Comparative Analysis of FCGR Gene Polymorphism in Pulmonary Sarcoidosis and Tuberculosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of the Studied Groups
2.2. Patients with Sarcoidosis
2.3. Patients with Tuberculosis
2.4. Healthy Control
2.5. Sample Collection and DNA Isolation
2.6. Polymerase Chain Reaction with Sequence-Specific Primers
2.7. Real-Time Polymerase Chain Reaction
2.8. Electrophoresis and DNA Staining
2.9. Statistical Analysis
3. Results
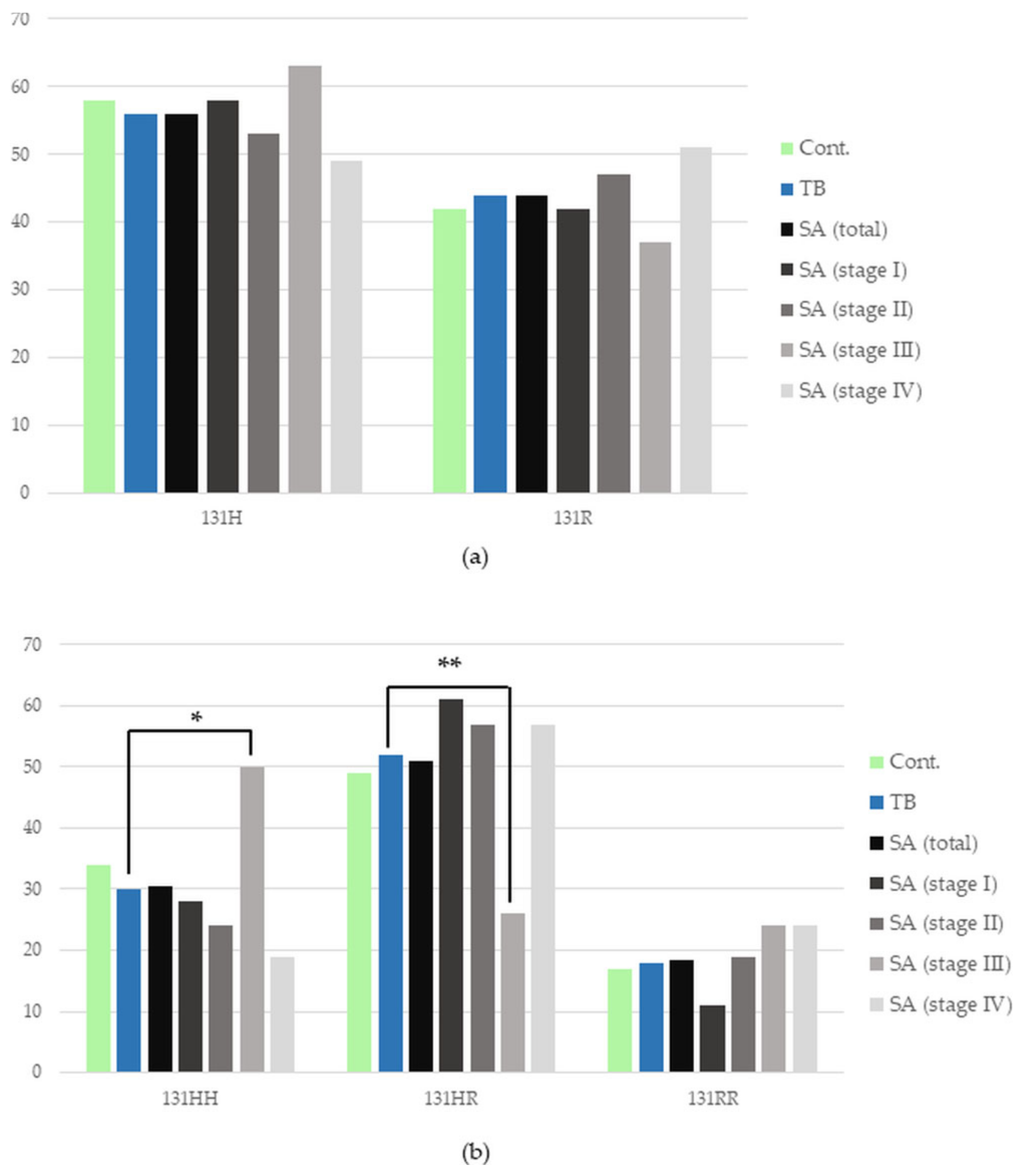
3.1. FCGR2A Gene Polymorphism
3.2. FCGR2B Gene Polymorphism
3.3. FCGR2C Gene Polymorphism
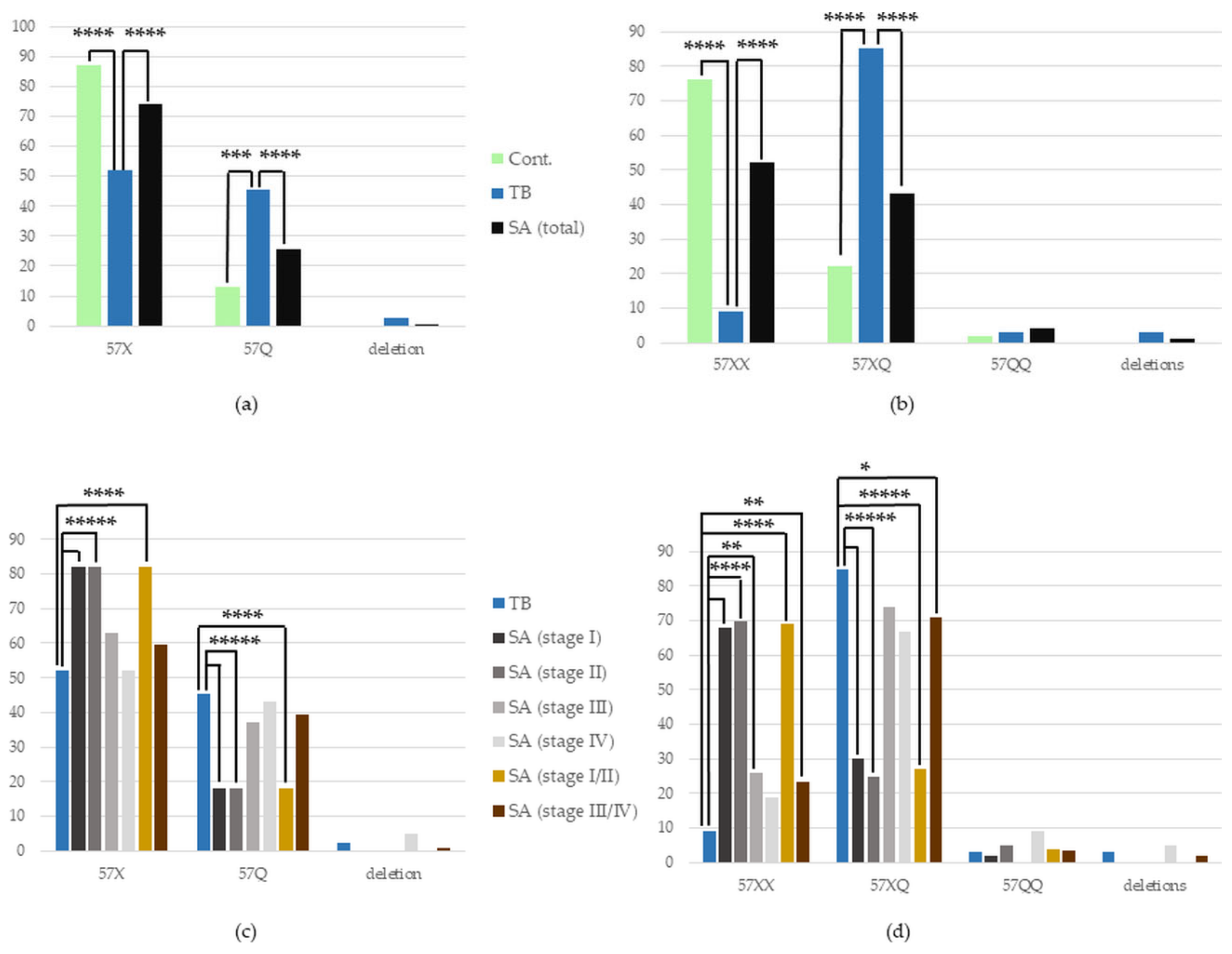
3.3.1. FCGR2C Polymorphism in the Whole SA Group vs. TB, and in TB vs. Control
3.3.2. FCGR2C Polymorphism in Certain Stages of SA vs. TB
3.4. FCGR3A Gene Polymorphism
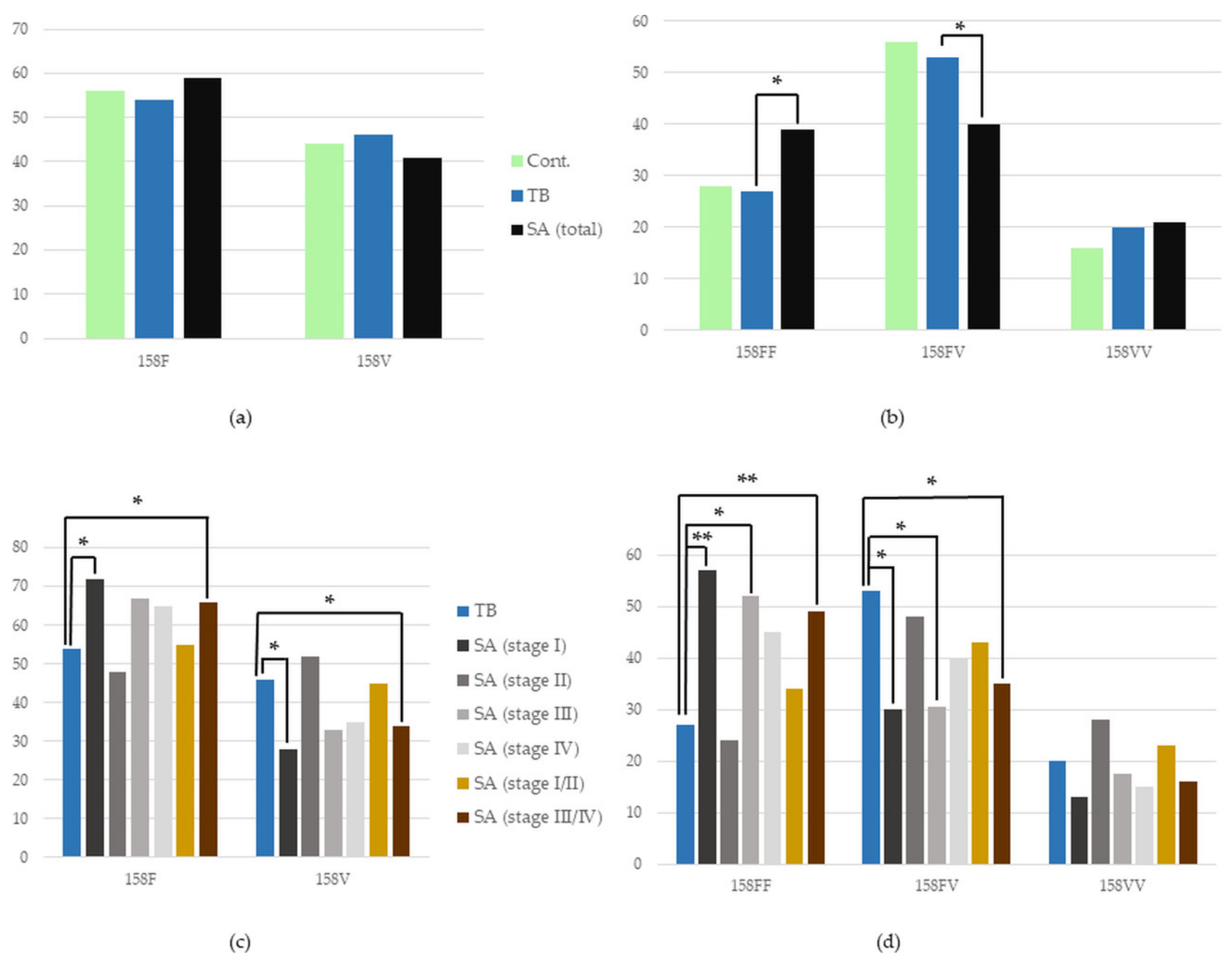
3.4.1. FCGR3A Polymorphism in the Whole SA Group vs. TB, and in TB vs. Control
3.4.2. FCGR3A Polymorphism in Certain Stages of SA vs. TB
3.5. FCGR3B Gene Polymorphism
3.5.1. FCGR3B Polymorphism in the Whole SA Group vs. TB, and in TB vs. Control
3.5.2. FCGR3B Polymorphism in Certain Stages of SA vs. TB
4. Discussion
| Location of Receptor | Receptor | Location of aa Change in the Receptor Protein | Gene | Gene Variant Responsible for | Reference | |
|---|---|---|---|---|---|---|
| Higher ICs Binding | Lower ICs Binding | |||||
| Mo, MΦ, N, less often on E, B, MC | FcγRIIa | second extracellular domain | FCGR2A | 131H | 131R | [14,35,36,37,38] |
| Almost all types of leukocytes (including B cells and memory CD8+ T cells), minority of NK; liver and aorta cells | FcγRIIb | transmembrane domain | FCGR2B | 232T * | 232I * | [14,35,36,38,39,40,41] |
| NK, less often on Mo, MΦ, N | FcγRIIc | first extracellular domain | FCGR2C | 57Q | 57X (truncated, nonfunctional receptor) | [14,35,36,38,42] |
| NK, Mo, MΦ, N, T γδ cells, DC, MC, E | FcγRIIIa | second extracellular domain | FCGR3A | 158V | 158F | [14,35,36,37,38] |
| N, less often on B, after induction on E | FcγRIIIb | first extracellular domain | FCGR3B | NA1 | NA2, SH | [14,35,36,38,43] |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Statement on Sarcoidosis. Joint Statement of the American Thoracic Society (ATS), the European Respiratory Society (ERS) and the World Association of Sarcoidosis and Other Granulomatous Disorders (WASOG) adopted by the ATS Board of Directors and by the ERS Executive Committee, February 1999. Am. J. Respir. Crit. Care Med. 1999, 160, 736–755. [CrossRef]
- Sève, P.; Pacheco, Y.; Durupt, F.; Jamilloux, Y.; Gerfaud-Valentin, M.; Isaac, S.; Boussel, L.; Calender, A.; Androdias, G.; Valeyre, D.; et al. Sarcoidosis: A Clinical Overview from Symptoms to Diagnosis. Cells 2021, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Dubaniewicz, A.; Zimmermann, A.; Smigielska, M.; Dubaniewicz-Wybieralska, M.; Moszkowska, G.; Wysocka, J.; Adamczyk-Bak, K.; Slominski, J.M.; Deeg, P. Sarcoidosis and tuberculosis: A connection to the human leukocyte antigen system. Adv. Exp. Med. Biol. 2013, 756, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Schupp, J.C.; Freitag-Wolf, S.; Bargagli, E.; Mihailović-Vučinić, V.; Rottoli, P.; Grubanovic, A.; Müller, A.; Jochens, A.; Tittmann, L.; Schnerch, J.; et al. Phenotypes of organ involvement in sarcoidosis. Eur. Respir. J. 2018, 51, 1700991. [Google Scholar] [CrossRef] [PubMed]
- Dubaniewicz, A.; Zimmermann, A.; Dudziak, M.; Typiak, M.; Skotarczak, M. Tuberculosis in the course of sarcoidosis treatment: Is genotyping necessary for personalized therapy? Expert Rev. Clin. Immunol. 2013, 9, 349–360. [Google Scholar] [CrossRef]
- Furin, J.; Cox, H.; Pai, M. Tuberculosis. Lancet 2019, 393, 1642–1656. [Google Scholar] [CrossRef]
- Dubaniewicz, A.; Dubaniewicz-Wybieralska, M.; Sternau, A.; Zwolska, Z.; Izycka-Swieszewska, E.; Augustynowicz-Kopec, E.; Skokowski, J.; Singh, M.; Zimnoch, L. Mycobacterium tuberculosis complex and mycobacterial heat shock proteins in lymph node tissue from patients with pulmonary sarcoidosis. J. Clin. Microbiol. 2006, 44, 3448–3451. [Google Scholar] [CrossRef]
- Dubaniewicz, A. Mycobacterium tuberculosis heat shock proteins and autoimmunity in sarcoidosis. Autoimmun. Rev. 2010, 9, 419–424. [Google Scholar] [CrossRef]
- Dubaniewicz, A. Microbial and human heat shock proteins as ’danger signals’ in sarcoidosis. Hum. Immunol. 2013, 74, 1550–1558. [Google Scholar] [CrossRef]
- Dubaniewicz, A.; Holownia, A.; Kalinowski, L.; Wybieralska, M.; Dobrucki, I.T.; Singh, M. Is mycobacterial heat shock protein 16 kDa, a marker of the dormant stage of Mycobacterium tuberculosis, a sarcoid antigen? Hum. Immunol. 2013, 74, 45–51. [Google Scholar] [CrossRef]
- Dubaniewicz, A.; Typiak, M.; Wybieralska, M.; Szadurska, M.; Nowakowski, S.; Staniewicz-Panasik, A.; Rogoza, K.; Sternau, A.; Deeg, P.; Trzonkowski, P. Changed phagocytic activity and pattern of Fcγ and complement receptors on blood monocytes in sarcoidosis. Hum. Immunol. 2012, 73, 788–794. [Google Scholar] [CrossRef]
- Typiak, M.; Rębała, K.; Dudziak, M.; Słomiński, J.M.; Dubaniewicz, A. Polymorphism of FCGR2A, FCGR2C, and FCGR3B Genes in the Pathogenesis of Sarcoidosis. Adv. Exp. Med. Biol. 2016, 905, 57–68. [Google Scholar] [CrossRef]
- Typiak, M.J.; Rębała, K.; Dudziak, M.; Dubaniewicz, A. Polymorphism of FCGR3A gene in sarcoidosis. Hum. Immunol. 2014, 75, 283–288. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]
- Iannuzzi, M.C.; Rybicki, B.A.; Teirstein, A.S. Sarcoidosis. N. Engl. J. Med. 2007, 357, 2153–2165. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Nurnberger, J.I., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991, 19, 5444. [Google Scholar] [CrossRef]
- Edberg, J.C.; Langefeld, C.D.; Wu, J.; Moser, K.L.; Kaufman, K.M.; Kelly, J.; Bansal, V.; Brown, W.M.; Salmon, J.E.; Rich, S.S.; et al. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum. 2002, 46, 2132–2140. [Google Scholar] [CrossRef]
- Su, K.; Wu, J.; Edberg, J.C.; McKenzie, S.E.; Kimberly, R.P. Genomic organization of classical human low-affinity Fcgamma receptor genes. Genes Immun. 2002, 3 (Suppl. 1), S51–S56. [Google Scholar] [CrossRef]
- Siriboonrit, U.; Tsuchiya, N.; Sirikong, M.; Kyogoku, C.; Bejrachandra, S.; Suthipinittharm, P.; Luangtrakool, K.; Srinak, D.; Thongpradit, R.; Fujiwara, K.; et al. Association of Fcgamma receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens 2003, 61, 374–383. [Google Scholar] [CrossRef]
- Van Den Berg, L.; Myhr, K.M.; Kluge, B.; Vedeler, C.A. Fcgamma receptor polymorphisms in populations in Ethiopia and Norway. Immunology 2001, 104, 87–91. [Google Scholar] [CrossRef]
- Fahsold, R.; Hoffmeyer, S.; Mischung, C.; Gille, C.; Ehlers, C.; Kücükceylan, N.; Abdel-Nour, M.; Gewies, A.; Peters, H.; Kaufmann, D.; et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 2000, 66, 790–818. [Google Scholar] [CrossRef] [PubMed]
- VassarStats. Vassar Stats Program; Lowry, R., Ed.; Vassar College: Poughkeepsie, NY, USA, 2022; Volume 2022, Available online: http://www.vassarstats.net/ (accessed on 23 February 2023).
- OEGE Software Organization. Odds Ratio Calculator. 2021. Available online: www.oege.org/software/orcalc.html (accessed on 15 December 2021).
- Rodriguez, S.; Gaunt, T.R.; Day, I.N. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009, 169, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Dubaniewicz, A.; Wybieralska, M.; Rogoza, K.; Typiak, M.; Sternau, A.; Slominski, J.M.; Trzonkowski, P. Phagocytosis by blood monocytes in differentiation between sarcoidosis and tuberculosis. Eur. Respir. J. 2012, 40, P2635. [Google Scholar]
- Dubaniewicz, A.; Rękawiecki, B.; Piprek, M.; Skotarczak, M.; Dubaniewicz-Wybieralska, M. Monocyte/neutrophil phagocytic activity ratio in differentiating tuberculosis from sarcoidosis. Eur. Respir. J. 2019, 54, PA1378. [Google Scholar] [CrossRef]
- Dubaniewicz, A.; Trzonkowski, P.; Dubaniewicz-Wybieralska, M.; Singh, M.; Myśliwski, A. Comparative analysis of mycobacterial heat shock proteins-induced apoptosis of peripheral blood mononuclear cells in sarcoidosis and tuberculosis. J. Clin. Immunol. 2006, 26, 243–250. [Google Scholar] [CrossRef]
- Dubaniewicz, A. Frequency of occurrence of circulating immune complexes in sera of patients with pulmonary sarcoidosis; preliminary report. Vestnik MCDC 2004, 3, 108–113. [Google Scholar]
- Dubaniewicz, A. Mycobacterial Heat Shock Proteins in Sarcoidosis and Tuberculosis. Int. J. Mol. Sci. 2023, 24, 5084. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Gordan, S.; Lux, A. FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015, 36, 325–336. [Google Scholar] [CrossRef]
- Sadki, K.; Lamsyah, H.; Rueda, B.; Akil, E.; Sadak, A.; Martin, J.; El Aouad, R. Analysis of MIF, FCGR2A and FCGR3A gene polymorphisms with susceptibility to pulmonary tuberculosis in Moroccan population. J. Genet. Genom. 2010, 37, 257–264. [Google Scholar] [CrossRef]
- Allen, M.; Bailey, C.; Cahatol, I.; Dodge, L.; Yim, J.; Kassissa, C.; Luong, J.; Kasko, S.; Pandya, S.; Venketaraman, V. Mechanisms of Control of Mycobacterium tuberculosis by NK Cells: Role of Glutathione. Front. Immunol. 2015, 6, 508. [Google Scholar] [CrossRef]
- Hilda, J.N.; Das, S.; Tripathy, S.P.; Hanna, L.E. Role of neutrophils in tuberculosis: A bird’s eye view. Innate Immun. 2020, 26, 240–247. [Google Scholar] [CrossRef]
- Typiak, M.; Rębała, K.; Haraś, A.; Skotarczak, M.; Słomiński, J.M.; Dubaniewicz, A. Copy number variation of FCGR genes in etiopathogenesis of sarcoidosis. PLoS ONE 2017, 12, e0177194. [Google Scholar] [CrossRef]
- Bournazos, S.; Woof, J.M.; Hart, S.P.; Dransfield, I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clin. Exp. Immunol. 2009, 157, 244–254. [Google Scholar] [CrossRef]
- Li, X.; Ptacek, T.S.; Brown, E.E.; Edberg, J.C. Fcgamma receptors: Structure, function and role as genetic risk factors in SLE. Genes Immun. 2009, 10, 380–389. [Google Scholar] [CrossRef]
- Spector, W.G.; Heesom, N. The production of granulomata by antigen-antibody complexes. J. Pathol. 1969, 98, 31–39. [Google Scholar] [CrossRef]
- Hogarth, P.M.; Pietersz, G.A. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug. Discov. 2012, 11, 311–331. [Google Scholar] [CrossRef]
- Takai, T. Fc receptors and their role in immune regulation and autoimmunity. J. Clin. Immunol. 2005, 25, 1–18. [Google Scholar] [CrossRef]
- Anderson, C.L.; Ganesan, L.P.; Robinson, J.M. The biology of the classical Fcγ receptors in non-hematopoietic cells. Immunol. Rev. 2015, 268, 236–240. [Google Scholar] [CrossRef]
- Starbeck-Miller, G.R.; Badovinac, V.P.; Barber, D.L.; Harty, J.T. Cutting edge: Expression of FcγRIIB tempers memory CD8 T cell function in vivo. J. Immunol. 2014, 192, 35–39. [Google Scholar] [CrossRef]
- Breunis, W.B.; van Mirre, E.; Bruin, M.; Geissler, J.; de Boer, M.; Peters, M.; Roos, D.; de Haas, M.; Koene, H.R.; Kuijpers, T.W. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood 2008, 111, 1029–1038. [Google Scholar] [CrossRef]
- Adu, B.; Dodoo, D.; Adukpo, S.; Hedley, P.L.; Arthur, F.K.; Gerds, T.A.; Larsen, S.O.; Christiansen, M.; Theisen, M. Fc γ receptor IIIB (FcγRIIIB) polymorphisms are associated with clinical malaria in Ghanaian children. PLoS ONE 2012, 7, e46197. [Google Scholar] [CrossRef] [PubMed]

| SA n = 154 (%) | TB n = 179 (%) | C n = 148 (%) | ||
|---|---|---|---|---|
| Age (years) | Mean ± SD | 44 ± 13 | 46 ± 15 | 42 ± 10 |
| Median | 44 | 45 | 40 | |
| Range | 21–89 | 20–80 | 18–79 | |
| Sex | Female | 70 (46) | 75 (42) | 67 (45) |
| Male | 84 (54) | 104 (58) | 81 (55) | |
| Smokers | 90 (58) | 145 (81) | 80 (54) | |
| BCG vaccinated | 154 (100) | 179 (100) | 148 (100) | |
| Positive PPD skin test | 0 | 179 (100) | 0 | |
| Löfgren’s syndrome | 28 (18) | 0 | 0 | |
| Relapse | 0 | 0 | 0 | |
| Symptoms | Cough | 71 (46) | 161 (90) | 0 |
| Dyspnea | 81 (53) | 45 (25) | 0 | |
| Increased body temperature | 21 (14) | 116 (65) | 0 | |
| Night sweats | 2 (<1) | 90 (50) | 0 | |
| Stage of SA | I | II | III | IV |
|---|---|---|---|---|
| Total | 38 | 60 | 35 | 21 |
| with Löfgren’s syndrome | 13 | 10 | 2 | 0 |
| with the duration of SA > 2 y | 14 (37) | 42 (70) | 35 (100) | 21 (100) |
| Gene Name | NCBI Reference Gene Sequence | Mutation in DNA with dbSNP ID | Amino Acid Change | Primer Names and Sequences for PCR-SSP (5′ to 3′) | Product Length (bp) | Reference |
|---|---|---|---|---|---|---|
| FCGR2A | NG_012066.2 | A519G, rs1801274 | H131R | 2A-gene: * TCAAAGTGAAACAACAGCCTGACT 2A-519A: GGAAAATCCCAGAAATTCACACA 2A-519G: GGAAAATCCCAGAAATTCACACG | 371 | [17] |
| FCGR2C | NG_011982.1 | T202C, rs10917661 | X57Q | 2C-gene: GAGATTCCCATTGTGGACCTACG 2C-202T: GGCTGTGCTGAAACTGGAGACCT 2C-202C: GGCTGTGCTGAAACTGGAGCCAC | 124 | [18] |
| FCGR3A | NG_009066.1 | T559G, rs396991 | V158F | 3A-gene: AGTTCATCATAATTCTGACTCCT 3A-559T: TGAAGACACATTTCTACTCCCTAA 3A-559G: TGAAGACACATTTCTACTCCCTAC | 100 | [17] |
| FCGR3B | NG_032926.2 | NA1/NA2 differences: C147T, rs447536 G141C, rs403016 A227G, rs448740 G277A, rs428888 G349A, rs2290834 SH/NA2 difference: C266A, rs5030738 | L38L R36S N65S D82N V106I A78D | 3B-gene: ATGGACTTCTAGCTGCAC 3B-NA1: CAGTGGTTTCACAATGAGAA 3B-NA2/SH: CAATGGTACAGCGTGCTT 3B-SH: TCGAGCTACTTCATTGACGA | NA1: 140 NA2/SH: 219 SH: 102 | [19] |
| hGH | NC_000017.11 | - | - | hGH-F: CAGTGCCTTCCCAACCATTCCCTTA hGH-R: ATCCACTCACGGATTTCTGTTGTGTTTC | 439 | [20] |
| NF1 | NG_009018.1 | - | - | NF1-exon4b-F: CTGTCCCCTAATACTTAATT NF1-exon4b-R: AATACTAGTTTTTGACCCAGT NF1-exon22-F: TCTTTAGCTTCCTACCTAAGAA NF1-exon22-R: AACACACATACACACAAAATGAA | 209 262 | [21] |
| Mutation in DNA with dbSNP ID | Amino Acid Change | Primer Names and Sequences (5′ to 3′) | Fluorescent Probes | Product Length (bp) |
|---|---|---|---|---|
| T695C, rs1050501 | I232T | 2B-SNP_F: CCTAGCTCCCAGCTCTTCAC 2B-SNP_R: CCACTACAGCAGCAACAATGG | 2B-SNP_695T-VIC: TCACTGGGATTGCTG 2B-SNP_695C-FAM: TCACTGGGACTGCTG | 36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Typiak, M.; Rękawiecki, B.; Rębała, K.; Dubaniewicz, A. Comparative Analysis of FCGR Gene Polymorphism in Pulmonary Sarcoidosis and Tuberculosis. Cells 2023, 12, 1221. https://doi.org/10.3390/cells12091221
Typiak M, Rękawiecki B, Rębała K, Dubaniewicz A. Comparative Analysis of FCGR Gene Polymorphism in Pulmonary Sarcoidosis and Tuberculosis. Cells. 2023; 12(9):1221. https://doi.org/10.3390/cells12091221
Chicago/Turabian StyleTypiak, Marlena, Bartłomiej Rękawiecki, Krzysztof Rębała, and Anna Dubaniewicz. 2023. "Comparative Analysis of FCGR Gene Polymorphism in Pulmonary Sarcoidosis and Tuberculosis" Cells 12, no. 9: 1221. https://doi.org/10.3390/cells12091221
APA StyleTypiak, M., Rękawiecki, B., Rębała, K., & Dubaniewicz, A. (2023). Comparative Analysis of FCGR Gene Polymorphism in Pulmonary Sarcoidosis and Tuberculosis. Cells, 12(9), 1221. https://doi.org/10.3390/cells12091221





