N-Terminal Processing and Modification of Ciliary Dyneins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Computational Methods
2.2. Chlamydomonas Strains and Growth Conditions
2.3. Preparation of Cilia Samples for Mass Spectrometry
2.4. Peptide Identification Using High-Resolution Mass Spectrometry
3. Results
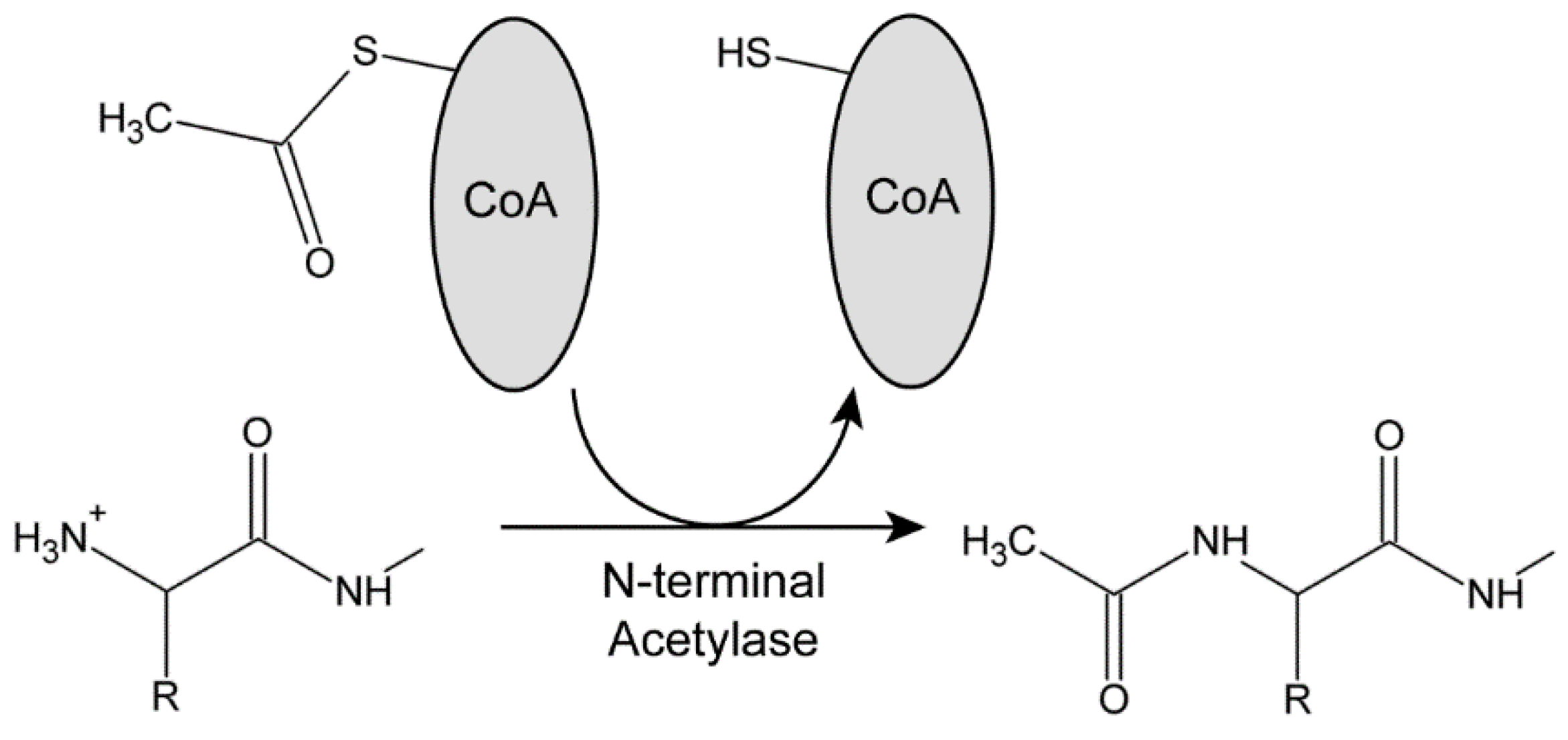
3.1. N-Terminal Acetylases in Chlamydomonas
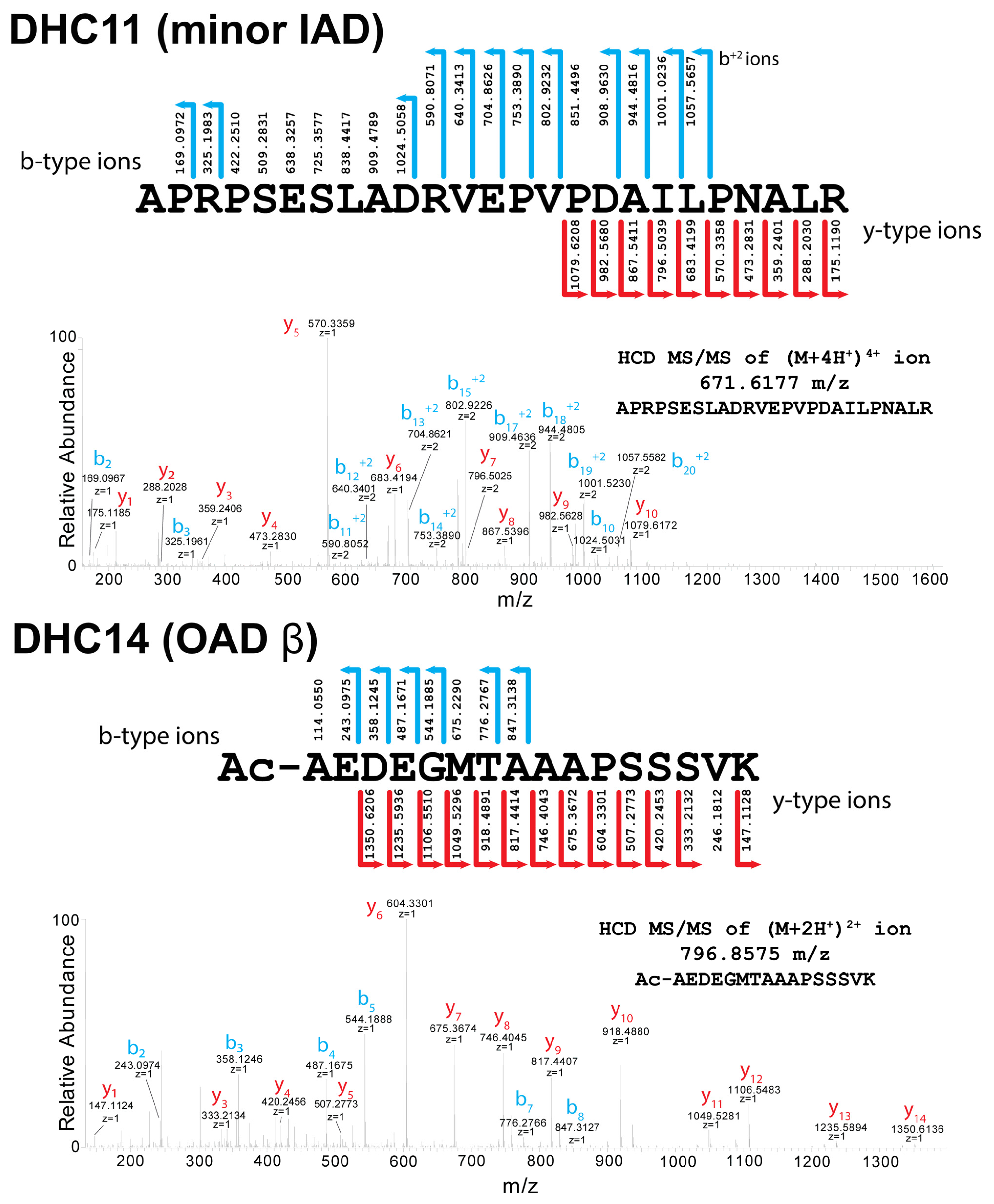
3.2. Acetylation of Dynein Heavy-Chain N-Termini
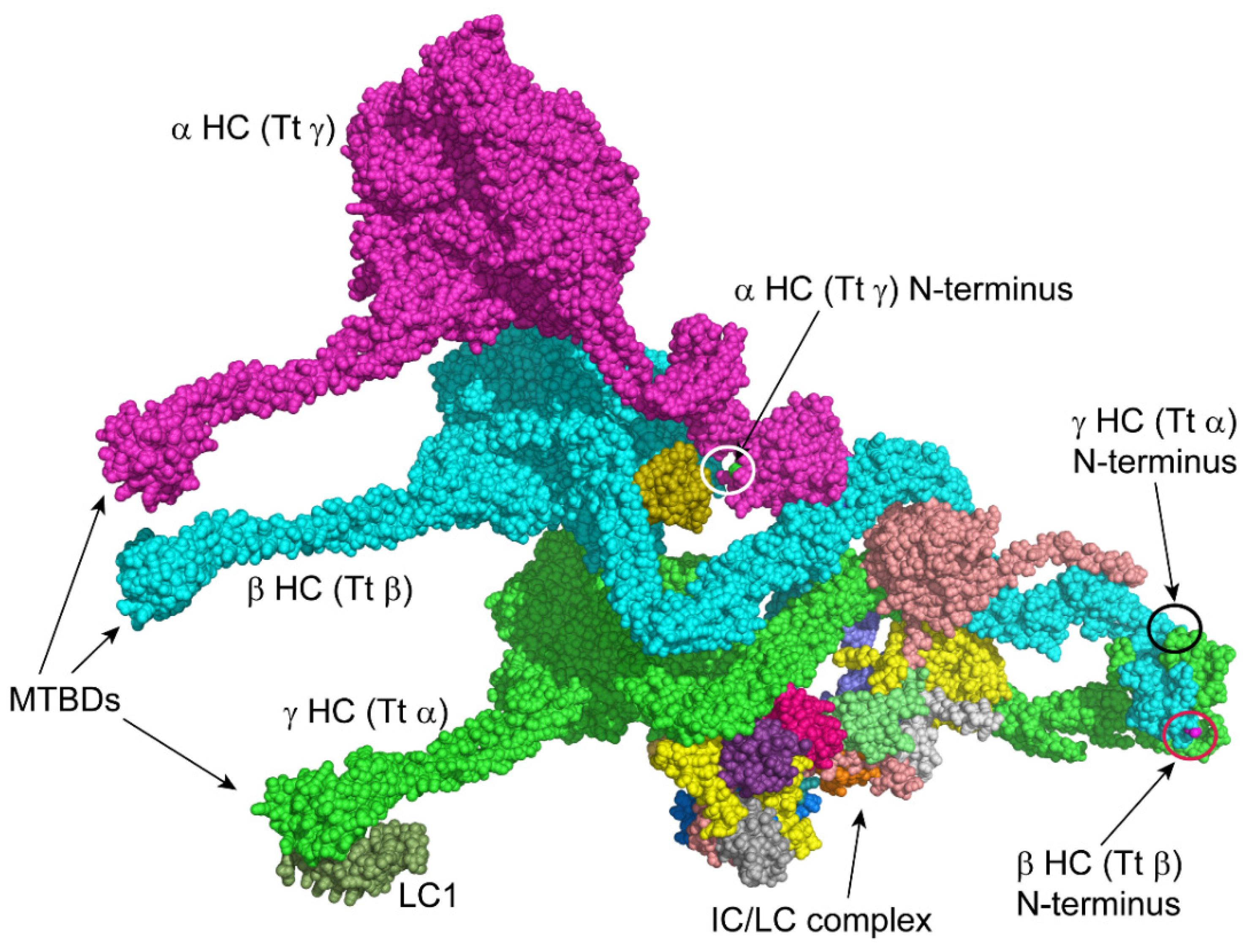
3.3. Acetylation of Other Axonemal and IFT Dynein Components
4. Discussion
4.1. Integrating N-Terminal Acetylation within the Axonemal Dynein Assembly Pathway
4.2. AcN-Degrons as a Potential Mechanism to Direct Dynein Subunit Stoichiometry
4.3. Recognition of Dynein AcN-Degrons
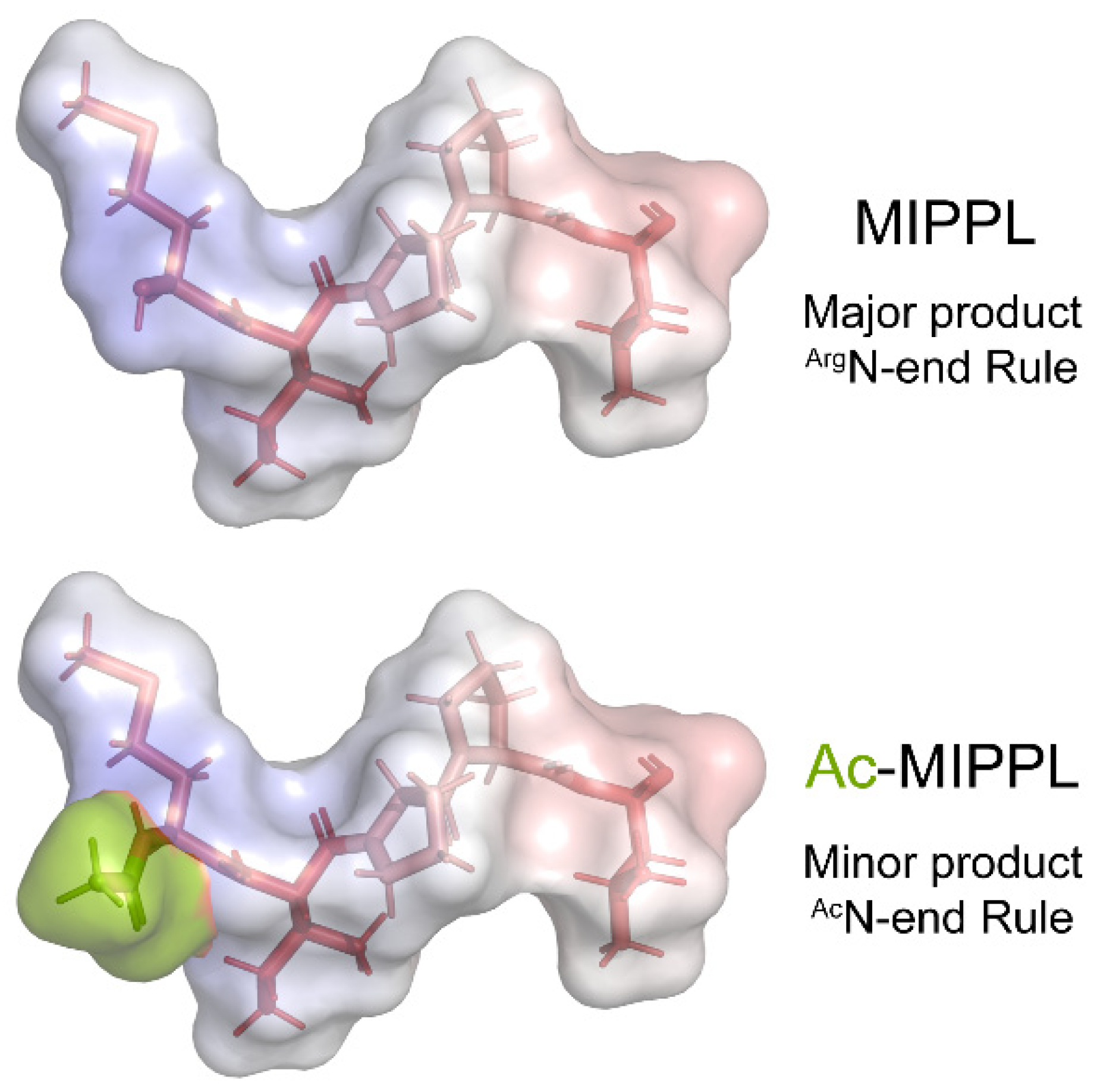
4.4. A Single Inner-Arm Dynein Protein Is Subject to the ArgN-End Rule Pathway
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, W.; Basto, R. Cilia; Cold Spring Harbor Perspectives in Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- King, S.M.; Sale, W.S. Fifty years of microtubule sliding in cilia. Mol. Biol. Cell 2018, 29, 698–701. [Google Scholar] [CrossRef] [PubMed]
- Wickstead, B. The evolutionary biology of dyneins. In Dyneins: Structure, Biology and Disease; Volume 1—The Biology of Dynein Motors; King, S.M., Ed.; Elsevier, Inc.: Oxford, UK, 2018; pp. 101–138. [Google Scholar]
- Fliegauf, M.; Benzing, T.; Omran, H. When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007, 8, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef]
- Li, Y.; Klena, N.T.; Gabriel, G.C.; Liu, X.; Kim, A.J.; Lemke, K.; Chen, Y.; Chatterjee, B.; Devine, W.; Damerla, R.R.; et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 2015, 521, 520–524. [Google Scholar] [CrossRef]
- Faubel, R.J.; Santos Canellas, V.S.; Gaesser, J.; Beluk, N.H.; Feinstein, T.N.; Wang, Y.; Yankova, M.; Karunakaran, K.B.; King, S.M.; Ganapathiraju, M.K.; et al. Flow blockage disrupts cilia-driven fluid transport in the epileptic brain. Acta Neuropathol. 2022, 144, 691–706. [Google Scholar] [CrossRef]
- King, S.M. Axonemal protofilament ribbons, DM10 domains and the link to juvenile myoclonic epilepsy. Cell Motil. Cytoskelet. 2006, 63, 245–253. [Google Scholar] [CrossRef]
- Ikeda, T.; Ikeda, K.; Enomoto, M.; Park, M.K.; Hirono, M.; Kamiya, R. The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 2005, 579, 819–822. [Google Scholar] [CrossRef] [PubMed]
- King, S.M. Composition and assembly of axonemal dyneins. In Dyneins: Structure, Biology and Disease; Volume 1—The Biology of Dynein Motors; King, S.M., Ed.; Elsevier, Inc: Oxford, UK, 2018; pp. 163–201. [Google Scholar]
- Carter, A.P.; Cho, C.; Jin, L.; Vale, R. Crystal structure of the dynein motor domain. Science 2011, 331, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Oyama, T.; Shimo-Kon, R.; Imamula, K.; Shima, T.; Sutoh, K.; Kurisu, G. The 2.8 Å crystal structure of the dynein motor domain. Nature 2012, 484, 345–350. [Google Scholar] [CrossRef]
- Burgess, S.A.; Walker, M.L.; Sakakibara, H.; Knight, P.J.; Oiwa, K. Dynein structure and power stroke. Nature 2003, 421, 715–718. [Google Scholar] [CrossRef]
- Schmidt, H.; Gleave, E.S.; Carter, A.P. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 2012, 19, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Carter, A.P. Mechanism and regulation of dynein motors. In Dyneins: Structure, Biology and Disease; Volume 2—Dynein Mechanics, Dysfunction and Disease; King, S.M., Ed.; Elsevier, Inc.: Oxford, UK, 2018; pp. 37–51. [Google Scholar]
- King, S.M. Regulatory mechanics of outer-arm dynein motors. In Dyneins: Structure, Biology and Disease; Volume 1—The Biology of Dynein Motors; King, S.M., Ed.; Elsevier, Inc.: Oxford, UK, 2018; pp. 251–269. [Google Scholar]
- Braschi, B.; Omran, H.; Witman, G.B.; Pazour, G.J.; Pfister, K.K.; Bruford, E.A.; King, S.M. Consensus nomenclature for dyneins and associated assembly factors. J. Cell Biol. 2022, 221, e202109014. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.R. Cytoplasmic preassembly and trafficking of axonemal dyneins. In Dyneins: Structure, Biology and Disease; Volume 1—The Biology of Dynein Motors; King, S.M., Ed.; Elsevier Inc.: Oxford, UK, 2018; pp. 141–161. [Google Scholar]
- King, S.M. Cytoplasmic factories for axonemal dynein assembly. J. Cell Sci. 2021, 134, jcs258626. [Google Scholar] [CrossRef] [PubMed]
- Barkalow, K.; Hamasaki, T.; Satir, P. Regulation of 22S dynein by a 29-kD light chain. J. Cell Biol. 1994, 126, 727–735. [Google Scholar] [CrossRef]
- Habermacher, G.; Sale, W.S. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 1997, 136, 167–176. [Google Scholar] [CrossRef]
- King, S.J.; Dutcher, S.K. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J. Cell Biol. 1997, 136, 177–191. [Google Scholar] [CrossRef]
- Piperno, G.; Luck, D.J. Inner arm dyneins from flagella of Chlamydomonas reinhardtii. Cell 1981, 27, 331–340. [Google Scholar] [CrossRef]
- King, S.M.; Witman, G.B. Multiple sites of phosphorylation within the α heavy chain of Chlamydomonas outer arm dynein. J. Biol. Chem. 1994, 269, 5452–5457. [Google Scholar] [CrossRef]
- Sakato-Antoku, M.; King, S.M. Developmental changes in ciliary composition during gametogenesis in Chlamydomonas. Mol. Biol. Cell 2022, 33, br10. [Google Scholar] [CrossRef]
- Wagner, V.; Gessner, G.; Heiland, I.; Kaminski, M.; Hawat, S.; Scheffler, K.; Mittag, M. Analysis of the phosphoproteome of Chlamydomonas reinhardtii provides new insights into various cellular pathways. Eukaryot. Cell 2006, 5, 457–468. [Google Scholar] [CrossRef]
- Aksnes, H.; Drazic, A.; Marie, M.; Arnesen, T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 2016, 41, 746–760. [Google Scholar] [CrossRef] [PubMed]
- Ree, R.; Varland, S.; Arnesen, T. Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.T.; Mun, S.H.; Lee, C.S.; Hwang, C.S. Control of protein degradation by N-terminal acetylation and the N-end rule pathway. Exp. Mol. Med. 2018, 50, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.M.; Shaw, J.M. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc. Natl. Acad. Sci. USA 2003, 100, 7644–7649. [Google Scholar] [CrossRef]
- Guzman, U.H.; Aksnes, H.; Ree, R.; Krogh, N.; Jakobsson, M.E.; Jensen, L.J.; Arnesen, T.; Olsen, J.V. Loss of N-terminal acetyltransferase A activity induces thermally unstable ribosomal proteins and increases their turnover in Saccharomyces cerevisiae. Nat. Commun. 2023, 14, 4517. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Muratore, T.L.; Schaner-Tooley, C.E.; Shabanowitz, J.; Hunt, D.F.; Macara, I.G. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 2007, 9, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Tooley, C.E.; Petkowski, J.J.; Muratore-Schroeder, T.L.; Balsbaugh, J.L.; Shabanowitz, J.; Sabat, M.; Minor, W.; Hunt, D.F.; Macara, I.G. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature 2010, 466, 1125–1128. [Google Scholar] [CrossRef]
- Tardif, M.; Atteia, A.; Specht, M.; Cogne, G.; Rolland, N.; Brugière, S.; Hippler, M.; Ferro, M.; Bruley, C.; Peltier, G.; et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012, 29, 3625–3639. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotech. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Aksnes, H.; McTiernan, N.; Arnesen, T. NATs at a glance. J. Cell Sci. 2023, 136, jcs260766. [Google Scholar] [CrossRef] [PubMed]
- Giglione, C.; Serero, A.; Pierre, M.; Boisson, B.; Meinnel, T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000, 19, 5916–5929. [Google Scholar] [CrossRef]
- King, S.M.; Yagi, T.; Kamiya, R. Axonemal dyneins: Assembly, structure and motor activity. In The Chlamydomonas Source Book, 3rd ed.; Volume 3: Cell Motility and Behavior; Dutcher, S.K., Ed.; Elsevier: San Diego, CA, USA, 2023; pp. 79–131. [Google Scholar]
- Yagi, T.; Uematsu, K.; Liu, Z.; Kamiya, R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J. Cell Sci. 2009, 122, 1306–1314. [Google Scholar] [CrossRef]
- Bui, K.H.; Yagi, T.; Yamamoto, R.; Kamiya, R.; Ishikawa, T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 2012, 198, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Walton, T.; Wu, H.; Brown, A. Structure of a microtubule-bound axonemal dynein. Nat. Commun. 2021, 12, 477. [Google Scholar] [CrossRef] [PubMed]
- Rao, Q.; Han, L.; Wang, Y.; Chai, P.; Kuo, Y.-w.; Yang, R.; Hu, F.; Yang, Y.; Howard, J.; Zhang, K. Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism. Nat. Struct. Mol. Biol. 2021, 28, 799–810. [Google Scholar] [CrossRef]
- Hom, E.; Witman, G.B.; Harris, E.H.; Dutcher, S.K.; Kamiya, R.; Mitchell, D.R.; Pazour, G.J.; Porter, M.E.; Sale, W.S.; Wirschell, M.; et al. A unified taxonomy for ciliary dyneins. Cytoskeleton 2011, 68, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Kubo, S.; Yang, S.K.; Black, C.S.; Dai, D.; Valente-Paterno, M.; Gaertig, J.; Ichikawa, M.; Bui, K.H. Remodeling and activation mechanisms of outer arm dyneins revealed by cryo-EM. EMBO Rep. 2021, 22, e52911. [Google Scholar] [CrossRef]
- Kato-Minoura, T.; Hirono, M.; Kamiya, R. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin- encoding gene. J. Cell Biol. 1997, 137, 649–656. [Google Scholar] [CrossRef]
- Kamiya, R.; Kurimoto, E.; Muto, E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 1991, 112, 441–447. [Google Scholar] [CrossRef] [PubMed]
- LeDizet, M.; Piperno, G. ida4-1, ida4-2, and ida4-3 are intron splicing mutations affecting the locus encoding p28, a light chain of Chlamydomonas axonemal inner dynein arms. Mol. Biol. Cell 1995, 6, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.-S.; Varshavsky, A. Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 19188–19193. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Vicente, J.; Sharma, G.; Abbas, M.; Zubrycka, A. The plant N-degron pathways of ubiquitin-mediated proteolysis. J. Integr. Plant Biol. 2020, 62, 70–89. [Google Scholar] [CrossRef]
- Owa, M.; Furuta, A.; Usukura, J.; Arisaka, F.; King, S.M.; Witman, G.B.; Kamiya, R.; Wakabayashi, K.-I. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc. Natl. Acad. Sci. USA 2014, 111, 9461–9466. [Google Scholar]
- Huizar, R.L.; Lee, C.; Boulgakov, A.A.; Horani, A.; Tu, F.; Marcotte, E.M.; Brody, S.L.; Wallingford, J.B. A liquid-like organelle at the root of motile ciliopathy. eLife 2018, 7, e38497. [Google Scholar] [CrossRef]
- Lee, C.; Cox, R.M.; Papoulas, O.; Horani, A.; Drew, K.; Devitt, C.C.; Brody, S.L.; Marcotte, E.M.; Wallingford, J.B. Functional partitioning of a liquid-like organelle during assembly of axonemal dyneins. eLife 2020, 9, e58662. [Google Scholar] [CrossRef]
- Yamamoto, R.; Obbineni, J.M.; Alford, L.M.; Ide, T.; Owa, M.; Hwang, J.; Kon, T.; Inaba, K.; James, N.; King, S.M.; et al. Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. PLoS Genet. 2017, 13, e1006996. [Google Scholar] [CrossRef]
- Kakihara, Y.; Houry, W.A. The R2TP complex: Discovery and functions. Biochim. Biophys. Acta 2012, 1823, 101–107. [Google Scholar]
- Yamamoto, R.; Yanagi, S.; Nagao, M.; Yamasaki, Y.; Tanaka, Y.; Sale, W.S.; Yagi, T.; Kon, T. Mutations in PIH proteins MOT48, TWI1 and PF13 define common and unique steps for preassembly of each, different ciliary dynein. PLoS Genet. 2020, 16, e1009126. [Google Scholar] [CrossRef]
- zur Lage, P.; Stefanopoulou, P.; Styczynska-Soczka, K.; Quinn, N.; Mali, G.; von Kriegsheim, A.; Mill, P.; Jarman, A.P. Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J. Cell Biol. 2018, 217, 2583–2598. [Google Scholar] [CrossRef] [PubMed]
- Maurizy, C.; Quinternet, M.; Abel, Y.; Verheggen, C.; Santo, P.E.; Bourguet, M.; Paiva, A.C.F.; Bragantini, B.; Chagot, M.-E.; Robert, M.-C.; et al. The RPAP3-C terminal domain identifies R2TP-like quaternary chaperones. Nat. Commun. 2018, 9, 2093. [Google Scholar] [CrossRef] [PubMed]
- Mali, G.R.; Yeyati, P.L.; Mizuno, S.; Dodd, D.O.; Tennant, P.A.; Keighren, M.A.; zur Lage, P.; Shoemark, A.; Garcia-Munoz, A.; Shimada, A.; et al. ZMYND10 functions in a chaperone relay during axonemal dynein assembly. eLife 2018, 7, e34389. [Google Scholar] [CrossRef]
- Patel-King, R.S.; Sakato-Antoku, M.; Yankova, M.; King, S.M. WDR92 is required for axonemal dynein heavy chain stability in cytoplasm. Mol. Biol. Cell 2019, 30, 1834–1845. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, L.; Pan, J. Chlamydomonas WDR92 in association with R2TP-like complex and multiple DNAAFs to regulate ciliary dynein preassembly. J. Mol. Cell Biol. 2019, 11, 770–780. [Google Scholar] [CrossRef]
- King, S.M. Inherently disordered regions of axonemal dynein assembly factors. Cytoskeleton 2023. [Google Scholar] [CrossRef]
- Remillard, S.P.; Witman, G.B. Synthesis, transport, and utilization of specific flagellar proteins during flagellar regeneration in Chlamydomonas. J. Cell Biol. 1982, 93, 615–631. [Google Scholar] [CrossRef]
- Stolc, V.; Samanta, M.P.; Tongprasit, W.; Marshall, W.F. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomons reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA 2005, 102, 3703–3707. [Google Scholar]
- Sakato-Antoku, M.; King, S.M. Outer arm dynein light chain LC1 is required for normal motor assembly kinetics, ciliary stability and motility. Mol. Biol. Cell 2023, 34, ar75. [Google Scholar]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar]
- Zhang, S.; Wang, L.; Tao, Y.; Bai, T.; Lu, R.; Zhang, T.; Chen, J.; Ding, J. Structural basis for the functional role of the Shu complex in homologous recombination. Nucleic Acids Res. 2017, 45, 13068–13079. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Diener, D.R.; Rosenbaum, J.L. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 2009, 186, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, H.-a.; Kamiya, R. Association between actin and light chains in Chlamydomonas flagellar inner-arm dyneins. Biochem. Biophys. Res. Commun. 2001, 288, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Wickstead, B.; Gull, K. Dyneins across eukaryotes: A comparative genomic analysis. Traffic 2007, 8, 1708–1721. [Google Scholar] [CrossRef] [PubMed]

| N-Terminal Acetylase Complex | Acetylase Catalytic Subunit | Chlamydomonas Gene Identifier | % Identity (Expect Value) | ||||
|---|---|---|---|---|---|---|---|
| Human | Arabidopsis | Chlamydomonas | Chlamydomonas/Human | Arabidopsis /Human | Chlamydomonas/Arabidopsis | ||
| NatA | NAA10 | NAA10 | NAT1 | Cre08.g364450 | 66% (4.88 × 10−68) | 67% (1.33 × 10−70) | 76% (1.02 × 10−84) |
| NatB | NAA20 | NAA20 (NAT3) | NAT3 | Cre08.g383150 | 63% (1.84 × 10−78) | 59% (3.38 × 10−74) | 64% (4.64 × 10−86) |
| NatC | NAA30 | ATMAK3 | NAT6 | Cre14.g623800 | 55% (1.39 × 10−53) | 57% (4.00 × 10−55) | 61% (3.51 × 10−59) |
| NatD | NAA40 | NAA40 | NAT24 | Cre10.g431450 | 33% (6.62 × 10−26) | 34% (1.25 × 10−30) | 30% (4.59 × 10−14) |
| NatE | NAA50 | NAA50 | NAT11 | Cre02.g101850 | 46% (9.06 × 10−41) | 55% (2.20 × 10−54) | 48% (1.32 × 10−51) |
| NatF | NAA60 | NAA60 | NAT29 | Cre13.g572150 | 29% (1.04 × 10−12) | 28% (1.18 × 10−17) | 40% (8.44 × 10−31) |
| NatH | NAA80 | ---- | ---- | ---- | ---- | ---- | ---- |
| Class | HC Protein (Dynein) | N-Terminal Sequence | Identified N-Terminal Residue | Predicted Terminus | Processing Requirements |
|---|---|---|---|---|---|
| I | DHC4 (IAD minor) | MSTSR… | ---- | Ac-Ser | Met AP + NatA |
| I | DHC7 (IAD g) | MASRE… | Ac-Ala | ----- | Met AP + NatA |
| I | DHC13 (OAD α) | MSIFW… | Ac-Ser | ----- | Met AP + NatA |
| I | DHC14 (OAD β) | MAEDE… | Ac-Ala | ----- | Met AP + NatA |
| I | DHC15 (OAD γ) | MALDN… | Ac-Ala | ----- | Met AP + NatA |
| I | DHC16 (IFT) | MSSDS… | Ac-Ser | ----- | Met AP + NatA |
| II | DHC11 (IAD minor) | MAPRP… | Ala | ----- | Met AP |
| III | DHC1 (IAD I1/f 1α) | MDRRL… | ---- | Ac-Met | NatB |
| III | DHC5 (IAD b) | MDRNR… | ---- | Ac-Met | NatB |
| III | DHC6 (IAD a) | MDWDD… | Ac-Met | ----- | NatB |
| III | DHC9 (IAD c) | MDFSM… | Ac-Met | ----- | NatB |
| III | DHC10 (IAD I1/f 1β) | MEPGD… | Ac-Met | ----- | NatB |
| III | DHC12 (IAD minor) | MEPQD… | ---- | Ac-Met | NatB |
| IV | DHC2 (IAD d) | MPGVA… | Pro | ----- | Met AP |
| IV | DHC3 (IAD minor) | MPTEL… | Pro | ----- | Met AP |
| IV | DHC8 (IAD e) | MPTSE… | Pro | ----- | Met AP |
| Dynein | Gene Symbol † | Protein | N-Terminal Sequence | Identified N-Terminal Residue | Predicted N-Terminal Residue | Processing Requirements |
|---|---|---|---|---|---|---|
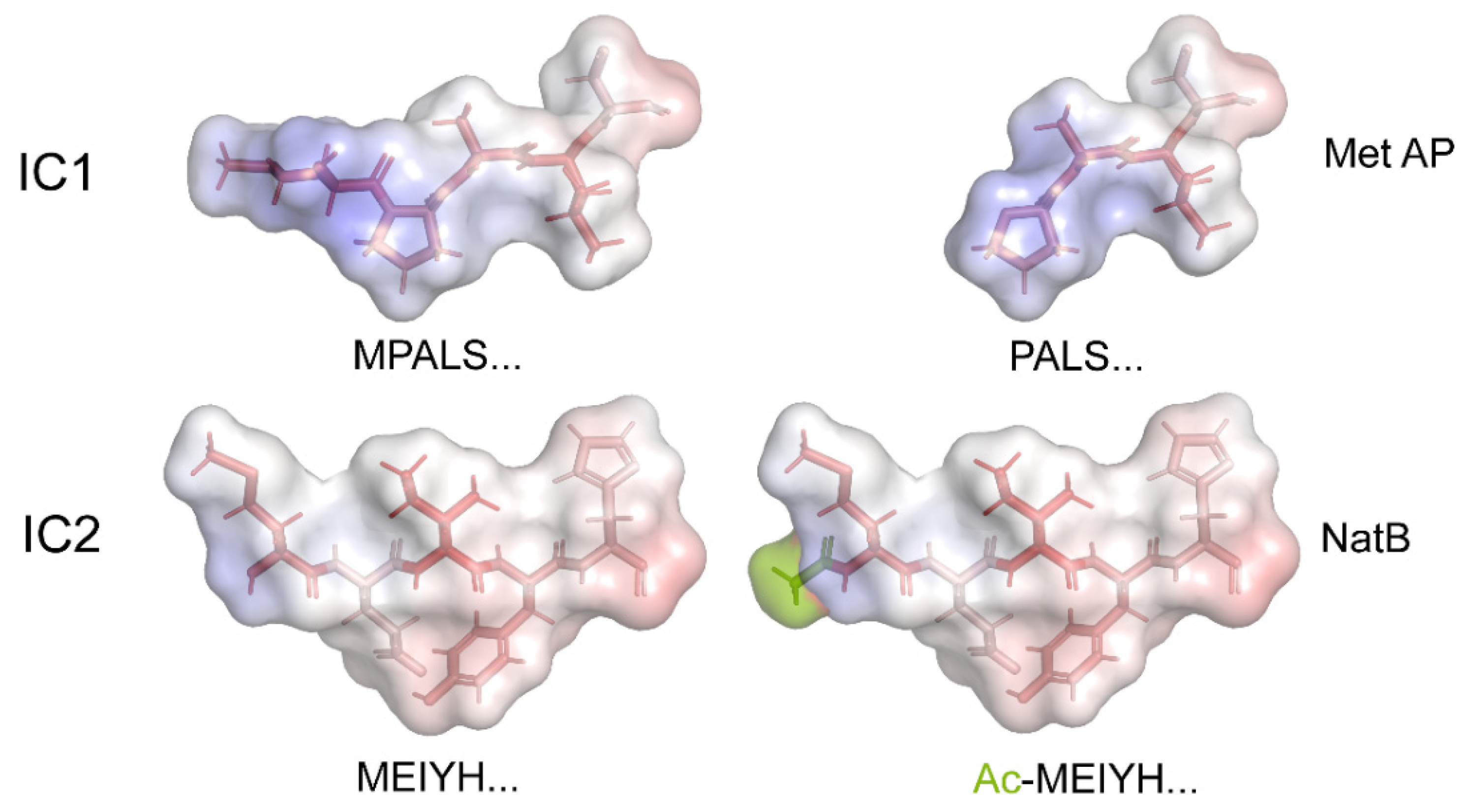
| OAD | DIC1 | IC1 | MPALS… | Pro | ---- | Met AP |
| DIC2 | IC2 | MEIYH… | Ac-Met | ---- | NatB | |
| DLU1 | LC1 | MAKAT… | Ac-Ala | ---- | Met AP + NatA | |
| DLT2 | LC2 | MDDMP… | Ac-Met | ---- | NatB | |
| DLX1 | LC3 | MAAGL… | Ac-Ala | ---- | Met AP + NatA | |
| DLE1 | LC4 | MAAKV… | ---- | Ac-Ala | Met AP + NatA | |
| DLX2 | LC5 | MAFIT… | Ac-Ala | ---- | Met AP + NatA | |
| DLL2 | LC6 | MADEK… | Ac-Ala | ---- | Met AP + NatA | |
| DLR1 | LC7a | MVDIA… | Ac-Val | ---- | Met AP + NatA | |
| DLR2 | LC7b | MSDIE… | Ac-Ser | ---- | Met AP + NatA | |
| DLL1 | LC8 | MASGS… | Ac-Ala | ---- | Met AP + NatA | |
| DLT1 | LC9 | MEDDT… | Ac-Met | ---- | NatB | |
| DLL3 | LC10 | MAEQQ… | Ac-Ala | ---- | Met AP + NatA | |
| DCC1 | DC1 | MAQKS… | Ac-Ala | ---- | Met AP + NatA | |
| DCC2 | DC2 | MPSAD… ‡ | Pro | ---- | Met AP | |
| DLE3 | DC3 | MASAA… | Ac-Ala | ---- | Met AP + NatA | |
| DOI1 | LIS1 | MSAEV… | Ac-Ser | ---- | Met AP + NatA | |
| IAD I1/f $ | DIC3 | IC140 | MEDAS… | ---- | Ac-Met | NatB |
| DIC4 | IC138 | MSDQK… | ---- | Ac-Ser | Met AP + NatA | |
| DII6 | IC97 | MAPKD… | ---- | Ac-Ala | Met AP + NatA | |
| DII7 | FAP120 | MDGEE… | Ac-Met | ---- | NatB | |
| DLT3 | TCTEX1 | MEGVD… | Ac-Met | ---- | NatB | |
| DLT4 | TCTEX2b | MAEAA… | Ac-Ala | ---- | Met AP + NatA | |
| DAU1 | ODA7 | MCACM… | Ac-Cys | Met AP + NatA | ||
| IAD | DII4 | Actin | MADEG… | Ac-Ala | ---- | Met AP + NatA |
| (monomeric) | DII5 | NAP | MTSGL… | Thr | ---- | Met AP |
| IDA4 | p28 | MIPPL… | Met Ac-Met @ | --- --- | ---- NatB | |
| DLE2 | Centrin | MSYKA… | Ac-Ser | ---- | Met AP + NatA | |
| DII2 | p38 | MATLT… | Ac-Ala | ---- | Met AP + NatA | |
| DII3 | p44 | MATLV… | Ac-Ala | ---- | Met AP + NatA | |
| IFT $ | DIC6 | D1bIC1 | MSDYE… | ---- | Ac-Ser | Met AP + NatA |
| DIC5 | D1bIC2 | MQEVP… | Ac-Met | ---- | NatB | |
| DLI1 | D1bLIC | MAAPA… | Ac-Ala | ---- | Met AP + NatA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakato-Antoku, M.; Balsbaugh, J.L.; King, S.M. N-Terminal Processing and Modification of Ciliary Dyneins. Cells 2023, 12, 2492. https://doi.org/10.3390/cells12202492
Sakato-Antoku M, Balsbaugh JL, King SM. N-Terminal Processing and Modification of Ciliary Dyneins. Cells. 2023; 12(20):2492. https://doi.org/10.3390/cells12202492
Chicago/Turabian StyleSakato-Antoku, Miho, Jeremy L. Balsbaugh, and Stephen M. King. 2023. "N-Terminal Processing and Modification of Ciliary Dyneins" Cells 12, no. 20: 2492. https://doi.org/10.3390/cells12202492
APA StyleSakato-Antoku, M., Balsbaugh, J. L., & King, S. M. (2023). N-Terminal Processing and Modification of Ciliary Dyneins. Cells, 12(20), 2492. https://doi.org/10.3390/cells12202492






