Isolation and Characterization of Cow-, Buffalo-, Sheep- and Goat-Milk-Derived Extracellular Vesicles
Abstract
:1. Introduction
2. Materials and Methods
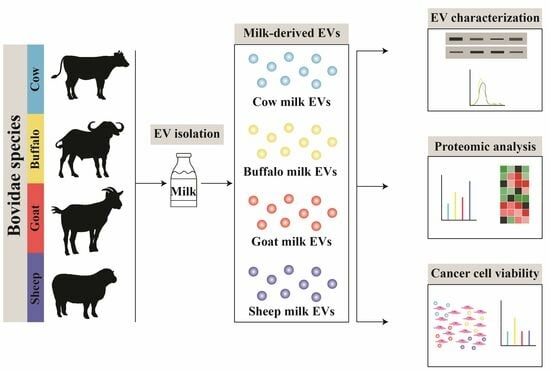
2.1. Milk Samples
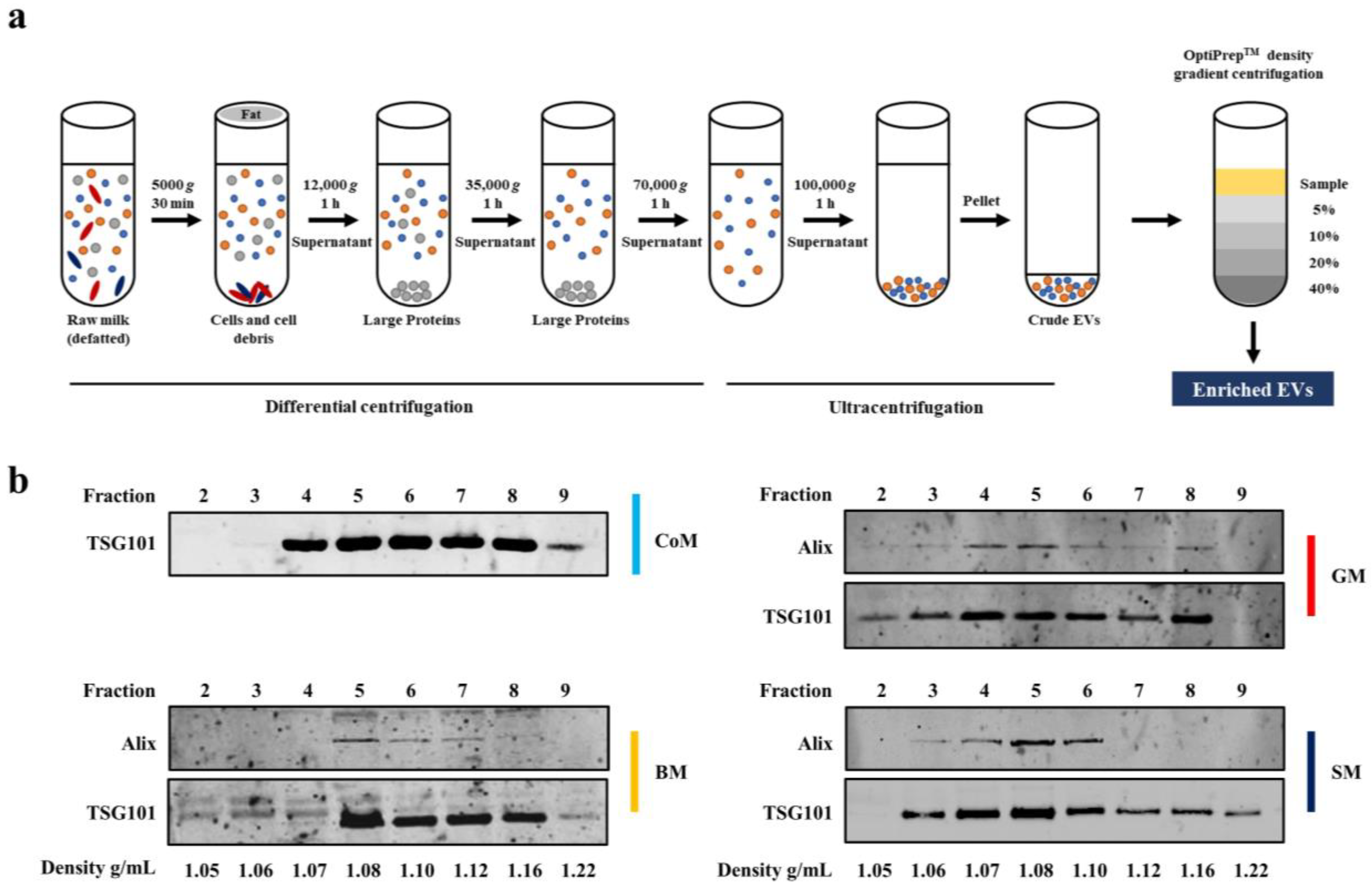
2.2. Isolation of EVs using Differential Centrifugation and Ultracentrifugation
2.3. Density Gradient Ultracentrifugation
2.4. Cell Culture
2.5. Western Blotting and Antibodies
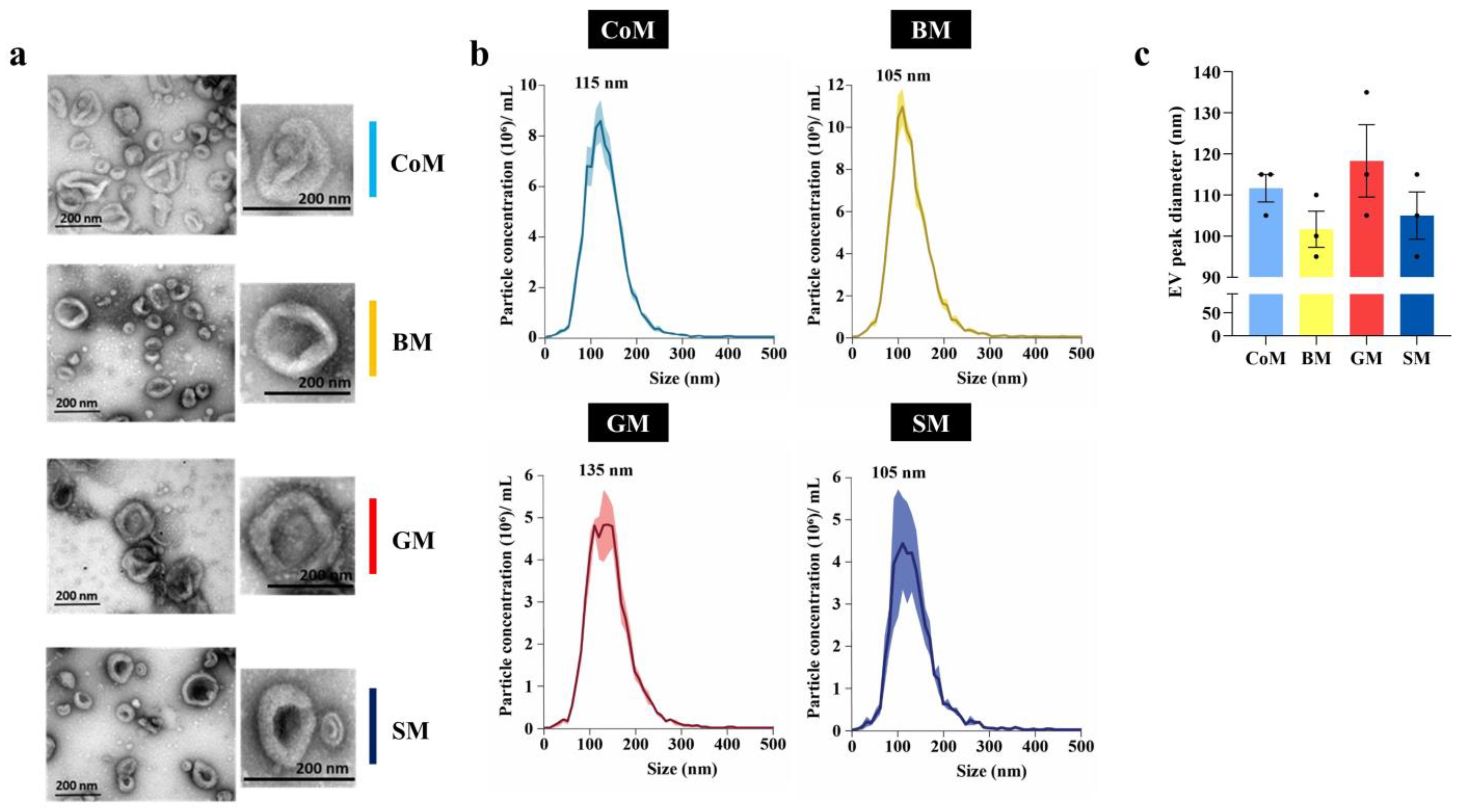
2.6. Transmission Electron Microscopy (TEM)
2.7. Nanoparticle Tracking Analysis (NTA)
2.8. FACS Cell Death Assay
2.9. Gram Staining
2.10. In-Gel Digestion
2.11. LC-MS/MS
2.12. Database Searching and Protein Identification
2.13. Label-Free Spectral Counting
2.14. Functional Enrichment Analysis
3. Results and Discussion
3.1. EV Markers Are Enriched in Fractions Corresponding to a Density of 1.08–1.22 g/mL
3.2. Biophysical Analysis of MEVs
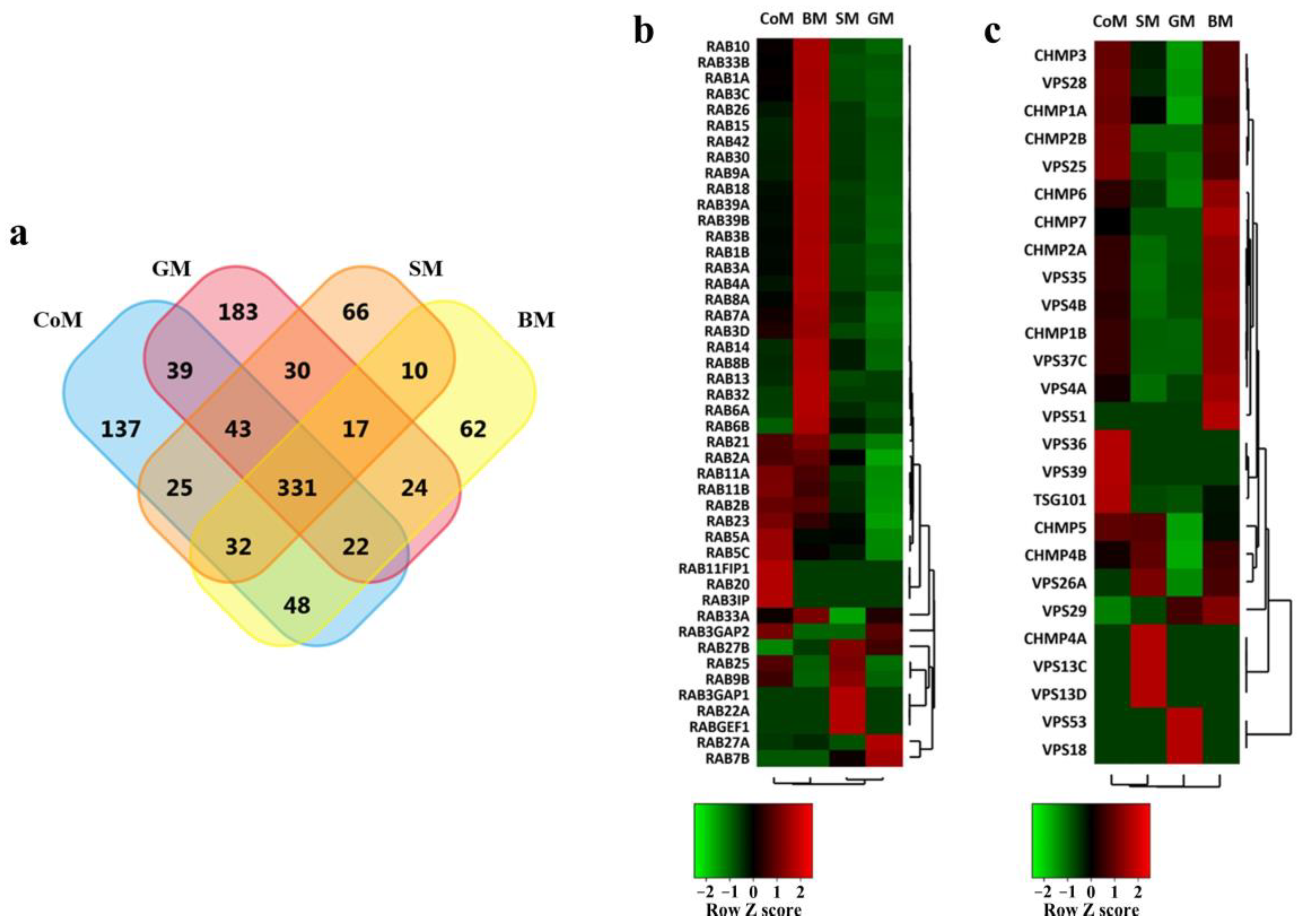
3.3. Proteomic Cargo Analysis of EVs Derived from the Milk of Bovidae Family Members
3.4. EV Markers Are Enriched in Samples Derived from All Species
3.5. Functional Analysis of Proteins Identified in the EVs
3.6. BM-Derived EVs Induced Higher Cell Death as Compared to MEVs from Other Bovine Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samuel, M.; Chisanga, D.; Liem, M.; Keerthikumar, S.; Anand, S.; Ang, C.-S.; Adda, C.G.; Versteegen, E.; Jois, M.; Mathivanan, S. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Sci. Rep. 2017, 7, 5933. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Provost, P. Milk MicroRNAs in Health and Disease. Compr. Rev. Food Sci. Food Saf. 2019, 18, 703–722. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk—A Nutrient System of Mammalian Evolution Promoting mTORC1-Dependent Translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef]
- Xu, R.; Sangild, P.T.; Zhang, Y.; Zhang, S. Bioactive compounds in porcine colostrum and milk and their effects on intestinal development in neonatal pigs1. Biol. Grow. Anim. 2002, 1, 169–192. [Google Scholar]
- German, J.B.; Dillard, C.J.; Ward, R.E. Bioactive components in milk. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 653–658. [Google Scholar] [CrossRef]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef]
- Ninkina, N.; Kukharsky, M.S.; Hewitt, M.V.; Lysikova, E.A.; Skuratovska, L.N.; Deykin, A.V.; Buchman, V.L. Stem cells in human breast milk. Hum. Cell 2019, 32, 223–230. [Google Scholar] [CrossRef]
- Chen, P.-W.; Lin, Y.-L.; Huang, M.-S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 2018, 26, 1235–1244. [Google Scholar] [CrossRef]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Kang, T.; Atukorala, I.; Mathivanan, S. Biogenesis of Extracellular Vesicles. In New Frontiers: Extracellular Vesicles; Mathivanan, S., Fonseka, P., Nedeva, C., Atukorala, I., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 19–43. [Google Scholar]
- Gilmore, W.J.; Bitto, N.J.; Kaparakis-Liaskos, M. Pathogenesis Mediated by Bacterial Membrane Vesicles. Subcell Biochem. 2021, 97, 101–150. [Google Scholar] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Cumba Garcia, L.M.; Peterson, T.E.; Cepeda, M.A.; Johnson, A.J.; Parney, I.F. Isolation and Analysis of Plasma-Derived Exosomes in Patients with Glioma. Front. Oncol. 2019, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, P.; Sabaratnam, R.; Jensen, B.L. Urinary extracellular vesicles: Origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiol. 2020, 228, e13346. [Google Scholar] [CrossRef]
- Welton, J.L.; Loveless, S.; Stone, T.; von Ruhland, C.; Robertson, N.P.; Clayton, A. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J. Extracell. Vesicles 2017, 6, 1369805. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Johnson, L.A.; Leone, D.A.; Majek, P.; Vaahtomeri, K.; Senfter, D.; Bukosza, N.; Schachner, H.; Asfour, G.; Langer, B.; et al. Lymphatic exosomes promote dendritic cell migration along guidance cues. J. Cell Biol. 2018, 217, 2205–2221. [Google Scholar] [CrossRef]
- Klingeborn, M.; Dismuke, W.M.; Bowes Rickman, C.; Stamer, W.D. Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye Res. 2017, 59, 158–177. [Google Scholar] [CrossRef]
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary Exosomes: Emerging Roles in Systemic Disease. Int. J. Biol. Sci. 2018, 14, 633–643. [Google Scholar] [CrossRef]
- Lässer, C.; O’Neil, S.E.; Ekerljung, L.; Ekström, K.; Sjöstrand, M.; Lötvall, J. RNA-containing exosomes in human nasal secretions. Am. J. Rhinol. Allergy 2011, 25, 89–93. [Google Scholar] [CrossRef]
- Peng, P.; Yan, Y.; Keng, S. Exosomes in the ascites of ovarian cancer patients: Origin and effects on anti-tumor immunity. Oncol. Rep. 2011, 25, 749–762. [Google Scholar] [PubMed]
- Madison, M.N.; Welch, J.L.; Okeoma, C.M. Isolation of Exosomes from Semen for in vitro Uptake and HIV-1 Infection Assays. Bio Protoc. 2017, 7, e2216. [Google Scholar] [CrossRef] [PubMed]
- van Herwijnen, M.J.C.; Zonneveld, M.I.; Goerdayal, S.; Nolte-’t Hoen, E.N.M.; Garssen, J.; Stahl, B.; Maarten Altelaar, A.F.; Redegeld, F.A.; Wauben, M.H.M. Comprehensive Proteomic Analysis of Human Milk-derived Extracellular Vesicles Unveils a Novel Functional Proteome Distinct from Other Milk Components. Mol. Cell. Proteom. MCP 2016, 15, 3412–3423. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, M.I.; van Herwijnen, M.J.C.; Fernandez-Gutierrez, M.M.; Giovanazzi, A.; de Groot, A.M.; Kleinjan, M.; van Capel, T.M.M.; Sijts, A.J.A.M.; Taams, L.S.; Garssen, J.; et al. Human milk extracellular vesicles target nodes in interconnected signalling pathways that enhance oral epithelial barrier function and dampen immune responses. J. Extracell. Vesicles 2021, 10, e12071. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2018, 47, D516–D519. [Google Scholar] [CrossRef]
- Samuel, M.; Fonseka, P.; Sanwlani, R.; Gangoda, L.; Chee, S.H.; Keerthikumar, S.; Spurling, A.; Chitti, S.V.; Zanker, D.; Ang, C.-S.; et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021, 12, 3950. [Google Scholar] [CrossRef]
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1440132. [Google Scholar] [CrossRef]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef]
- Fonseka, P.; Kang, T.; Chee, S.; Chitti, S.V.; Sanwlani, R.; Ang, C.-S.; Mathivanan, S. Temporal Quantitative Proteomics Analysis of Neuroblastoma Cells Treated with Bovine Milk-Derived Extracellular Vesicles Highlights the Anti-Proliferative Properties of Milk-Derived Extracellular Vesicles. Cells 2021, 10, 750. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Sanwlani, R.; Kang, T.; Gummadi, S.; Nedeva, C.; Ang, C.-S.; Mathivanan, S. Bovine milk-derived extracellular vesicles enhance doxorubicin sensitivity in triple negative breast cancer cells by targeting metabolism and STAT signalling. Proteomics 2023, 23, 2200482. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Agrawal, A.K.; Mudd, A.M.; Kyakulaga, A.H.; Singh, I.P.; Vadhanam, M.V.; Gupta, R.C. Exosomal formulation of anthocyanidins against multiple cancer types. Cancer Lett. 2017, 393, 94–102. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Gangoda, L.; Liem, M.; Fonseka, P.; Atukorala, I.; Ozcitti, C.; Mechler, A.; Adda, C.G.; Ang, C.-S.; Mathivanan, S. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget 2015, 6, 15375. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Tauro, B.J.; Chen, Y.-S.; Simpson, R.J. Identifying mutated proteins secreted by colon cancer cell lines using mass spectrometry. J. Proteom. 2012, 76, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, A.C.; Washburn, M.P. Quantitation in proteomic experiments utilizing mass spectrometry. Biotechnol. Genet. Eng. Rev. 2006, 22, 1–20. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.-S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Sanwlani, R.; Fonseka, P.; Chitti, S.V.; Mathivanan, S. Milk-Derived Extracellular Vesicles in Inter-Organism, Cross-Species Communication and Drug Delivery. Proteomes 2020, 8, 11. [Google Scholar] [CrossRef]
- An, J.; Ha, E.M. Extracellular vesicles derived from Lactobacillus plantarum restore chemosensitivity through the PDK2-mediated glucose metabolic pathway in 5-FU-resistant colorectal cancer cells. J. Microbiol. 2022, 60, 735–745. [Google Scholar] [CrossRef]
- Behzadi, E.; Mahmoodzadeh Hosseini, H.; Imani Fooladi, A.A. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb. Pathog. 2017, 110, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Zhang, X.; Tong, L.; Liu, Q.; Liang, X.; Bu, Y.; Gong, P.; Liu, T.; Zhang, L.; Xia, Y.; et al. Effect of Extracellular Vesicles Derived from Lactobacillus plantarum Q7 on Gut Microbiota and Ulcerative Colitis in Mice. Front. Immunol. 2021, 12, 777147. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Yang, C.; Liu, L.; Mai, G.; Li, H.; Wu, L.; Jin, M.; Chen, Y. Commensal bacteria-derived extracellular vesicles suppress ulcerative colitis through regulating the macrophages polarization and remodeling the gut microbiota. Microb. Cell Fact. 2022, 21, 88. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Zhang, X.; Hao, H.; Liu, Q.; Zhou, Z.; Liang, X.; Liu, T.; Gong, P.; Zhang, L.; Zhai, Z.; et al. Lactobacillus rhamnosus GG Derived Extracellular Vesicles Modulate Gut Microbiota and Attenuate Inflammatory in DSS-Induced Colitis Mice. Nutrients 2021, 13, 3319. [Google Scholar] [CrossRef] [PubMed]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2017, 9, 95–106. [Google Scholar] [CrossRef]
- Henrick, B.M.; Nag, K.; Yao, X.-D.; Drannik, A.G.; Aldrovandi, G.M.; Rosenthal, K.L. Milk matters: Soluble Toll-like receptor 2 (sTLR2) in breast milk significantly inhibits HIV-1 infection and inflammation. PLoS ONE 2012, 7, e40138. [Google Scholar] [CrossRef]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef]
- de Oliviera Nascimento, L.; Massari, P.; Wetzler, L. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef]
- Kavanaugh, N.L.; Zhang, A.Q.; Nobile, C.J.; Johnson, A.D.; Ribbeck, K. Mucins suppress virulence traits of Candida albicans. mBio 2014, 5, e01911. [Google Scholar] [CrossRef]
- Woodman, T.; Strunk, T.; Patole, S.; Hartmann, B.; Simmer, K.; Currie, A. Effects of lactoferrin on neonatal pathogens and Bifidobacterium breve in human breast milk. PLoS ONE 2018, 13, e0201819. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Karmaus, W.; Davis, S.; Gangur, V. Immune markers in breast milk and fetal and maternal body fluids: A systematic review of perinatal concentrations. J. Hum. Lact. 2011, 27, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Henkel, T. Function and activation of NF-kappaB in the immune system. Annu. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef]
- El-Salam, M.H.A.; El-Shibiny, S. Bioactive Peptides of Buffalo, Camel, Goat, Sheep, Mare, and Yak Milks and Milk Products. Food Rev. Int. 2013, 29, 1–23. [Google Scholar] [CrossRef]
- Mosca, F.; Giannì, M.L. Human milk: Composition and health benefits. Pediatr. Medica Chir. Med. Surg. Pediatr. 2017, 39, 155. [Google Scholar] [CrossRef] [PubMed]
- Górska-Warsewicz, H.; Rejman, K.; Laskowski, W.; Czeczotko, M. Milk and Dairy Products and Their Nutritional Contribution to the Average Polish Diet. Nutrients 2019, 11, 1771. [Google Scholar] [CrossRef]
- Gateway to Dairy Production and Products. Available online: http://www.fao.org/dairy-production-products/products/milk-composition/en/ (accessed on 20 July 2023).
- Ong, S.L.; Blenkiron, C.; Haines, S.; Acevedo-Fani, A.; Leite, J.A.S.; Zempleni, J.; Anderson, R.C.; McCann, M.J. Ruminant Milk-Derived Extracellular Vesicles: A Nutritional and Therapeutic Opportunity? Nutrients 2021, 13, 2505. [Google Scholar] [CrossRef]
- Buratta, S.; Urbanelli, L.; Tognoloni, A.; Latella, R.; Cerrotti, G.; Emiliani, C.; Chiaradia, E. Protein and Lipid Content of Milk Extracellular Vesicles: A Comparative Overview. Life 2023, 13, 401. [Google Scholar] [CrossRef]
- Sedykh, S.E.; Purvinish, L.V.; Monogarov, A.S.; Burkova, E.E.; Grigor’eva, A.E.; Bulgakov, D.V.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Purified horse milk exosomes contain an unpredictable small number of major proteins. Biochim. Open 2017, 4, 61–72. [Google Scholar] [CrossRef]
- Reinhardt, T.A.; Lippolis, J.D.; Nonnecke, B.J.; Sacco, R.E. Bovine milk exosome proteome. J. Proteom. 2012, 75, 1486–1492. [Google Scholar] [CrossRef]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Pieters, B.C.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.; van Caam, A.P.; Koenders, M.I.; van Lent, P.L.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M.; et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk: An exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J. Transl. Med. 2014, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Sanwlani, R.; Fonseka, P.; Mathivanan, S. Are Dietary Extracellular Vesicles Bioavailable and Functional in Consuming Organisms? Sub-Cell. Biochem. 2021, 97, 509–521. [Google Scholar]
- Medhammar, E.; Wijesinha-Bettoni, R.; Stadlmayr, B.; Nilsson, E.; Charrondiere, U.R.; Burlingame, B. Composition of milk from minor dairy animals and buffalo breeds: A biodiversity perspective. J. Sci. Food Agric. 2012, 92, 445–474. [Google Scholar] [CrossRef]
- Yang, T.X.; Wang, F.; Li, H.; Liu, Q.S.; Li, Q.Y. The Nutrition of Buffalo Milk: A Comparison with Cow Milk. Adv. Mater. Res. 2013, 781, 1460–1463. [Google Scholar] [CrossRef]

| Top 20 Most Abundant Proteins Identified | |||
|---|---|---|---|
| CoM | BM | GM | SM |
| BTN1A1 | BTN1A1 | BTN1A1 | BTN1A1 |
| XDH | XDH | XDH | XDH |
| MFGE8 | CD36 | FASN | PAEP |
| CD36 | MFGE8 | CSN1S1 | CSN1S1 |
| ABCG2 | CSN1S1 | PLIN2 | LOC101115115 |
| FASN | CSN3 | ABCG2 | ABCG2 |
| SLC34A2 | FABP3 | ACTG1 | ALB |
| ALB | ALB | CSN3 | PLIN2 |
| FABP3 | RAB1A | ACTB | EZR |
| EZR | PLIN2 | ALB | MFGE8 |
| PLIN2 | ABCG2 | CSN1S2 | HSPA8 |
| ENPP3 | SLC34A2 | EZR | ACTB |
| RAB1A | FASN | MFGE8 | ACTG1 |
| IDH1 | RAB1B | PAEP | STOM |
| HSPA8 | RAB18 | STOM | CD36 |
| RAB18 | EZR | CD36 | MSN |
| MSN | LOC102406615 | MSN | RAB18 |
| ACTB | PAEP | ACTC1 | CSN2 |
| ACTG1 | ENPP3 | ACTA1 | FASN |
| PAEP | MSN | ACTG2 | FABP3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samuel, M.; Sanwlani, R.; Pathan, M.; Anand, S.; Johnston, E.L.; Ang, C.-S.; Kaparakis-Liaskos, M.; Mathivanan, S. Isolation and Characterization of Cow-, Buffalo-, Sheep- and Goat-Milk-Derived Extracellular Vesicles. Cells 2023, 12, 2491. https://doi.org/10.3390/cells12202491
Samuel M, Sanwlani R, Pathan M, Anand S, Johnston EL, Ang C-S, Kaparakis-Liaskos M, Mathivanan S. Isolation and Characterization of Cow-, Buffalo-, Sheep- and Goat-Milk-Derived Extracellular Vesicles. Cells. 2023; 12(20):2491. https://doi.org/10.3390/cells12202491
Chicago/Turabian StyleSamuel, Monisha, Rahul Sanwlani, Mohashin Pathan, Sushma Anand, Ella L. Johnston, Ching-Seng Ang, Maria Kaparakis-Liaskos, and Suresh Mathivanan. 2023. "Isolation and Characterization of Cow-, Buffalo-, Sheep- and Goat-Milk-Derived Extracellular Vesicles" Cells 12, no. 20: 2491. https://doi.org/10.3390/cells12202491
APA StyleSamuel, M., Sanwlani, R., Pathan, M., Anand, S., Johnston, E. L., Ang, C.-S., Kaparakis-Liaskos, M., & Mathivanan, S. (2023). Isolation and Characterization of Cow-, Buffalo-, Sheep- and Goat-Milk-Derived Extracellular Vesicles. Cells, 12(20), 2491. https://doi.org/10.3390/cells12202491