The Material Properties of the Cell Nucleus: A Matter of Scale
Abstract
1. Introduction
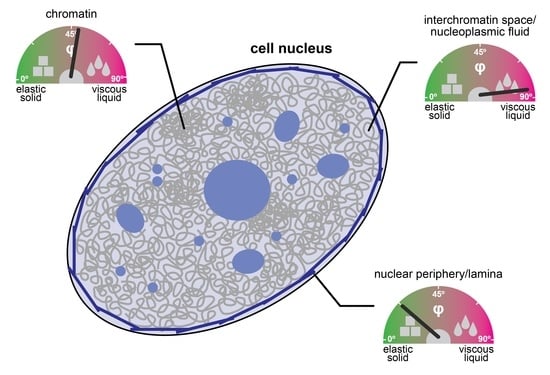
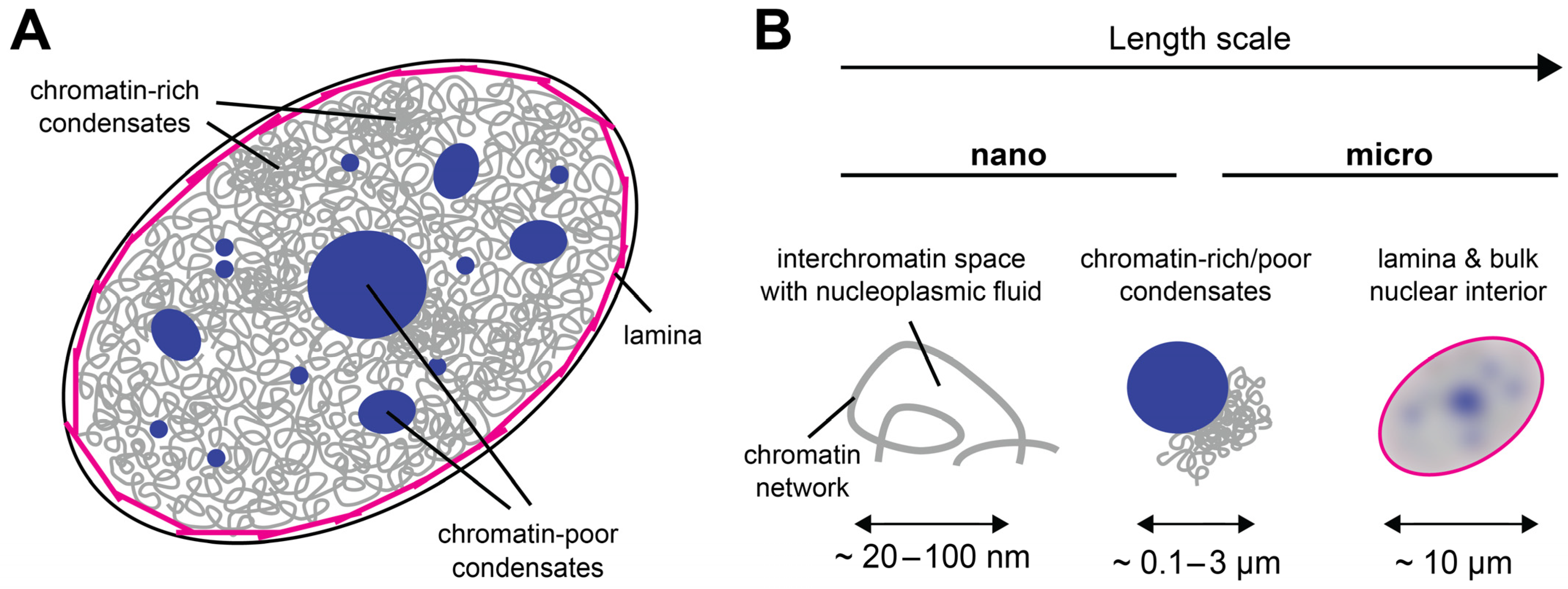
2. The Cell Nucleus as a Multiscale Porous Medium
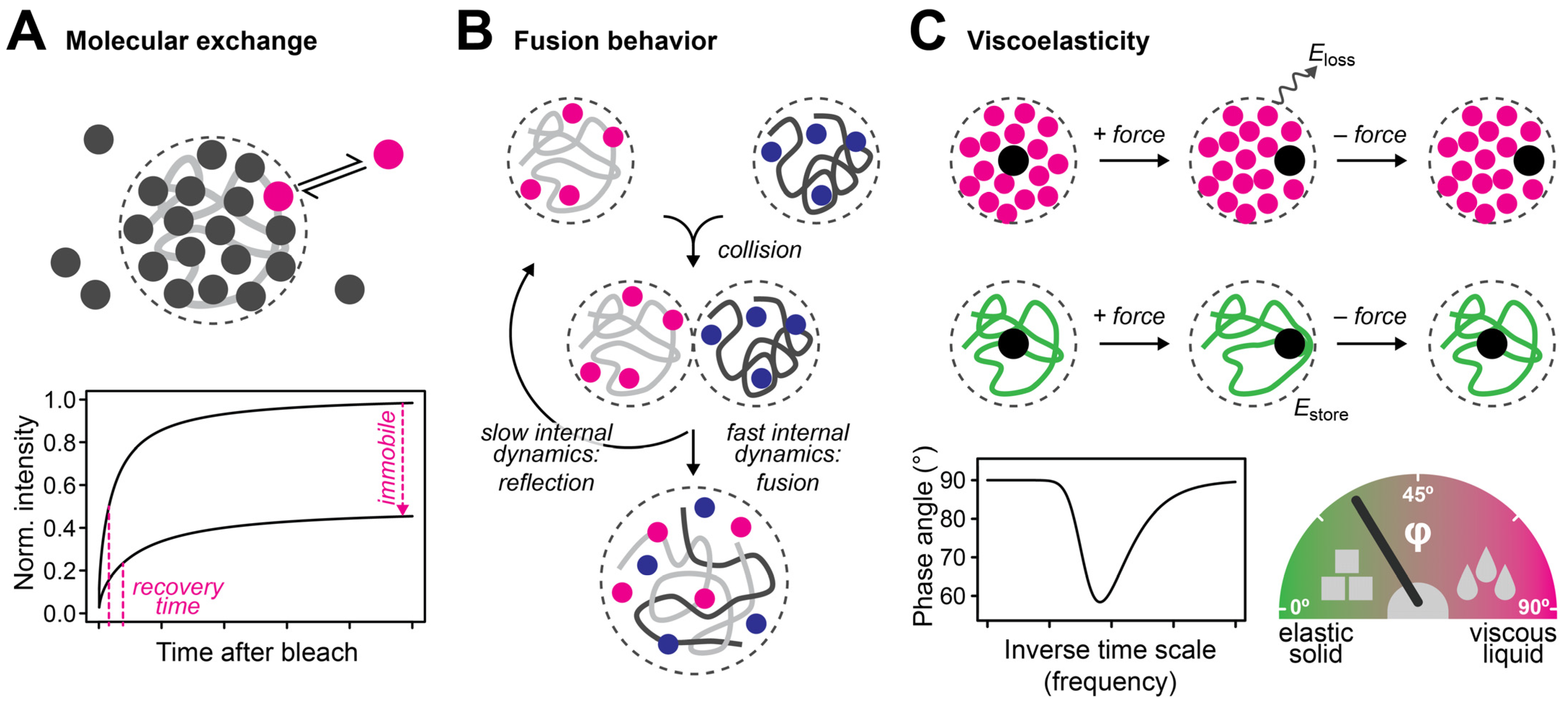
3. Liquid- and Solid-like Features of Multiscale Porous Media
4. Scale-Dependent Material Properties of the Nuclear Interior
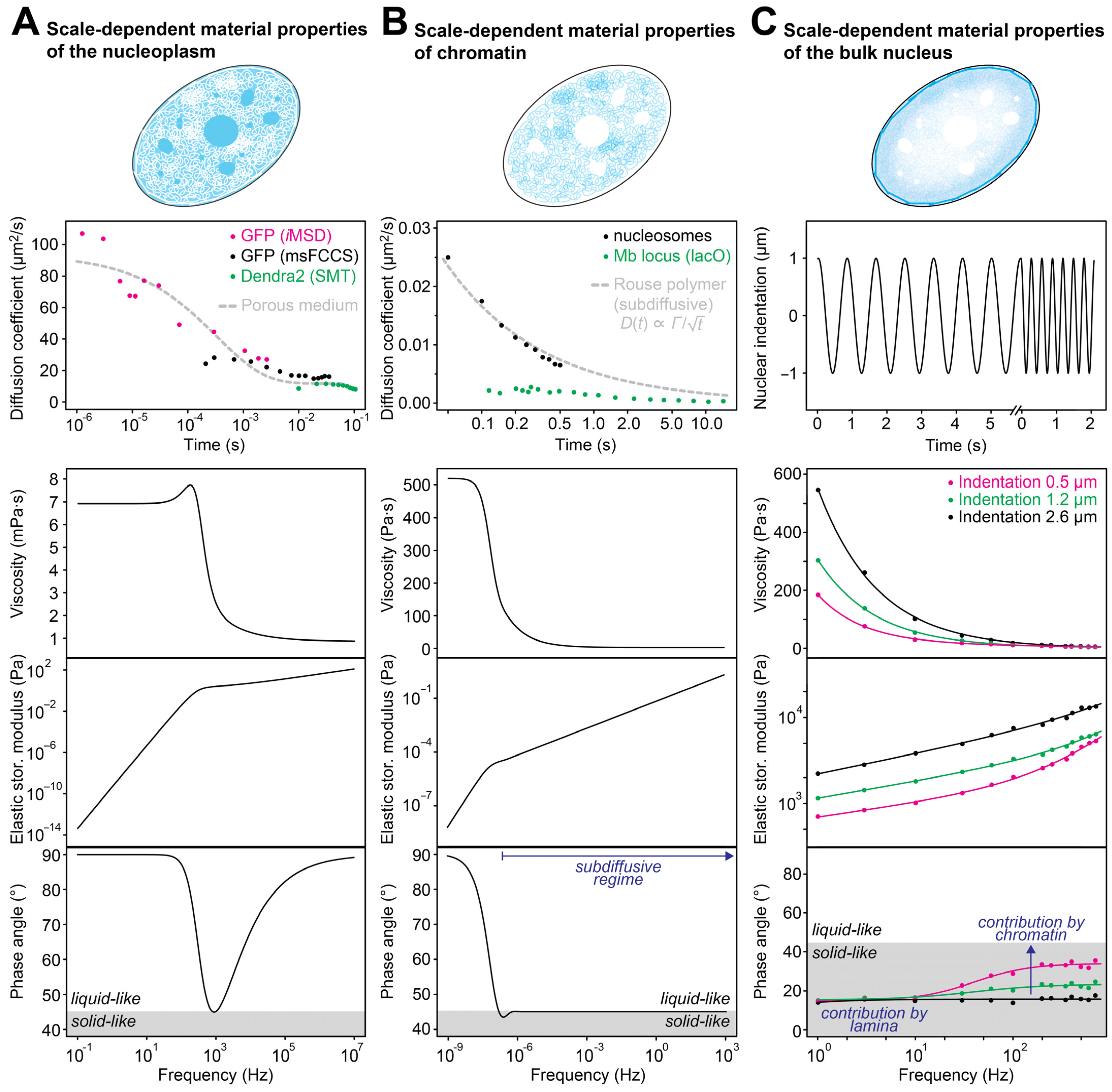
4.1. Nanoscale Properties of the Nucleoplasmic Fluid
4.2. Scale-Dependent Transport across the Nucleoplasm
4.3. Scale-Dependent Dynamics of Chromatin
4.4. Molecular Dynamics in Nuclear Condensates
5. Bulk Mechanical Properties of the Cell Nucleus
6. The Nucleus as a Mechanosensory and Mechanoregulatory Hub
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell 2017, 67, 282–293.e7. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Chang, Y.-C.; Lee, D.S.W.; Berry, J.; Sanders, D.W.; Ronceray, P.; Wingreen, N.S.; Haataja, M.; Brangwynne, C.P. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 2018, 175, 1481–1491.e13. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.A.; Imakaev, M.; Abdennur, N. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 2019, 58, 142–152. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Guttman, M. Essential roles for RNA in shaping nuclear organization. Cold Spring Harb. Perspect. Biol. 2022, 14, a039719. [Google Scholar] [CrossRef]
- Lee, D.S.W.; Strom, A.R.; Brangwynne, C.P. The mechanobiology of nuclear phase separation. APL Bioeng. 2022, 6, 021503. [Google Scholar] [CrossRef]
- Broedersz, C.P.; MacKintosh, F.C. Modeling semiflexible polymer networks. Rev. Mod. Phys. 2014, 86, 995–1036. [Google Scholar] [CrossRef]
- Strom, A.R.; Biggs, R.J.; Banigan, E.J.; Wang, X.; Chiu, K.; Herman, C.; Collado, J.; Yue, F.; Ritland Politz, J.C.; Tait, L.J.; et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Elife 2021, 10, e63972. [Google Scholar] [CrossRef] [PubMed]
- Milster, S.; Kim, W.K.; Kanduč, M.; Dzubiella, J. Tuning the permeability of regular polymeric networks by the cross-link ratio. J. Chem. Phys. 2021, 154, 154902. [Google Scholar] [CrossRef]
- Quail, T.; Golfier, S.; Elsner, M.; Ishihara, K.; Murugesan, V.; Renger, R.; Jülicher, F.; Brugués, J. Force generation by protein–DNA co-condensation. Nat. Phys. 2021, 17, 1007–1012. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.L.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Meaburn, K.J.; Misteli, T. Cell biology: Chromosome territories. Nature 2007, 445, 379–781. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, C.C.; Langowski, J. Anomalous diffusion in the interphase cell nucleus: The effect of spatial correlations of chromatin. J. Chem. Phys. 2010, 133, 025101. [Google Scholar] [CrossRef]
- Ou, H.D.; Phan, S.; Deerinck, T.J.; Thor, A.; Ellisman, M.H.; O’Shea, C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357, eaag0025. [Google Scholar] [CrossRef]
- Valet, M.; Rosa, A. Viscoelasticity of model interphase chromosomes. J. Chem. Phys. 2014, 141, 245101. [Google Scholar] [CrossRef]
- Erickson, H.P. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 2009, 11, 32–51. [Google Scholar] [CrossRef]
- Hyeon, C.; Dima, R.I.; Thirumalai, D. Size, shape, and flexibility of RNA structures. J. Chem. Phys. 2006, 125, 194905. [Google Scholar] [CrossRef]
- Richmond, T.J.; Finch, J.T.; Rushton, B.; Rhodes, D.; Klug, A. Structure of the nucleosome core particle at 7 A resolution. Nature 1984, 311, 532–537. [Google Scholar] [CrossRef]
- Forman-Kay, J.D.; Ditlev, J.A.; Nosella, M.L.; Lee, H.O. What are the distinguishing features and size requirements of biomolecular condensates and their implications for RNA-containing condensates? RNA 2022, 28, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rademacher, A.; Vlijm, R.; Tünnermann, J.; Frank, L.; Weinmann, R.; Schweigert, E.; Yserentant, K.; Hummert, J.; Bauer, C.; et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol. Cell 2020, 78, 236–249.e7. [Google Scholar] [CrossRef]
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Nucleolar dynamics and interactions with nucleoplasm in living cells. Elife 2019, 8, e47533. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V.; de Thé, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: Where form meets function. Wiley Interdiscip. Rev. RNA 2013, 4, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Caracausi, M.; Antonaros, F.; Pelleri, M.C.; Vitale, L. GeneBase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016, 2016, baw153. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F. Biophysical mechanisms of chromatin patterning. Curr. Opin. Genet. Dev. 2020, 61, 62–68. [Google Scholar] [CrossRef]
- Chen, D.; Huang, S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001, 153, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Phair, R.D.; Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature 2000, 404, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Strickfaden, H.; Zunhammer, A.; van Koningsbruggen, S.; Köhler, D.; Cremer, T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus 2010, 1, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Shinkai, S.; Ide, S.; Higashi, K.; Tamura, S.; Shimazoe, M.A.; Nakagawa, M.; Suzuki, Y.; Okada, Y.; Sasai, M.; et al. Condensed but liquid-like domain organization of active chromatin regions in living human cells. Sci. Adv. 2023, 9, eadf1488. [Google Scholar] [CrossRef]
- Wiesmeijer, K.; Krouwels, I.M.; Tanke, H.J.; Dirks, R.W. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation 2008, 76, 83–90. [Google Scholar] [CrossRef]
- Baum, M.; Erdel, F.; Wachsmuth, M.; Rippe, K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 2014, 5, 4494. [Google Scholar] [CrossRef]
- Görisch, S.M.; Wachsmuth, M.; Tóth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118 Pt 24, 5825–5834. [Google Scholar] [CrossRef]
- Mason, T.G.; Weitz, D.A. Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids. Phys. Rev. Lett. 1995, 74, 1250–1253. [Google Scholar] [CrossRef]
- Berry, J.; Brangwynne, C.P.; Haataja, M. Physical principles of intracellular organization via active and passive phase transitions. Rep. Prog. Phys. 2018, 81, 046601. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Zhou, H.-X. Determinants for fusion speed of biomolecular droplets. Angew. Chem. Int. Ed. 2020, 59, 20837–20840. [Google Scholar] [CrossRef]
- Tanaka, H. Viscoelastic phase separation. J. Phys. Condens. Matter 2000, 12, R207–R264. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, D.S.W.; Meir, Y.; Brangwynne, C.P.; Wingreen, N.S. Mechanical frustration of phase separation in the cell nucleus by chromatin. Phys. Rev. Lett. 2021, 126, 258102. [Google Scholar] [CrossRef]
- Bartolini, A.; Tempesti, P.; Ghobadi, A.F.; Berti, D.; Smets, J.; Aouad, Y.G.; Baglioni, P. Liquid-liquid phase separation of polymeric microdomains with tunable inner morphology: Mechanistic insights and applications. J. Colloid Interface Sci. 2019, 556, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Dundr, M.; Misteli, T. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000711. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Bubak, G.; Kwapiszewska, K.; Kalwarczyk, T.; Bielec, K.; Andryszewski, T.; Iwan, M.; Bubak, S.; Hołyst, R. Quantifying Nanoscale Viscosity and Structures of Living Cells Nucleus from Mobility Measurements. J. Phys. Chem. Lett. 2021, 12, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, C.C.; Langowski, J. Chromosome dynamics, molecular crowding, and diffusion in the interphase cell nucleus: A Monte Carlo lattice simulation study. Chromosome Res. 2011, 19, 63–81. [Google Scholar] [CrossRef][Green Version]
- Seksek, O.; Biwersi, J.; Verkman, A.S. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 1997, 138, 131–142. [Google Scholar] [CrossRef]
- Dross, N.; Spriet, C.; Zwerger, M.; Müller, G.; Waldeck, W.; Langowski, J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS ONE 2009, 4, e5041. [Google Scholar] [CrossRef]
- Di Rienzo, C.; Piazza, V.; Gratton, E.; Beltram, F.; Cardarelli, F. Probing short-range protein Brownian motion in the cytoplasm of living cells. Nat. Commun. 2014, 5, 5891. [Google Scholar] [CrossRef]
- Fushimi, K.; Verkman, A.S. Low viscosity in the aqueous domain of cell cytoplasm measured by picosecond polarization microfluorimetry. J. Cell Biol. 1991, 112, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Yoo, J.; Do, S.; Hwang, D.S.; Park, Y.; Shin, Y. RNA-mediated demixing transition of low-density condensates. Nat. Commun. 2023, 14, 2425. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Nessling, M.; Lichter, P. Macromolecular crowding and its potential impact on nuclear function. Biochim. Biophys. Acta 2008, 1783, 2100–2107. [Google Scholar] [CrossRef]
- Bancaud, A.; Huet, S.; Daigle, N.; Mozziconacci, J.; Beaudouin, J.; Ellenberg, J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009, 28, 3785–3798. [Google Scholar] [CrossRef]
- Izeddin, I.; Récamier, V.; Bosanac, L.; Cissé, I.I.; Boudarene, L.; Dugast-Darzacq, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Bensaude, O.; et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife 2014, 3, e02230. [Google Scholar] [CrossRef] [PubMed]
- Kues, T.; Peters, R.; Kubitscheck, U. Visualization and tracking of single protein molecules in the cell nucleus. Biophys. J. 2001, 80, 2954–2967. [Google Scholar] [CrossRef]
- Mazza, D.; Abernathy, A.; Golob, N.; Morisaki, T.; McNally, J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012, 40, e119. [Google Scholar] [CrossRef]
- Tseng, Y.; Lee, J.S.H.; Kole, T.P.; Jiang, I.; Wirtz, D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 2004, 117 Pt 10, 2159–2167. [Google Scholar] [CrossRef]
- Platani, M.; Goldberg, I.; Lamond, A.I.; Swedlow, J.R. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002, 4, 502–508. [Google Scholar] [CrossRef]
- Görisch, S.M.; Wachsmuth, M.; Ittrich, C.; Bacher, C.P.; Rippe, K.; Lichter, P. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc. Natl. Acad. Sci. USA 2004, 101, 13221–13226. [Google Scholar] [CrossRef]
- Erdel, F.; Baum, M.; Rippe, K. The viscoelastic properties of chromatin and the nucleoplasm revealed by scale-dependent protein mobility. J. Phys. Condens. Matter 2015, 27, 064115. [Google Scholar] [CrossRef]
- Rouse, P.E. A theory of the linear viscoelastic properties of dilute solutions of coiling polymers. J. Chem. Phys. 1953, 21, 1272–1280. [Google Scholar] [CrossRef]
- Doi, M.; Edwards, S. The Theory of Polymer Dynamics; Oxford University Press: Oxford, UK, 1986. [Google Scholar]
- Levi, V.; Ruan, Q.; Plutz, M.; Belmont, A.S.; Gratton, E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005, 89, 4275–4285. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Everaers, R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008, 4, e1000153. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Banigan, E.J.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 2017, 28, 1984–1996. [Google Scholar] [CrossRef] [PubMed]
- Lherbette, M.; Dos Santos, Á.; Hari-Gupta, Y.; Fili, N.; Toseland, C.P.; Schaap, I.A.T. Atomic Force Microscopy micro-rheology reveals large structural inhomogeneities in single cell-nuclei. Sci. Rep. 2017, 7, 8116. [Google Scholar] [CrossRef]
- Hinde, E.; Cardarelli, F.; Digman, M.A.; Gratton, E. In vivo pair correlation analysis of EGFP intranuclear diffusion reveals DNA-dependent molecular flow. Proc. Natl. Acad. Sci. USA 2010, 107, 16560–16565. [Google Scholar] [CrossRef]
- Muzzopappa, F.; Hummert, J.; Anfossi, M.; Tashev, S.A.; Herten, D.-P.; Erdel, F. Detecting and quantifying liquid-liquid phase separation in living cells by model-free calibrated half-bleaching. Nat. Commun. 2022, 13, 7787. [Google Scholar] [CrossRef]
- Itoh, Y.; Woods, E.J.; Minami, K.; Maeshima, K.; Collepardo-Guevara, R. Liquid-like chromatin in the cell: What can we learn from imaging and computational modeling? Curr. Opin. Struct. Biol. 2021, 71, 123–135. [Google Scholar] [CrossRef]
- Jerkovic, I.; Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 2021, 22, 511–528. [Google Scholar] [CrossRef]
- Gómez-García, P.A.; Portillo-Ledesma, S.; Neguembor, M.V.; Pesaresi, M.; Oweis, W.; Rohrlich, T.; Wieser, S.; Meshorer, E.; Schlick, T.; Cosma, M.P.; et al. Mesoscale Modeling and Single-Nucleosome Tracking Reveal Remodeling of Clutch Folding and Dynamics in Stem Cell Differentiation. Cell Rep. 2021, 34, 108614. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Kamijo, K.; Fox, P.D.; Oda, H.; Morisaki, T.; Sato, Y.; Kimura, H.; Stasevich, T.J. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat. Commun. 2019, 10, 2947. [Google Scholar] [CrossRef]
- Socol, M.; Wang, R.; Jost, D.; Carrivain, P.; Vaillant, C.; Le Cam, E.; Dahirel, V.; Normand, C.; Bystricky, K.; Victor, J.-M.; et al. Rouse model with transient intramolecular contacts on a timescale of seconds recapitulates folding and fluctuation of yeast chromosomes. Nucleic Acids Res. 2019, 47, 6195–6207. [Google Scholar] [CrossRef]
- Vivante, A.; Bronshtein, I.; Garini, Y. Chromatin viscoelasticity measured by local dynamic analysis. Biophys. J. 2020, 118, 2258–2267. [Google Scholar] [CrossRef]
- de Vries, A.H.B.; Krenn, B.E.; van Driel, R.; Subramaniam, V.; Kanger, J.S. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett. 2007, 7, 1424–1427. [Google Scholar] [CrossRef]
- Keizer, V.I.P.; Grosse-Holz, S.; Woringer, M.; Zambon, L.; Aizel, K.; Bongaerts, M.; Delille, F.; Kolar-Znika, L.; Scolari, V.F.; Hoffmann, S.; et al. Live-cell micromanipulation of a genomic locus reveals interphase chromatin mechanics. Science 2022, 377, 489–495. [Google Scholar] [CrossRef]
- Gouveia, B.; Kim, Y.; Shaevitz, J.W.; Petry, S.; Stone, H.A.; Brangwynne, C.P. Capillary forces generated by biomolecular condensates. Nature 2022, 609, 255–264. [Google Scholar] [CrossRef]
- Michieletto, D.; Marenda, M. Rheology and viscoelasticity of proteins and nucleic acids condensates. JACS Au 2022, 2, 1506–1521. [Google Scholar] [CrossRef]
- Wang, H.; Kelley, F.M.; Milovanovic, D.; Schuster, B.S.; Shi, Z. Surface tension and viscosity of protein condensates quantified by micropipette aspiration. Biophys. Rep. 2021, 1, 100011. [Google Scholar] [CrossRef]
- Jawerth, L.; Fischer-Friedrich, E.; Saha, S.; Wang, J.; Franzmann, T.; Zhang, X.; Sachweh, J.; Ruer, M.; Ijavi, M.; Saha, S.; et al. Protein condensates as aging Maxwell fluids. Science 2020, 370, 1317–1323. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Pack, C.; Saito, K.; Tamura, M.; Kinjo, M. Microenvironment and effect of energy depletion in the nucleus analyzed by mobility of multiple oligomeric EGFPs. Biophys. J. 2006, 91, 3921–3936. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Han, S.-S.; Sako, Y.; Pack, C.-G. Dynamic and unique nucleolar microenvironment revealed by fluorescence correlation spectroscopy. FASEB J. 2015, 29, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef]
- Lee, D.A.; Knight, M.M.; Campbell, J.J.; Bader, D.L. Stem cell mechanobiology. J. Cell. Biochem. 2011, 112, 1–9. [Google Scholar] [CrossRef]
- Wong, X.; Stewart, C.L. The Laminopathies and the Insights They Provide into the Structural and Functional Organization of the Nucleus. Annu. Rev. Genom. Hum. Genet. 2020, 21, 263–288. [Google Scholar] [CrossRef]
- Davidson, P.M.; Fedorchak, G.R.; Mondésert-Deveraux, S.; Bell, E.S.; Isermann, P.; Aubry, D.; Allena, R.; Lammerding, J. High-throughput microfluidic micropipette aspiration device to probe time-scale dependent nuclear mechanics in intact cells. Lab Chip 2019, 19, 3652–3663. [Google Scholar] [CrossRef]
- Hobson, C.M.; Kern, M.; O’Brien, E.T.; Stephens, A.D.; Falvo, M.R.; Superfine, R. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 2020, 31, 1788–1801. [Google Scholar] [CrossRef]
- Schreiner, S.M.; Koo, P.K.; Zhao, Y.; Mochrie, S.G.J.; King, M.C. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 2015, 6, 7159. [Google Scholar] [CrossRef] [PubMed]
- Kalukula, Y.; Stephens, A.D.; Lammerding, J.; Gabriele, S. Mechanics and functional consequences of nuclear deformations. Nat. Rev. Mol. Cell Biol. 2022, 23, 583–602. [Google Scholar] [CrossRef]
- Dupont, S.; Wickström, S.A. Mechanical regulation of chromatin and transcription. Nat. Rev. Genet. 2022, 23, 624–643. [Google Scholar] [CrossRef] [PubMed]
- Samwer, M.; Schneider, M.W.G.; Hoefler, R.; Schmalhorst, P.S.; Jude, J.G.; Zuber, J.; Gerlich, D.W. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170, 956–972.e23. [Google Scholar] [CrossRef]
- Hsia, C.-R.; McAllister, J.; Hasan, O.; Judd, J.; Lee, S.; Agrawal, R.; Chang, C.-Y.; Soloway, P.; Lammerding, J. Confined migration induces heterochromatin formation and alters chromatin accessibility. iScience 2022, 25, 104978. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Dreger, M.; Madrazo, E.; Williams, C.J.; Samaniego, R.; Hodson, N.W.; Monroy, F.; Baena, E.; Sánchez-Mateos, P.; Hurlstone, A.; et al. WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc. Natl. Acad. Sci. USA 2018, 115, 8581–8586. [Google Scholar] [CrossRef] [PubMed]
- Davidson, P.M.; Denais, C.; Bakshi, M.C.; Lammerding, J. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 2014, 7, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Nava, M.M.; Miroshnikova, Y.A.; Biggs, L.C.; Whitefield, D.B.; Metge, F.; Boucas, J.; Vihinen, H.; Jokitalo, E.; Li, X.; García Arcos, J.M.; et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 2020, 181, 800–817.e22. [Google Scholar] [CrossRef] [PubMed]
- Tajik, A.; Zhang, Y.; Wei, F.; Sun, J.; Jia, Q.; Zhou, W.; Singh, R.; Khanna, N.; Belmont, A.S.; Wang, N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016, 15, 1287–1296. [Google Scholar] [CrossRef]

| Subcomponent | Size | System | References |
|---|---|---|---|
| Typical protein | 3–10 nm | mammalian cells | [19] |
| Typical RNA 1 | 2–8 nm | human cells | [20] |
| Nucleosome | 10 nm | mammalian cells | [21] |
| Transcriptional condensate | 50–500 nm | human/mouse cells | [22] |
| Compact chromatin domain | 160 nm | human cells (HeLa) | [2] |
| Polycomb body | 0.5–1 μm | mouse embryonic stem cells | [23] |
| Chromocenter | 0.5–1 μm | mouse fibroblasts | [24] |
| Chromosome territory | 1–2 μm | human/mouse cells | [15] |
| Nucleolus | 0.1–5.5 μm | human cells (HeLa) | [25] |
| Nuclear speckle | 1 to a few μm | mammalian cells | [26] |
| PML body 2 | 0.1–1 μm | mammalian cells | [27] |
| Cajal body | 0.5–1 μm | cells of higher eukaryotes | [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hertzog, M.; Erdel, F. The Material Properties of the Cell Nucleus: A Matter of Scale. Cells 2023, 12, 1958. https://doi.org/10.3390/cells12151958
Hertzog M, Erdel F. The Material Properties of the Cell Nucleus: A Matter of Scale. Cells. 2023; 12(15):1958. https://doi.org/10.3390/cells12151958
Chicago/Turabian StyleHertzog, Maud, and Fabian Erdel. 2023. "The Material Properties of the Cell Nucleus: A Matter of Scale" Cells 12, no. 15: 1958. https://doi.org/10.3390/cells12151958
APA StyleHertzog, M., & Erdel, F. (2023). The Material Properties of the Cell Nucleus: A Matter of Scale. Cells, 12(15), 1958. https://doi.org/10.3390/cells12151958