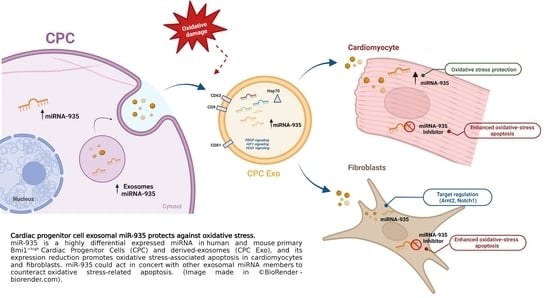
Cardiac Progenitor Cell Exosomal miR-935 Protects against Oxidative Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Culture Conditions
2.1.1. Primary Cells
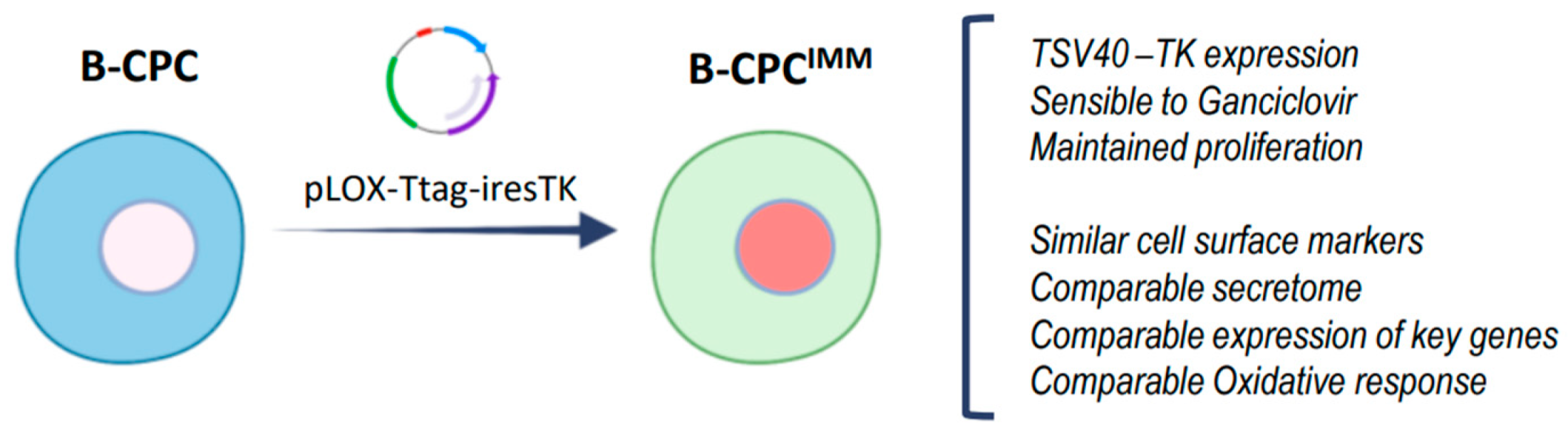
2.1.2. Cell Lines
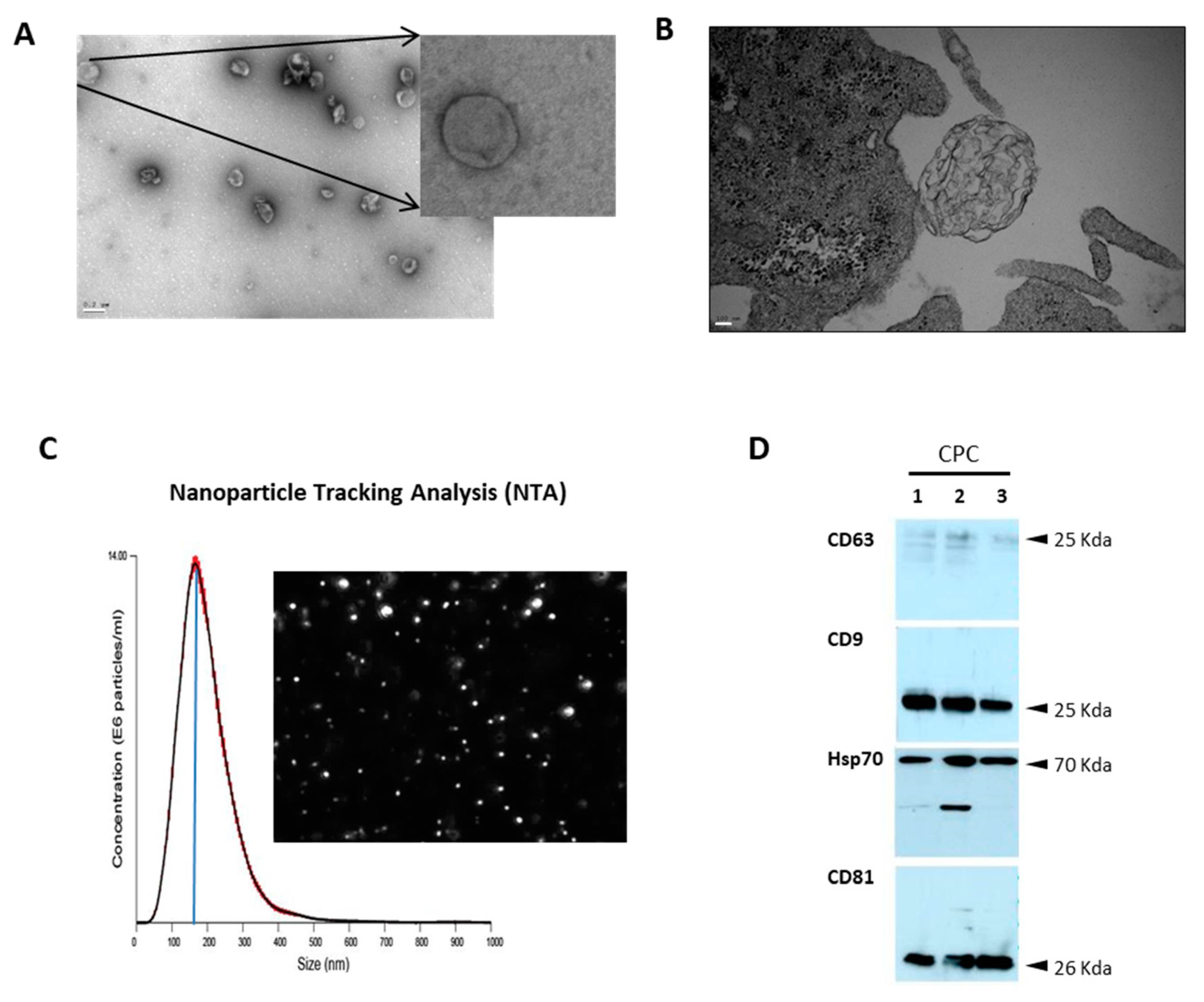
2.2. Exosome Isolation and Characterization
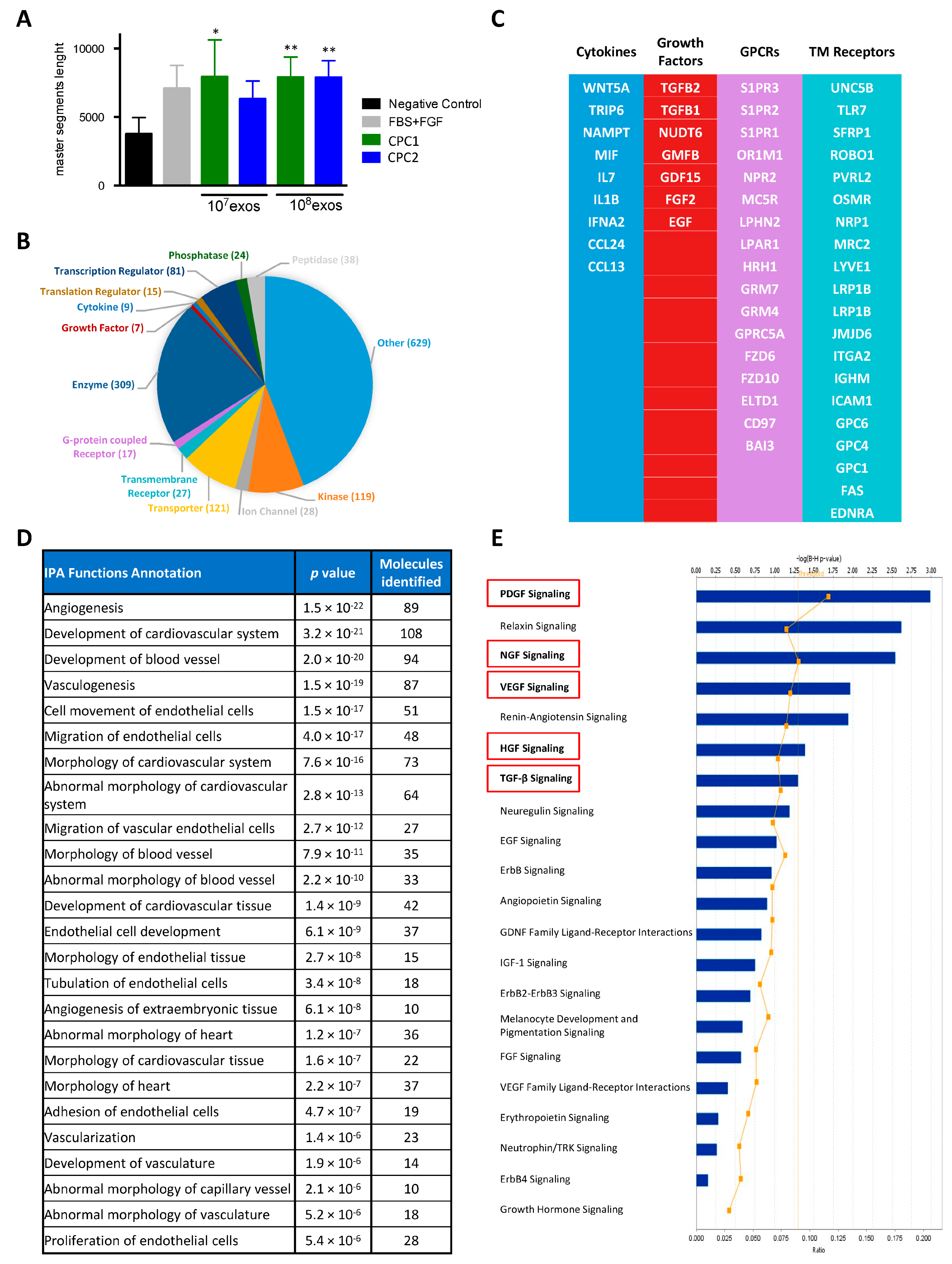
2.3. Label-Free Proteomics and Bioinformatics Analysis
2.4. Exosomal miRNA RNAseq and System Biology Analysis
2.5. Angiogenic Activity Determination
2.6. Co-Cultures Experiments
2.7. Flow Cytometry
2.8. Viability, Proliferation and Apoptosis Assays
2.9. Gene Silencing Assays
2.10. miRNA Vector Cloning and AAV Viral Vector Production
2.11. RT-qPCR Analyses
2.12. Western Blotting
2.13. Myocardial Infarction and AAV Administration
2.14. Echocardiographic Studies
2.15. Statistics
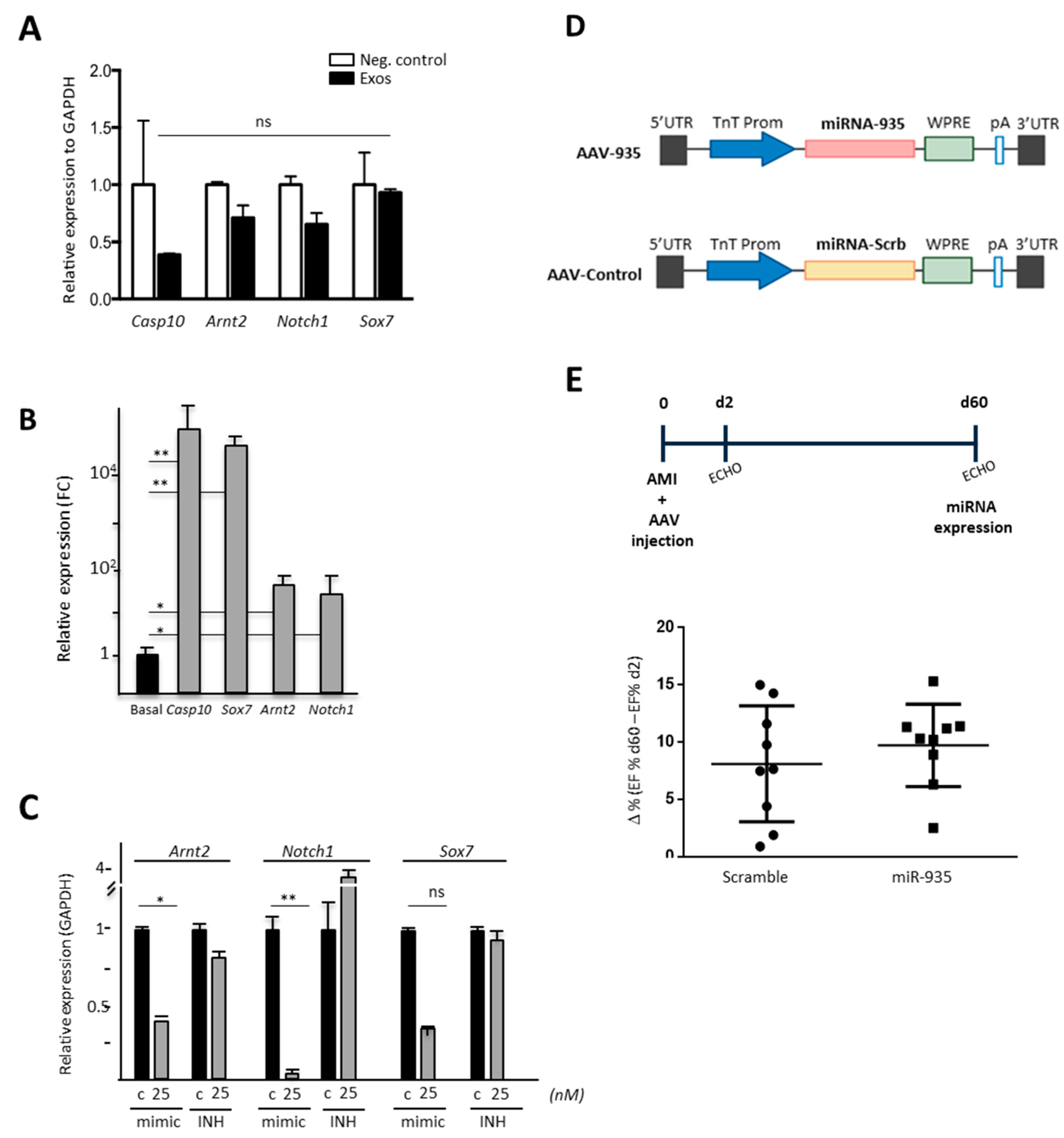
3. Results
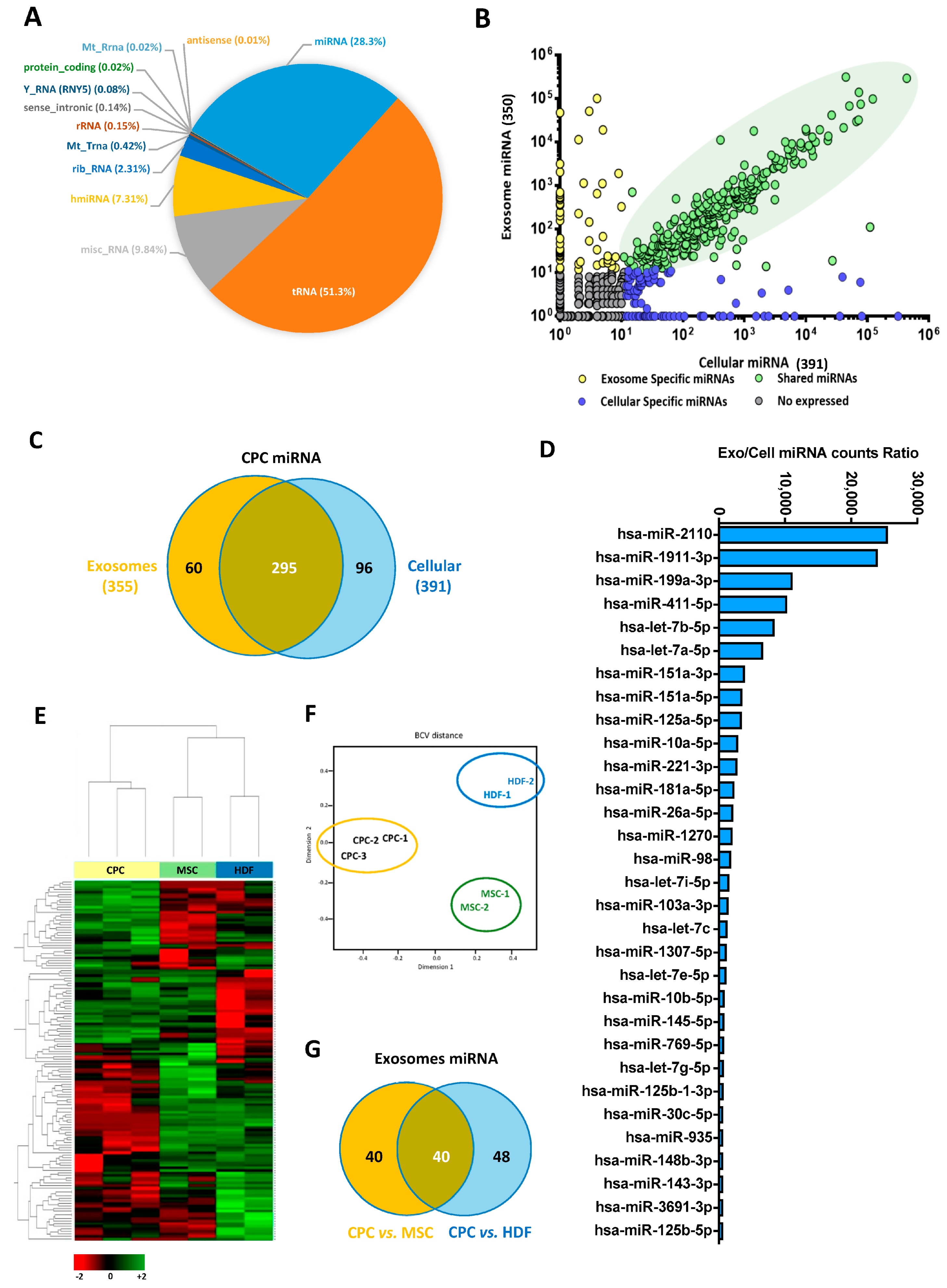
3.1. Comparative Proteomics Analysis of Adult Human Cardiac Progenitor Cell (CPC) Exosomal Compartment
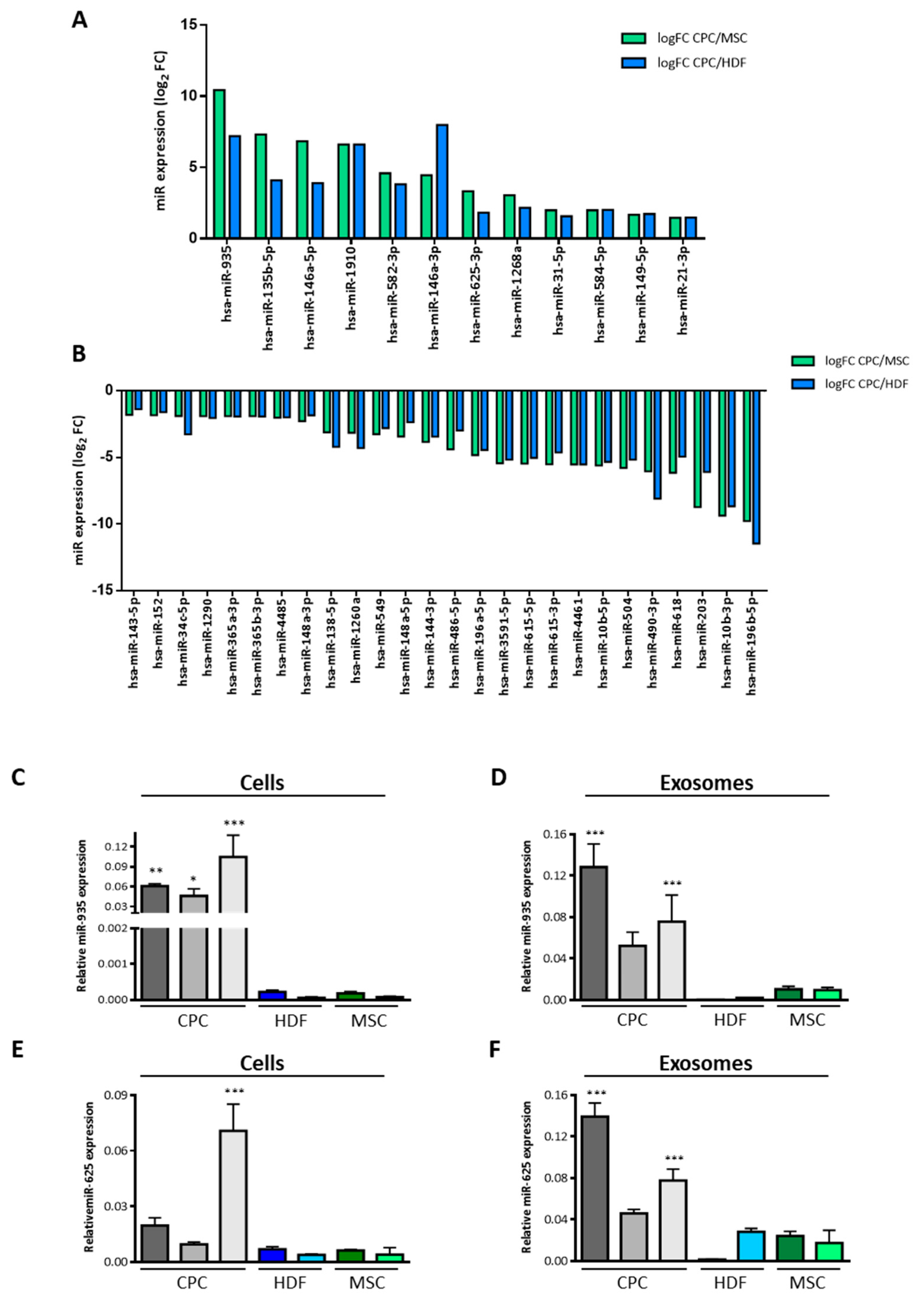
3.2. Specific miRNA Repertoire of Human CPC Exosomal Compartment
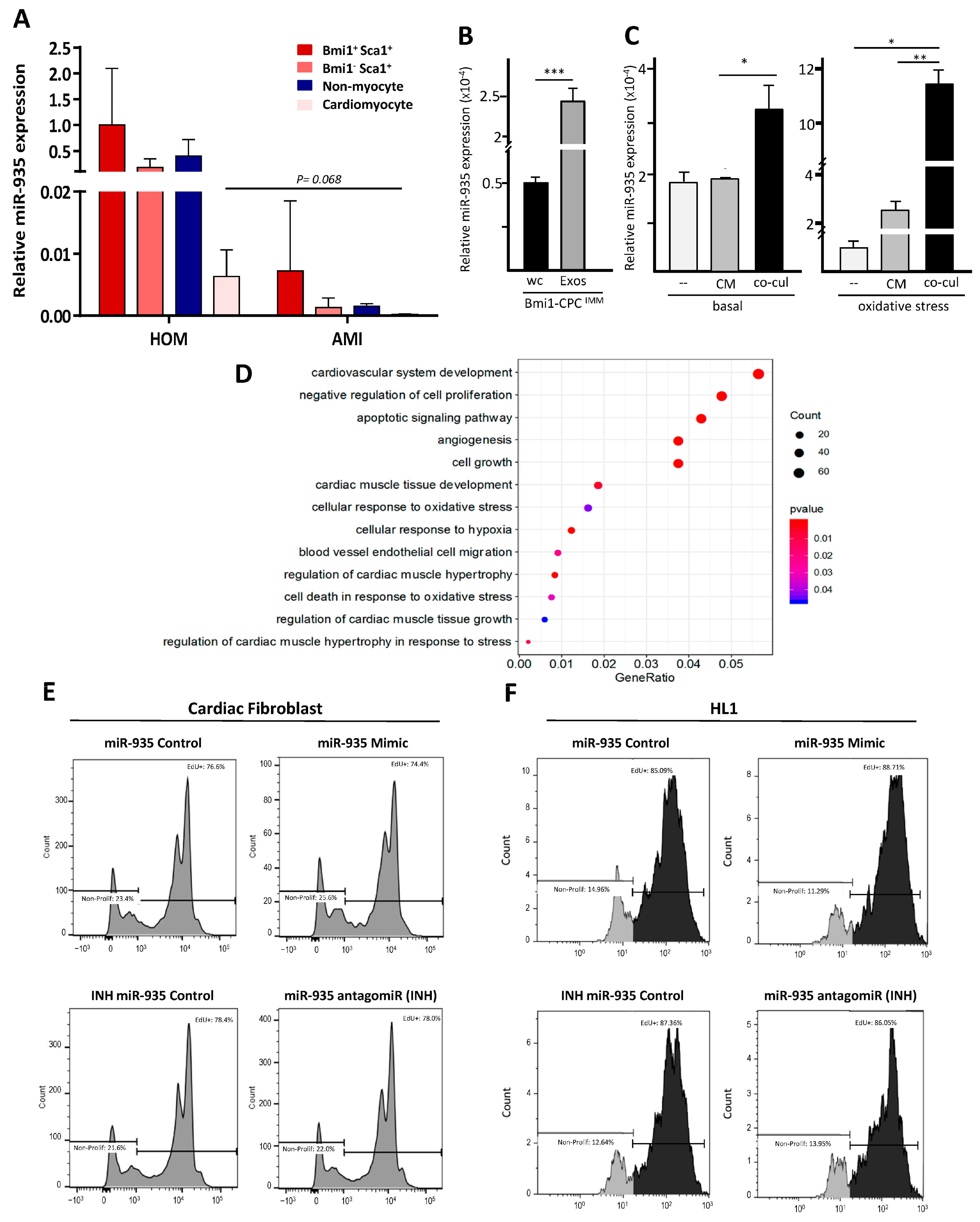
3.3. miR-935 in the Exosomal miRNA Repertoire of Murine Cardiac Progenitors
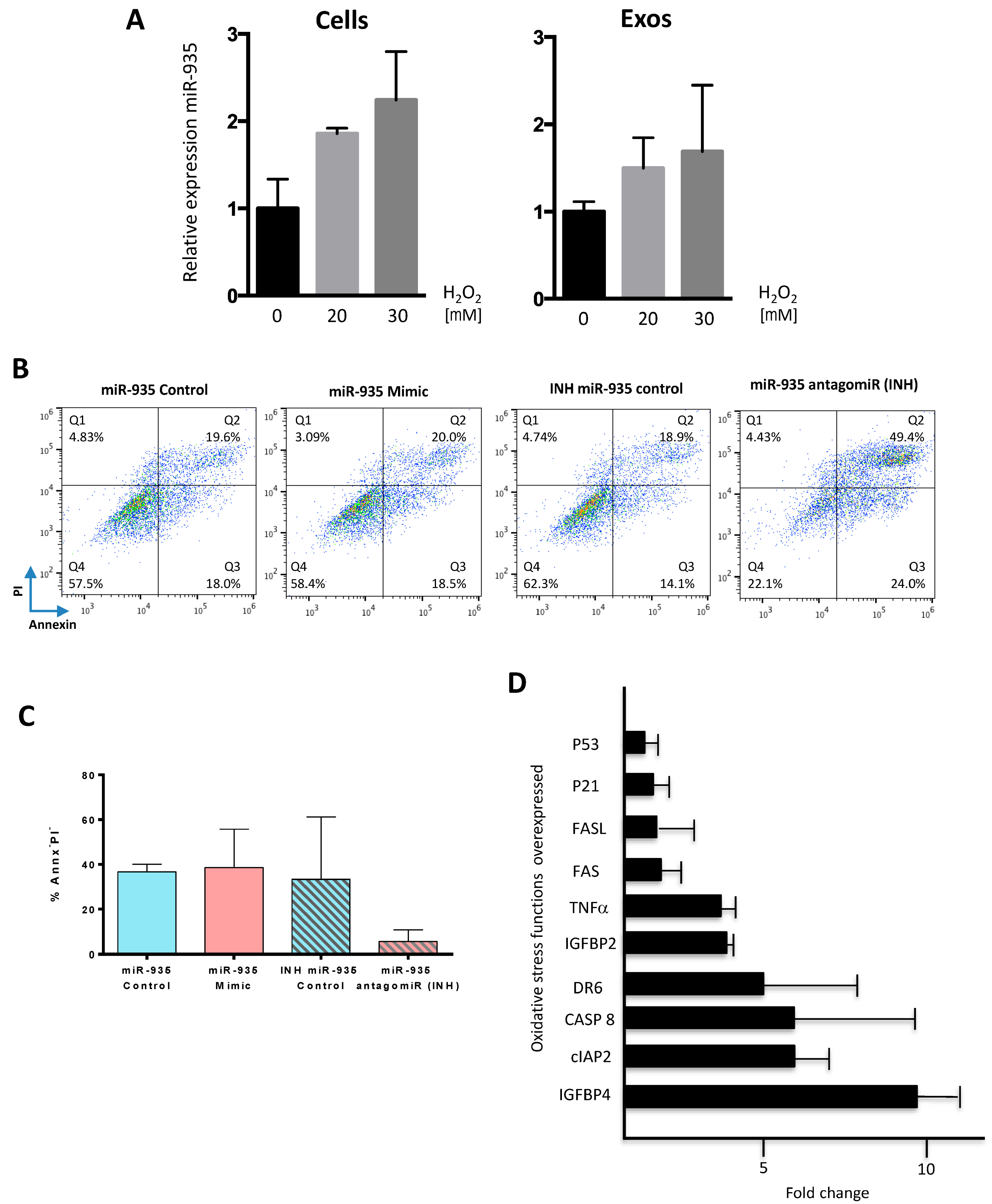
3.4. miR-935 Does Not Regulate Proliferation but It Is Involved in Antioxidative Damage Response
3.5. Evaluation of Putative miR-935 Targets in the Oxidative Damage Context
3.6. Evaluation of Cardioprotection Activity of miR-935 in a Mouse Model of Acute Myocardial Infarct
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno associated vectors |
| AMI | Acute Myocardial Infarct |
| Annex | Annexin |
| B-CPC | Murine cardiac progenitor cells Bmi1+ high |
| Bmi1-CPCIMM | Conditional SV40-immortalized B-CPC cells |
| BrdU | 5-bromo-2-desoxiuridina |
| CDC | Cardiosphere-derived cells |
| mCF | mouse Cardiac Fibroblasts |
| CM | Cardiomyocytes |
| CPC | Cardiac progenitor cells |
| Edu | 5-Ethynyl-2′-deoxyuridine |
| EF | Ejection Fraction |
| EV | Extracellular Vesicles |
| miRNA | microRNA |
| MSC | Mesenchymal stem cells |
| HDF | Human dermal fibroblasts |
| H2O2 | Hydrogen peroxide |
| HUVEC | Human Umbilical Vain Endothelial Cells |
| IPA | Ingenuity Pathway Analysis |
| LVEF | Left Ventricle Ejection Fraction |
| LogFC | Log2 fold-change |
| Non-CM | Non cardiomyocytes |
| PI | Propidium Iodide |
| RT-qPCR | Quantitative polymerase chain reaction coupled to reverse transcription |
| siRNA | Small interfering RNA |
| Tx | Tamoxifen |
References
- Bolli, R.; Solankhi, M.; Tang, X.L.; Kahlon, A. Cell Therapy in Patients with Heart Failure: A Comprehensive Review and Emerging Concepts. Cardiovasc. Res. 2021, 118, 951–976. [Google Scholar] [CrossRef]
- Eschenhagen, T.; Bolli, R.; Braun, T.; Field, L.J.; Fleischmann, B.K.; Frisén, J.; Giacca, M.; Hare, J.M.; Houser, S.R.; Lee, R.T. Cardiomyocyte Regeneration: A Consensus Statement. Circulation 2017, 36, 680–686. [Google Scholar] [CrossRef]
- Crisostomo, V.; Baez, C.; Abad, J.L.; Sanchez, B.; Alvarez, V.; Rosado, R.; Gómez-Mauricio, G.; Gheysens, O.; Blanco-Blazquez, V.; Blazquez, R.; et al. Dose-Dependent Improvement of Cardiac Function in a Swine Model of Acute Myocardial Infarction after Intracoronary Administration of Allogeneic Heart-Derived Cells. Stem Cell Res. Ther. 2019, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Crisostomo, V.; Baez-Diaz, C.; Maestre, J.; Garcia-Lindo, M.; Sun, F.; Casado, J.G.; Blazquez, R.; Abad, J.L.; Palacios, I.; Rodriguez-Borlado, L.; et al. Delayed Administration of Allogeneic Cardiac Stem Cell Therapy for Acute Myocardial Infarction could Ameliorate Adverse Remodeling: Experimental Study in Swine. J. Transl. Med. 2015, 13, 156. [Google Scholar] [CrossRef]
- Malliaras, K.; Smith, R.R.; Kanazawa, H.; Yee, K.; Seinfeld, J.; Tseliou, E.; Dawkins, J.F.; Kreke, M.; Cheng, K.; Luthringer, D. Validation of Contrast-Enhanced Magnetic Resonance Imaging to Monitor Regenerative Efficacy after Cell Therapy in a Porcine model of Convalescent Myocardial Infarction. Circulation 2013, 128, 2764–2775. [Google Scholar] [CrossRef]
- Malliaras, K.; Li, T.S.; Luthringer, D.; Terrovitis, J.; Cheng, K.; Chakravarty, T.; Galang, G.; Zhang, Y.; Schoenhoff, F.; Van Eyk, J.; et al. Safety and Efficacy of Allogeneic Cell Therapy in Infarcted Rats Transplanted with Mismatched Cardiosphere-Derived Cells. Circulation 2012, 125, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, S.; Ohtsuki, S.; Tarui, S.; Ousaka, D.; Eitoku, T.; Kondo, M.; Okuyama, M.; Kobayashi, J.; Baba, K.; Arai, S.; et al. Intracoronary Autologous Cardiac Progenitor Cell Transfer in Patients with Hypoplastic Left Heart Syndrome: The TICAP Prospective Phase 1 Controlled Trial. Circ. Res. 2015, 116, 653–664. [Google Scholar] [CrossRef]
- Malliaras, K.; Makkar, R.R.; Smith, R.R.; Cheng, K.; Wu, E.; Bonow, R.O.; Marbán, L.; Mendizabal, A.; Cingolani, E.; Johnston, P.V. Intracoronary Cardiosphere-Derived Cells after Myo-Cardial Infarction: Evidence for Therapeutic Regeneration in the Final 1-year Results of the CADUCEUS Trial. J. Am. Coll. Cardiol. 2014, 63, 110–122. [Google Scholar] [CrossRef]
- Hirai, K.; Ousaka, D.; Fukushima, Y.; Kondo, M.; Oh, H. Cardiosphere-Derived Exosomal microRNAs for Myocardial Repair in Pediatric Dilated Cardiomyopathy. Sci. Transl. Med. 2020, 12, eabb3336. [Google Scholar] [CrossRef]
- Chakravarty, T.; Makkar, R.R.; Ascheim, D.D.; Traverse, J.H.; Schatz, R.; Demaria, A.; Francis, G.S.; Povsic, T.J.; Smith, R.R.; Lima, J.A.; et al. ALLogeneic Heart STem Cells to Achieve Myocardial Regeneration (ALLSTAR) Trial: Rationale and Design. Cell Transplant. 2017, 26, 205–214. [Google Scholar] [CrossRef]
- Fernández-Avilés, F.; Sanz-Ruiz, R.; Bogaert, J.; Plasencia, A.C.; Gilaberte, I.; Belmans, A.; Fernández-Santos, M.E.; Charro, D.; Mulet, M.; Yotti, R.; et al. Safety and Efficacy of Intra-coronary Infusion of Allogeneic Human Cardiac Stem Cells in Patients With ST-Segment Elevation Myocardial Infarction and Left Ventricular Dysfunction. Circ. Res. 2018, 123, 579–589. [Google Scholar] [CrossRef]
- Maxeiner, H.; Krehbiehl, N.; Müller, A.; Woitasky, N.; Akintürk, H.; Müller, M.; Weigand, M.A.; Abdallah, Y.; Kasseckert, S.; Schreckenberg, R.; et al. New Insights into Paracrine Mechanisms of Human Cardiac Progenitor Cells. Eur. J. Heart Fail. 2010, 12, 730–737. [Google Scholar] [CrossRef]
- Pavo, N.; Zimmermann, M.; Pils, D.; Mildner, M.; Petrási, Z.; Petneházy, O.; Fuzik, J.; Jakab, A.; Gabriel, C.; Sipos, W.; et al. Long-Acting Beneficial Effect of Percutaneously Intramyocardially Delivered Secretome of Apoptotic Peripheral Blood Cells on Porcine Chronic Ischemic Left Ventricular Dysfunction. Biomaterials 2014, 35, 3541–3550. [Google Scholar] [CrossRef]
- Vagnozzi, R.J.; Maillet, M.; Sargent, M.A.; Khalil, H.; Johansen, A.K.Z.; Schwanekamp, J.A.; York, A.J.; Huang, V.; Nahrendorf, M.; Sadayappan, S.; et al. An Acute Immune Response Under-Lies the Benefit of Cardiac Stem Cell Therapy. Nature 2018, 577, 405–409. [Google Scholar] [CrossRef]
- Gaceb, A.; Martinez, M.C.; Andriantsitohaina, R. Extracellular Vesicles: New Players in Cardiovascular Diseases. Int. J. Biochem. Cell Biol. 2014, 50, 24–28. [Google Scholar] [CrossRef][Green Version]
- Estébanez, B.; Jiménez-Pavón, D.; Huang, C.J.; Cuevas, M.J.; González-Gallego, J. Effects of exercise on exosome release and cargo in in vivo and ex vivo models: A systematic review. J. Cell. Physiol. 2021, 236, 3336–3353. [Google Scholar] [CrossRef]
- Qiao, L.; Hu, S.; Liu, S.; Zhang, H.; Ma, H.; Huang, K.; Li, Z.; Su, T.; Vandergriff, A.; Tang, J.; et al. microRNA-21-5p Dysregulation in Exosomes Derived from Heart Failure Patients Impairs Regenerative Potential. J. Clin. Investig. 2019, 129, 2237–2250. [Google Scholar] [CrossRef]
- Kita, S.; Maeda, N.; Shimomura, I. Interorgan Communication by Exosomes, Adipose Tissue, and Adiponectin in Metabolic Syndrome. J. Clin. Investig. 2019, 129, 4041–4049. [Google Scholar] [CrossRef]
- Ibrahim, A.; Marbán, E. Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu. Rev. Physiol. 2016, 78, 67–83. [Google Scholar] [CrossRef]
- Huang, P.; Wang, L.; Li, Q.; Xu, J.; Xu, J.; Xiong, Y.; Chen, G.; Qian, H.; Jin, C.; Yu, Y.; et al. Combinatorial Treatment of Acute Myocardial Infarction Using Stem Cells and their Derived Exosomes Resulted in Improved Heart Performance. Stem Cell Res. Ther. 2019, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, S.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Xu, J.; Xia, K.; Chang, Y.; et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer 2018, 17, 82. [Google Scholar] [CrossRef]
- Ibrahim, A.G.-E.; Cheng, K.; Marbán, E. Exosomes as Critical Agents of Cardiac Regeneration Triggered by Cell Therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef]
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by Cardiac Progenitor Cell-Secreted Exosomes: Role of Pregnancy-Associated Plasma Protein-A. Cardiovasc. Res. 2018, 114, 992–1005. [Google Scholar] [CrossRef]
- Gallet, R.; Dawkins, J.; Valle, J.; Simsolo, E.; De Couto, G.; Middleton, R.; Tseliou, E.; Luthringer, D.; Kreke, M.; Smith, R.R.; et al. Exosomes Secreted by Cardiosphere-Derived Cells Reduce Scarring, Attenuate Adverse Remodelling, and Improve Function in Acute and Chronic Porcine Myocardial Infarction. Eur. Heart J. 2017, 38, 201–211. [Google Scholar] [CrossRef]
- Barile, L.; Moccetti, T.; Marbán, E.; Vassalli, G. Roles of Exosomes in Cardioprotection. Eur. Heart J. 2016, 38, 1372–1379. [Google Scholar] [CrossRef]
- Huang, C.K.; Kafert-Kasting, S.; Thum, T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardio-vascular Disease. Circ. Res. 2020, 26, 663–678. [Google Scholar] [CrossRef]
- Täubel, J.; Hauke, W.; Rump, S.; Viereck, J.; Batkai, S.; Poetzsch, J.; Laura, R.; Henning, W.; Celina, G.; Ulrike, L.; et al. Novel Antisense Therapy Targeting microRNA-132 in Patients with Heart Failure: Results of a First-in-Human Phase 1b Randomized, Double-Blind, Placebo-Controlled Study. Eur. Heart. J. 2021, 42, 178–188. [Google Scholar] [CrossRef]
- Lauden, L.; Boukouaci, W.; Borlado, L.R.; López, I.P.; Sepúlveda, P.; Tamouza, R.; Charron, D.; Al-Daccak, R. Allogenicity of Human Cardiac Stem/Progenitor Cells Orchestrated by Programmed Death Ligand 1. Circ. Res. 2013, 112, 451–464. [Google Scholar] [CrossRef]
- Torán, J.L.; Aguilar, S.; López, J.A.; Torroja, C.; Quintana, J.A.; Santiago, C.; Abad, J.L.; Gomes-Alves, P.; Gonzalez, A.; Bernal, J.A.; et al. CXCL6 is an Important Paracrine Factor in the Pro-angiogenic Human Cardiac Progenitor-Like Cell Secretome. Sci. Rep. 2017, 7, 12490. [Google Scholar] [CrossRef]
- Torán, J.L.; López, J.A.; Gomes-Alves, P.; Aguilar, S.; Torroja, C.; Trevisan-Herraz, M.; Moscoso, I.; Sebastião, M.J.; Serra, M.; Brito, C.; et al. Definition of a Cell Surface Signature for Human Cardiac Progenitor Cells after Comprehensive Comparative Transcriptomic and Proteomic Characterization. Sci. Rep. 2019, 9, 4647. [Google Scholar] [CrossRef]
- Moscoso, I.; Tejados, N.; Barreiro, O.; Sepúlveda, P.; Izarra, A.; Calvo, E.; Dorronsoro, A.; Salcedo, J.M.; Sádaba, R.; Díez-Juan, A. Podocalyxin-like Protein 1 is a Relevant Marker for Human c-kit(pos) Cardiac Stem Cells. J. Tissue Eng. Regen. Med. 2016, 10, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Herrero, D.; Cañón, S.; Albericio, G.; Carmona, R.M.; Aguilar, S.; Mañes, S.; Bernad, A. Age-Related Oxidative Stress Confines Damage-Responsive Bmi1+ Cells to Perivascular Regions in the Murine Adult Heart. Redox Biol. 2019, 22, 101156. [Google Scholar] [CrossRef]
- Valiente-Alandi, I.; Albo-Castellanos, C.; Herrero, D.; Arza, E.; Garcia-Gomez, M.; Segovia, J.C.; Capecchi, M.; Bernad, A. Cardiac Bmi1+ Cells Contribute to Myocardial Renewal in the Murine Adult Heart. Stem Cell Res. Ther. 2015, 6, 205. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Albericio, G.; Aguilar, S.; Torán, J.L.; Yañez, R.; López, J.A.; Vázquez, J.; Mora, C.; Bernad, A. Comparative Proteomic Analysis of Nuclear and Cyto-plasmic Compartments in Human Cardiac Progenitor Cells. Sci. Rep. 2022, 12, 146. [Google Scholar] [CrossRef]
- Valiente-Alandi, I.; Albo-Castellanos, C.; Herrero, D.; Sanchez, I.; Bernad, A. Bmi1+ Cardiac Progenitor Cells Contribute to Myo-cardial Repair Following Acute Injury. Stem Cell Res. Ther. 2016, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.F.; Lin, C.T.; Chen, W.C.; Yang, C.T.; Chen, C.C.; Liao, S.K.; Liu, J.M.; Lu, C.H.; Lee, K.D. The Sensitivity of Human Mesenchymal Stem Cells to Ionizing Radiation. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 244–253. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Zeng, Y.; Wagner, E.J.; Cullen, B.R. Both Natural and Designed Micro RNAs Can Inhibit the Expression of Cognate mRNAs When Expressed in Human Cells. Mol. Cell 2002, 9, 1327–1333. [Google Scholar] [CrossRef]
- García-Olloqui, P.; Rodriguez-Madoz, J.R.; Di Scala, M.; Abizanda, G.; Vales, A.; Olagüe, C.; Iglesias-García, O.; Larequi, E.; Aguado-Alvaro, L.P.; Ruiz-Villalba, A.; et al. Effect of Heart Ischemia and Administration Route on Biodistribution and Transduction Efficiency of AAV9 Vectors. J. Tissue Eng. Regen. Med. 2019, 14, 123–134. [Google Scholar] [CrossRef]
- Zufferey, R.; Donello, J.E.; Trono, D.; Hope, T.J. Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances Expression of Transgenes Delivered by Retroviral Vectors. J. Virol. 1999, 73, 2886–2892. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Villalba, A.; Romero, J.P.; Hernández, S.C.; Vilas-Zornoza, A.; Fortelny, N.; Castro-Labrador, L.; Martin-Uriz, P.S.; Lorenzo-Vivas, E.; García-Olloqui, P.; Palacio, M.; et al. Single-Cell RNA Sequencing Analysis Reveals a Crucial Role for CTHRC1 (Collagen Triple Helix Repeat Containing 1) Cardiac Fibroblasts After Myocardial Infarction. Circulation 2020, 142, 1831–1847. [Google Scholar] [CrossRef]
- Benavides-Vallve, C.; Corbacho, D.; Iglesias-Garcia, O.; Pelacho, B.; Albiasu, E.; Castaño, S.; Muñoz-Barrutia, A.; Prosper, F.; Ortiz-de-Solorzano, C. New Strategies for Echocardio-Graphic Evaluation of Left Ventricular Function in a Mouse Model of Long-Term Myocardial Infarction. PLoS ONE 2012, 7, e41691. [Google Scholar] [CrossRef]
- Tseliou, E.; Pollan, S.; Malliaras, K.; Terrovitis, J.; Sun, B.; Galang, G.; Marbán, L.; Luthringer, D.; Marbán, E. Allogeneic Cardiospheres Safely Boost Cardiac Function and Attenuate Adverse Remodeling After Myocardial Infarction in Immunologically Mismatched Rat Strains. J. Am. Coll. Cardiol. 2013, 61, 1108–1119. [Google Scholar] [CrossRef][Green Version]
- Li, T.-S.; Cheng, K.; Malliaras, K.; Matsushita, N.; Sun, B.; Marbán, L.; Zhang, Y.; Marbán, E. Expansion of Human Cardiac Stem Cells in Physiological Oxygen Improves Cell Production Efficiency and Potency for Myocardial Repair. Cardiovasc. Res. 2010, 89, 157–165. [Google Scholar] [CrossRef]
- Dougherty, J.A.; Patel, N.; Kumar, N.; Rao, S.G.; Angelos, M.G.; Singh, H.; Cai, C.; Khan, M. Human Cardiac Progenitor Cells Enhance Exosome Release and Promote Angiogenesis Under Physoxia. Front. Cell Dev. Biol. 2020, 8, 130. [Google Scholar] [CrossRef]
- Xu, H.; Wang, Z.; Liu, L.; Zhang, B.; Li, B. Exosomes Derived from Adipose Tissue, Bone Marrow, and Umbilical Cord Blood for Cardioprotection after Myocardial Infarction. J. Cell. Biochem. 2020, 121, 2089–2102. [Google Scholar] [CrossRef]
- Ferguson, S.W.; Wang, J.; Lee, C.J.; Liu, M.; Neelamegham, S.; Canty, J.M.; Nguyen, J. The microRNA Regulatory Landscape of MSC-Derived Exosomes: A Systems View. Sci. Rep. 2018, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Herrero, D.; Cañón, S.; Pelacho, B.; Salvador-Bernáldez, M.; Aguilar, S.; Pogontke, C.; Carmona, R.M.; Salvador, J.M.; Perez-Pomares, J.M.; Klein, O.D.; et al. Bmi1-Progenitor Cell Ablation Impairs the Angiogenic Response to Myocardial Infarction. Arter. Thromb. Vasc. Biol. 2018, 38, 2160–2173. [Google Scholar] [CrossRef]
- Cruz, F.M.; Tomé, M.; Bernal, J.A.; Bernad, A. miR-300 mediates Bmi1 Function and Regulates Differentiation in Primitive Cardiac Progenitors. Cell Death Dis. 2015, 6, e1953. [Google Scholar] [CrossRef] [PubMed]
- Albericio, G.; Higuera, M.; Herrero, D.; García-Brenes, M.A.; Torres, M.; Moreno, I. Bidirectional Regulation Between Adult Cardiac Progenitor Cells and Cardiac Endothelium Defines a Minimal Vascular Niche. 2023; in preparation. [Google Scholar]
- Zhang, X.; Li, F.; Zhou, Y.; Mao, F.; Lin, Y.; Shen, S.; Li, Y.; Zhang, S.; Sun, Q. Long noncoding RNA AFAP1-AS1 Promotes Tumor Progression and Invasion by Regulating the miR-2110/Sp1 Axis in Triple-negative Breast Cancer. Cell Death Dis. 2021, 12, 627. [Google Scholar] [CrossRef]
- Mendonça, D.B.; Nguyen, J.T.; Haidar, F.; Fox, A.L.; Ray, C.; Amatullah, H.; Liu, F.; Kim, J.K.; Krebsbach, P.H. MicroRNA-1911-3p Targets mEAK-7 to Suppress mTOR Signaling in Human Lung Cancer Cells. Heliyon 2020, 6, e05734. [Google Scholar] [CrossRef]
- Taheri, M.; Safarzadeh, A.; Hussen, B.M.; Ghafouri-Fard, S.; Baniahmad, A. LncRNA/miRNA/mRNA Network Introduces Novel Biomarkers in Prostate Cancer. Cells 2022, 11, 3776. [Google Scholar] [CrossRef]
- Milano, G.; Biemmi, V.; Lazzarini, E.; Balbi, C.; Ciullo, A.; Bolis, S.; Ameri, P.; Di Silvestre, D.; Mauri, P.; Barile, L.; et al. Intravenous Administration of Cardiac Progenitor Cell-Derived Exosomes Protects against Doxorubicin/Trastuzumab-Induced Cardiac Toxicity. Cardiovasc. Res. 2020, 116, 383–392. [Google Scholar] [CrossRef]
- Raitoharju, E.; Seppälä, I.; Oksala, N.; Lyytikäinen, L.P.; Raitakari, O.; Viikari, J.; Ala-Korpela, M.; Soininen, P.; Kangas, A.J.; Waldenberger, M.; et al. Blood microRNA Profile Associates with the Levels of Serum Lipids and Metabolites Associated with Glucose Metabolism and Insulin Resistance and Pinpoints Pathways Underlying Metabolic Syndrome: The Cardiovascular Risk in Young Finns Study. Mol. Cell. Endocrinol. 2014, 391, 41–49. [Google Scholar] [CrossRef]
- Verma, K.; Jyotsana, N.; Buenting, I.; Luther, S.; Pfanne, A.; Thum, T.; Ganser, A.; Heuser, M.; Weissinger, E.M.; Hambach, L. miR-625-3p is Upregulated in CD8+ T cells During Early Immune Reconstitution after Allogeneic Stem Cell Transplantation. PLoS ONE 2017, 12, e0183828. [Google Scholar] [CrossRef]
- Zhang, M.; Xiong, F.; Zhang, S.; Guo, W.; He, Y. Crucial Roles of miR-625 in Human Cancer. Front. Med. 2022, 9, 845094. [Google Scholar] [CrossRef]
- Lu, M.; Huang, Y.; Sun, W.; Li, P.; Li, L.; Li, L. miR-135b-5p promotes gastric cancer progression by targeting CMTM3. Int. J. Oncol. 2017, 52, 589–598. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA Sequence Codes for Small Extracellular Vesicle Release and Cellular Retention. Nature 2021, 601, 446–451. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Xu, J.; Niu, W. microRNA-935 is Reduced in Non-small Cell Lung Cancer Tissue, is Linked to Poor Outcome, and Acts on Signal Transduction Mediator E2F7 and the AKT Pathway. Br. J. Biomed. Sci. 2018, 76, 17–23. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Q.; Zhao, X.; Cui, B.; Zhang, L.; Wang, Q. MicroRNA-935 Inhibits Proliferation and Invasion of Osteosarcoma Cells by Directly Targeting High Mobility Group Box 1. Oncol. Res. 2018, 26, 1439–1446. [Google Scholar] [CrossRef]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Beckler, M.D.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-Dependent Sorting of miRNA to Exosomes. eLife 2015, 4, e07197. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, X.Y.; Li, P.F.; Fan, Y.N.; Zhang, L.L.; Ma, X.H.; Sun, R.B.; Liu, Y.W.; Li, W.Y. MicroRNA-935-Modified Bone Marrow Mesenchymal Stem Cells-Derived Exosomes Enhance Osteoblast Proliferation and Differentiation in Osteoporotic Rats. Life Sci. 2021, 272, 119204. [Google Scholar] [CrossRef]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef]
- Zhou, X.-L.; Wan, L.; Xu, Q.-R.; Zhao, Y.; Liu, J.-C. Notch Signaling Activation Contributes to Cardioprotection Provided by Ischemic Preconditioning and Postconditioning. J. Transl. Med. 2013, 11, 251. [Google Scholar] [CrossRef]
- Yang, R.S.; Wang, Y.H.; Ding, C.; Su, X.H.; Gong, X.B. MiR-146 Regulates the Repair and Regeneration of Intervertebral Nucleus Pulposus Cells via Notch1 Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4591–4598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.K.; Du, Y.; He, Y.Q.; Liu, Y.W.; Zhang, G.L.; Yang, C.X.; Gao, F. INT-HA Induces M2-Like Macrophage Differentiation of Human Monocytes via TLR4-miR-935 Pathway. Cancer Immunol. Immunother. 2019, 68, 189–200. [Google Scholar] [CrossRef]
- Beischlag, T.V.; Morales, J.L.; Brett, D.; Perdew, G.H.; Hollingshead, B.D. The Aryl Hydrocarbon Receptor Complex and the Control of Gene Expression. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 207–250. [Google Scholar] [CrossRef]
- Mandl, M.; Depping, R. Hypoxia-Inducible Aryl Hydrocarbon Receptor nuclear Translocator (ARNT) (HIF-1β): Is it a Rare Exception? Mol. Med. 2014, 20, 215–220. [Google Scholar] [CrossRef]
- Bi, Y.; Yang, Q.; Li, Z.; Wang, Y.; Wang, Y.; Jia, A.; Pan, Z.; Yang, R.; Liu, G. Aryl hydrocarbon Receptor Nuclear Translocator Limits the Recruitment and Function of Regulatory Neutrophils against Colorectal Cancer by Regulating the Gut Microbiota. J. Exp. Clin. Cancer Res. 2023, 42, 53. [Google Scholar] [CrossRef] [PubMed]
- Bogeas, A.; Morvan-Dubois, G.; El-Habr, E.A.; Lejeune, F.-X.; Defrance, M.; Narayanan, A.; Kuranda, K.; Burel-Vandenbos, F.; Sayd, S.; Delaunay, V.; et al. Changes in Chromatin State Reveal ARNT2 at a Node of a tumorigenic Transcription Factor Signature Driving Glioblastoma Cell Aggressiveness. Acta Neuropathol. 2017, 135, 267–283. [Google Scholar] [CrossRef]
- Weidenbusch, M.; Rodler, S.; Song, S.; Romoli, S.; Marschner, J.A.; Kraft, F.; Holderied, A.; Kumar, S.; Mulay, S.R.; Honarpisheh, M. Gene Expression Profiling of the Notch-AhR-IL22 Axis at Homeostasis and in Response to Tissue Injury. Biosci. Rep. 2017, 37, BSR20170099. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Yang, J.J.; Zhao, X.Q.; Zhang, E.; Zeng, Q.T.; Yu, Y.; Yang, L.; Wu, B.W.; Yi, G.W.; Mao, X.B.; et al. Circulating Myocardial microRNAs from Infarcted Hearts are Carried in Exosomes and Mobilise Bone Marrow Progenitor Cells. Nat. Commun. 2019, 10, 959. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar, S.; García-Olloqui, P.; Amigo-Morán, L.; Torán, J.L.; López, J.A.; Albericio, G.; Abizanda, G.; Herrero, D.; Vales, Á.; Rodríguez-Diaz, S.; et al. Cardiac Progenitor Cell Exosomal miR-935 Protects against Oxidative Stress. Cells 2023, 12, 2300. https://doi.org/10.3390/cells12182300
Aguilar S, García-Olloqui P, Amigo-Morán L, Torán JL, López JA, Albericio G, Abizanda G, Herrero D, Vales Á, Rodríguez-Diaz S, et al. Cardiac Progenitor Cell Exosomal miR-935 Protects against Oxidative Stress. Cells. 2023; 12(18):2300. https://doi.org/10.3390/cells12182300
Chicago/Turabian StyleAguilar, Susana, Paula García-Olloqui, Lidia Amigo-Morán, José Luis Torán, Juan Antonio López, Guillermo Albericio, Gloria Abizanda, Diego Herrero, África Vales, Saray Rodríguez-Diaz, and et al. 2023. "Cardiac Progenitor Cell Exosomal miR-935 Protects against Oxidative Stress" Cells 12, no. 18: 2300. https://doi.org/10.3390/cells12182300
APA StyleAguilar, S., García-Olloqui, P., Amigo-Morán, L., Torán, J. L., López, J. A., Albericio, G., Abizanda, G., Herrero, D., Vales, Á., Rodríguez-Diaz, S., Higuera, M., García-Martín, R., Vázquez, J., Mora, C., González-Aseguinolaza, G., Prosper, F., Pelacho, B., & Bernad, A. (2023). Cardiac Progenitor Cell Exosomal miR-935 Protects against Oxidative Stress. Cells, 12(18), 2300. https://doi.org/10.3390/cells12182300