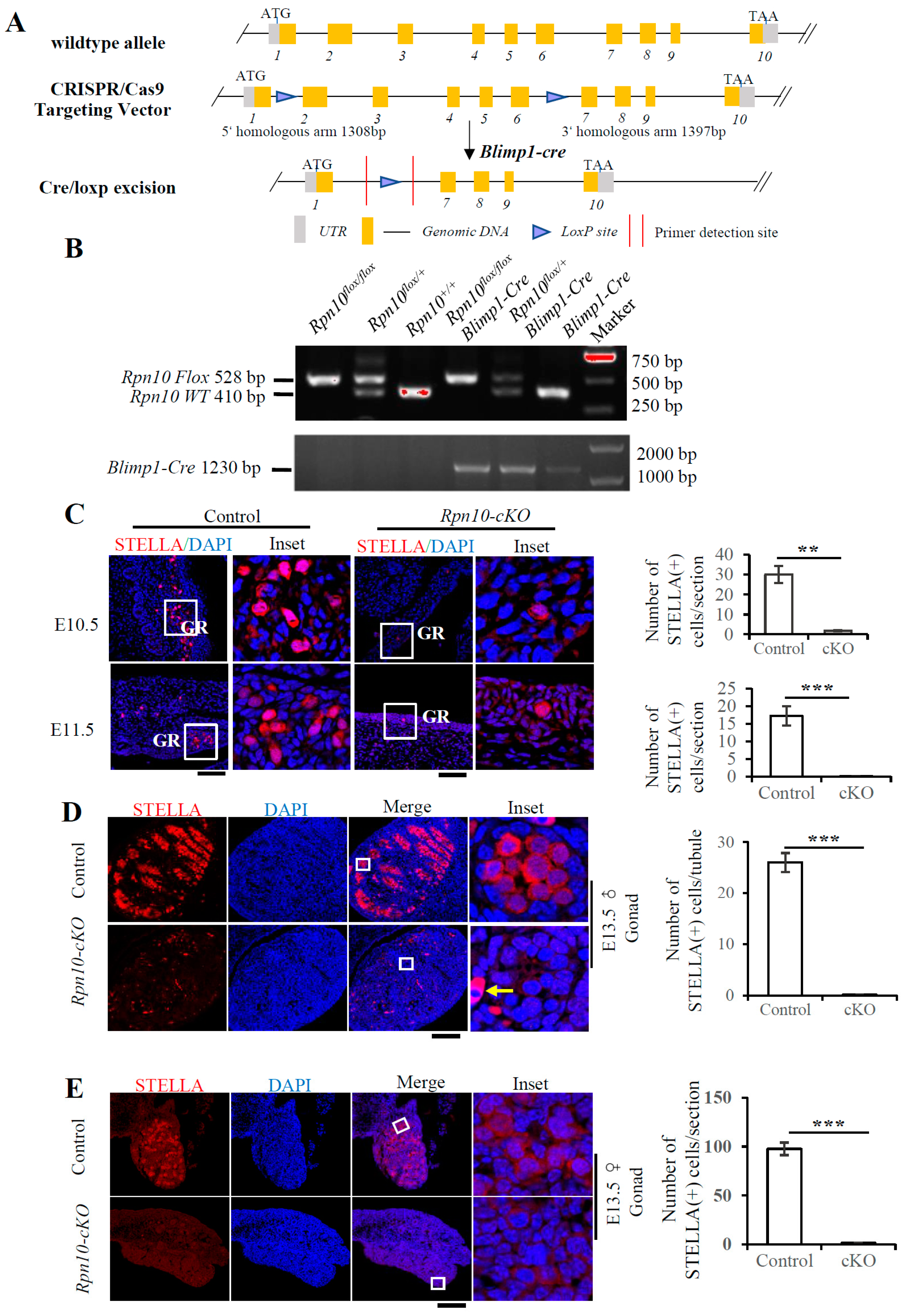
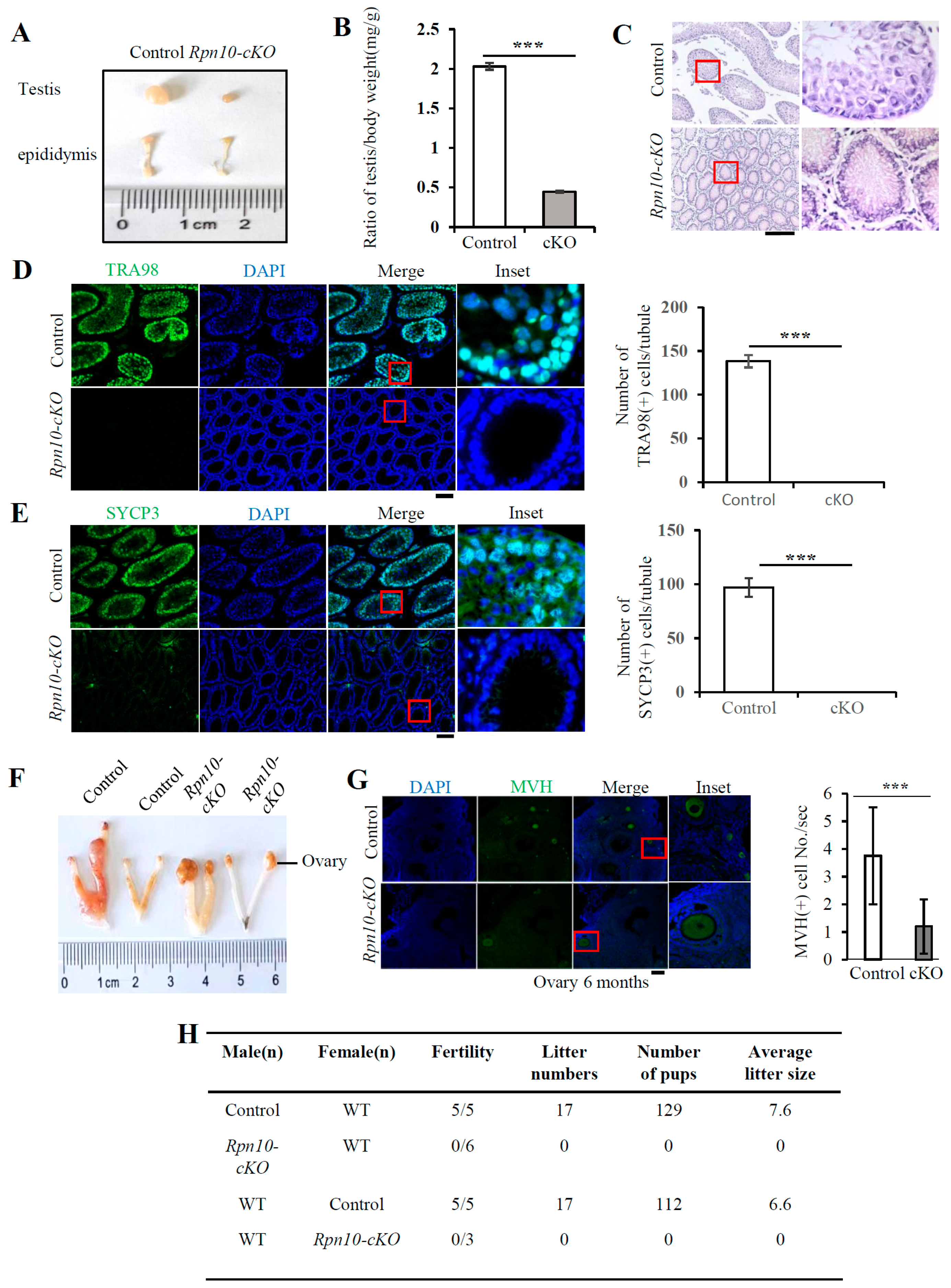
3.5. Deletion of Rpn10 in PGCs Sharply Reduces PGC Migration and Leads to Infertility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PGC | Primordial gem cell |
| GVBD | Breakdown of germinal vesicle |
| CP | Core particle |
| RP | Regulatory particle |
| cKO | Conditional knockout |
| WT | Wild type |
| MVH | Mouse vasa homolog |
| H & E | Hematoxylin and eosin |
| P | Postnatal day |
| PLZF | Promyelocytic leukemia zinc-finger protein |
| SYCP | Synaptonemal complex protein |
| AMC | Amino-4-methylcoumarin |
References
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Western, P.S.; Miles, D.C.; van den Bergen, J.A.; Burton, M.; Sinclair, A.H. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 2008, 26, 339–347. [Google Scholar] [CrossRef]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front. Cell Dev. Biol. 2021, 9, 654028. [Google Scholar] [CrossRef] [PubMed]
- de Rooij, D.G.; Russell, L.D. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000, 21, 776–798. [Google Scholar] [CrossRef]
- McLaren, A. Meiosis and differentiation of mouse germ cells. Symp. Soc. Exp. Biol. 1984, 38, 7–23. [Google Scholar]
- Jiang, T.-X.; Zhao, M.; Qiu, X.-B. Substrate receptors of proteasomes. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1765–1777. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.-X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.-Y.; Ji, D.-Y.; Wang, G.-F.; Zhu, Q.-Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef]
- Hamazaki, J.; Hirayama, S.; Murata, S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 2015, 11, e1005401. [Google Scholar] [CrossRef]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, Q.; Ehlinger, A.; Randles, L.; Lary, J.W.; Kang, Y.; Haririnia, A.; Storaska, A.J.; Cole, J.L.; Fushman, D.; et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol. Cell 2009, 35, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.-H.; Shi, Y.; Zhang, N.; de Poot, S.A.H.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef]
- Al-Shami, A.; Jhaver, K.G.; Vogel, P.; Wilkins, C.; Humphries, J.; Davis, J.J.; Xu, N.; Potter, D.G.; Gerhardt, B.; Mullinax, R.; et al. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS ONE 2010, 5, e13654. [Google Scholar] [CrossRef]
- Hamazaki, J.; Sasaki, K.; Kawahara, H.; Hisanaga, S.-I.; Tanaka, K.; Murata, S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell Biol. 2007, 27, 6629–6638. [Google Scholar] [CrossRef]
- Qiu, X.B.; Ouyang, S.Y.; Li, C.J.; Miao, S.; Wang, L.; Goldberg, A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006, 25, 5742–5753. [Google Scholar] [CrossRef] [PubMed]
- Hamazaki, J.; Iemura, S.; Natsume, T.; Yashiroda, H.; Tanaka, K.; Murata, S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006, 25, 4524–4536. [Google Scholar] [CrossRef]
- Yao, T.; Song, L.; Xu, W.; DeMartino, G.N.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Cohen, R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006, 8, 994–1002. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Jiang, T.-X.; Chen, L.-B.; Zhou, W.; Liu, Y.; Gao, F.; Qiu, X.-B. Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J. Biol. Chem. 2021, 296, 100130. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Bikoff, E.K.; Morgan, M.A.; Robertson, E.J. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 2009, 19, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Saitou, M.; Barton, S.C.; Surani, M.A. A molecular programme for the specification of germ cell fate in mice. Nature 2002, 418, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Hsu, T.-H.; Yen, P.H. Mouse sperm acquire a new structure on the apical hook during epididymal maturation. Asian J. Androl. 2013, 15, 523–528. [Google Scholar] [CrossRef]
- Teves, M.E.; Roldan, E.R.S. Sperm bauplan and function and underlying processes of sperm formation and selection. Physiol. Rev. 2022, 102, 7–60. [Google Scholar] [CrossRef]
- Carmell, M.A.; Dokshin, G.A.; Skaletsky, H.; Hu, Y.-C.; van Wolfswinkel, J.C.; Igarashi, K.J.; Bellott, D.W.; Nefedov, M.; Reddien, P.W.; Enders, G.C.; et al. A widely employed germ cell marker is an ancient disordered protein with reproductive functions in diverse eukaryotes. eLife 2016, 5, e19993. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, T.; Shirley, L.; John, G.B.; Castrillon, D.H. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007, 45, 413–417. [Google Scholar] [CrossRef]
- Oatley, J.M.; Brinster, R.L. Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 2008, 24, 263–286. [Google Scholar] [CrossRef]
- He, Z.; Kokkinaki, M.; Dym, M. Signaling molecules and pathways regulating the fate of spermatogonial stem cells. Microsc. Res. Tech. 2009, 72, 586–595. [Google Scholar] [CrossRef]
- Hermo, L.; Pelletier, R.M.; Cyr, D.G.; Smith, C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: Background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 2010, 73, 241–278. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, W.-Y.; Zhang, Y.; Jiang, T.-X.; Qiu, X.-B. Non-Redundant Essential Roles of Proteasomal Ubiquitin Receptors Rpn10 and Rpn13 in Germ Cell Formation and Fertility. Cells 2025, 14, 696. https://doi.org/10.3390/cells14100696
Yue W-Y, Zhang Y, Jiang T-X, Qiu X-B. Non-Redundant Essential Roles of Proteasomal Ubiquitin Receptors Rpn10 and Rpn13 in Germ Cell Formation and Fertility. Cells. 2025; 14(10):696. https://doi.org/10.3390/cells14100696
Chicago/Turabian StyleYue, Wan-Yu, Yi Zhang, Tian-Xia Jiang, and Xiao-Bo Qiu. 2025. "Non-Redundant Essential Roles of Proteasomal Ubiquitin Receptors Rpn10 and Rpn13 in Germ Cell Formation and Fertility" Cells 14, no. 10: 696. https://doi.org/10.3390/cells14100696
APA StyleYue, W.-Y., Zhang, Y., Jiang, T.-X., & Qiu, X.-B. (2025). Non-Redundant Essential Roles of Proteasomal Ubiquitin Receptors Rpn10 and Rpn13 in Germ Cell Formation and Fertility. Cells, 14(10), 696. https://doi.org/10.3390/cells14100696






