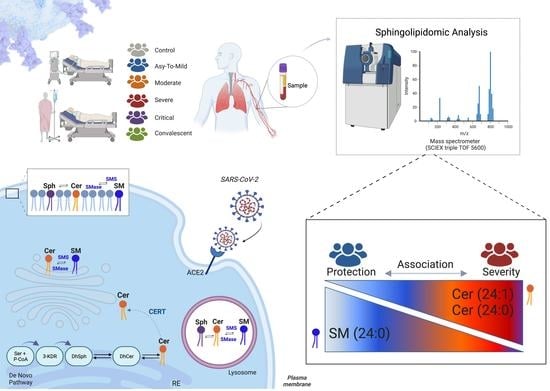
Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design and Blood Collection
2.2. Ethical Considerations
2.3. Laboratory and Data Collection
2.4. Cytokine Measurements
2.5. Lipid Extraction and Sample Preparation for LC-MS/MS
2.6. Sphingolipid Quantification by LC-MS/MS
2.7. RNA Extraction and Analysis
2.8. Transcriptome Profiling
2.8.1. Bioinformatic Analysis of Transcriptome Data
2.8.2. Validation of Microarray Data by Reverse Transcription Quantitative Real-Time PCR
2.9. Statistical Data Analysis
3. Results
3.1. Characterization of Study Participants
3.2. COVID-19 Severity Increased Gene Expression of Key Enzymes Involved in SM and Cer Synthesis
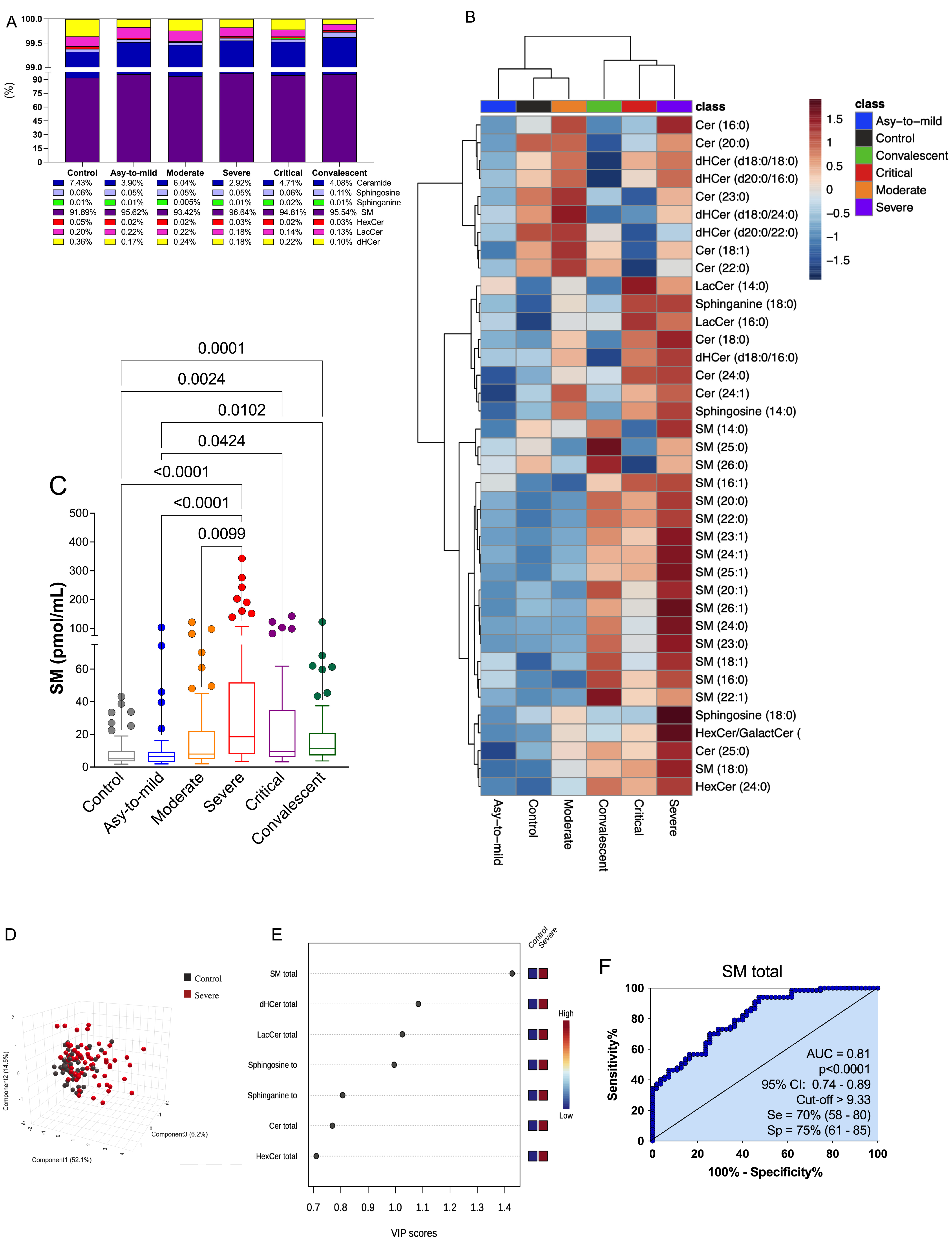
3.3. Plasma SM Profile Is Associated with COVID-19 and Can Be a Potential Biomarker for Assessing Severity of Disease
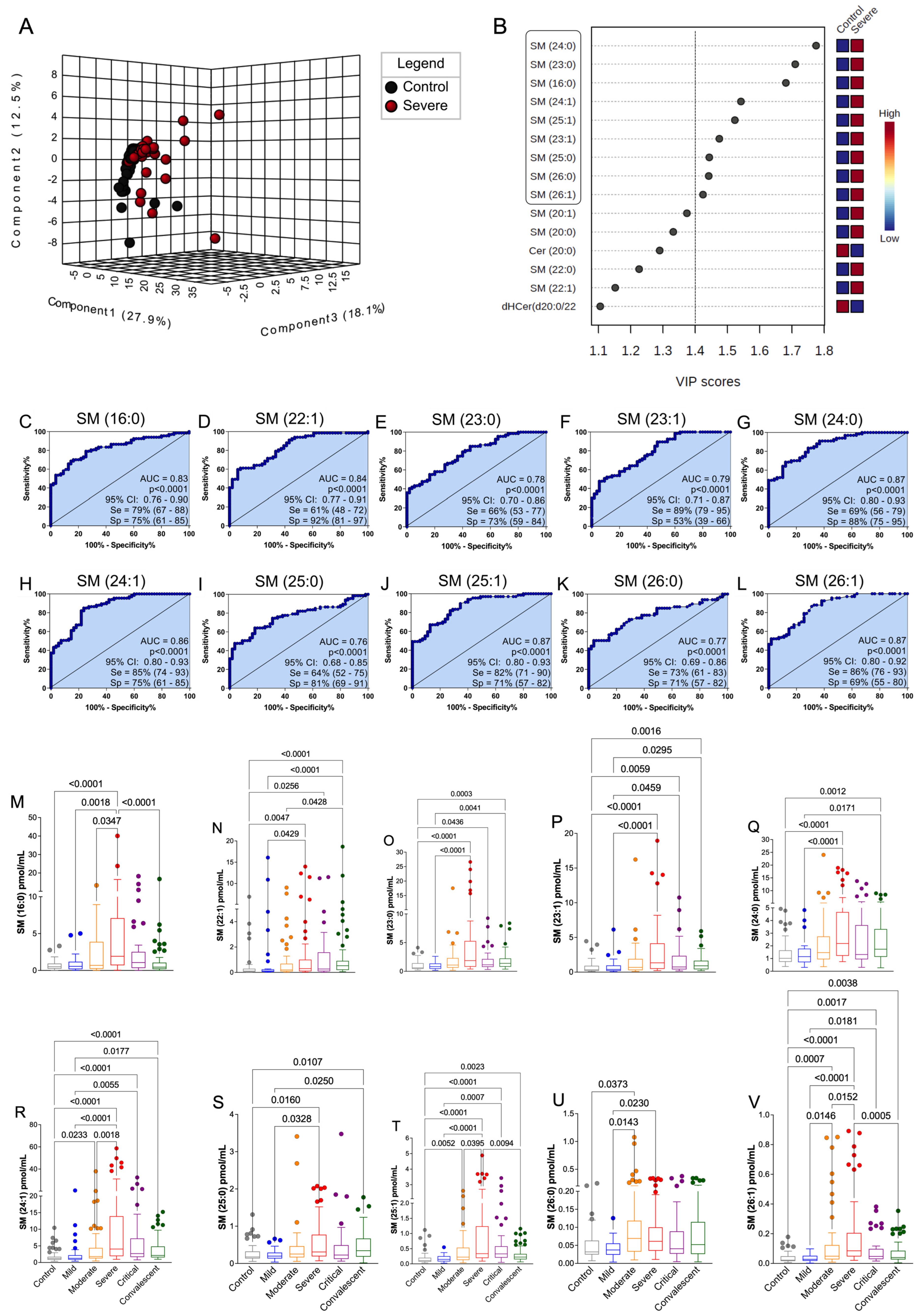
3.4. The Discovered Plasma SM Species Panel Effectively Distinguished Severe COVID-19
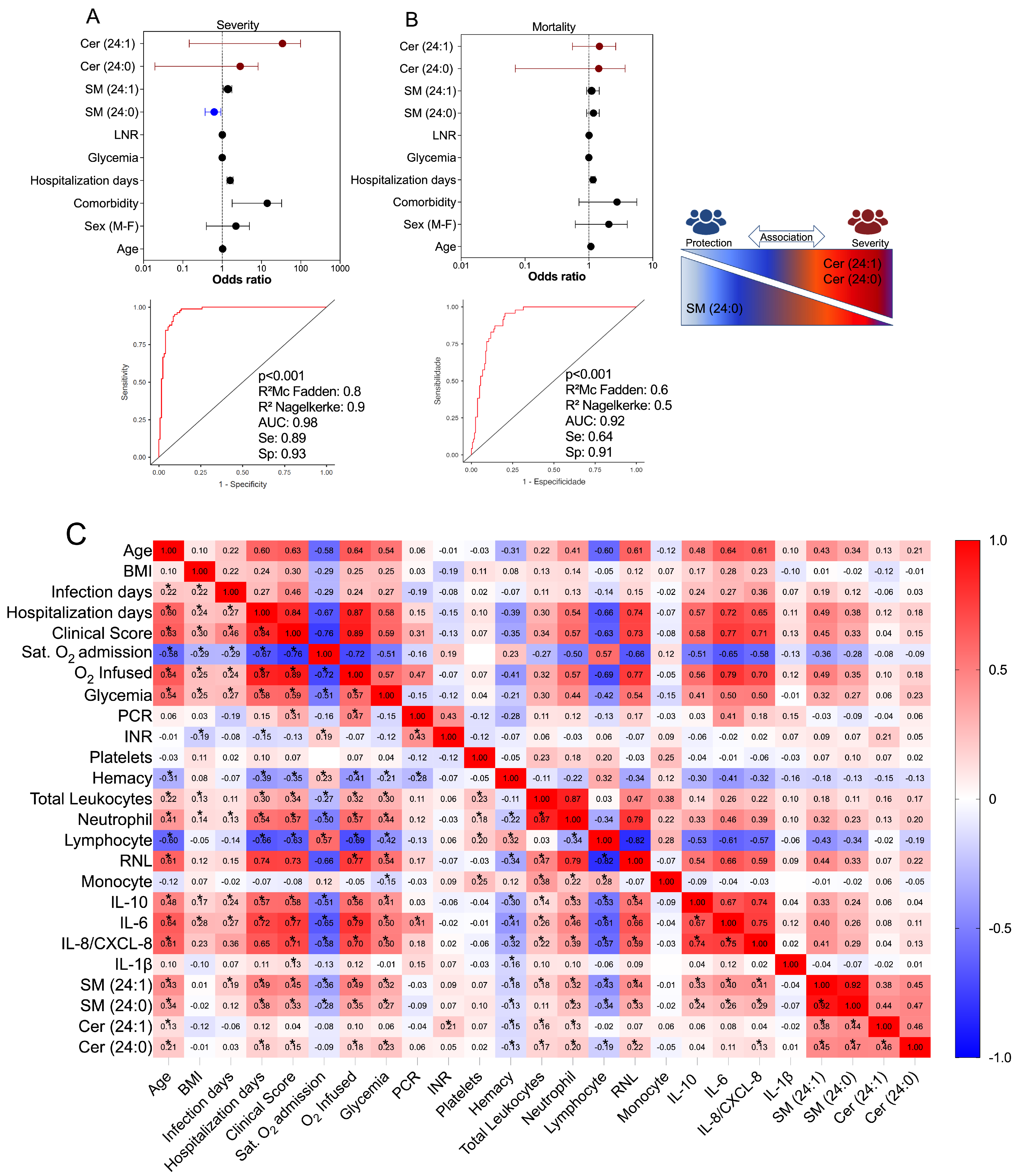
3.5. Multivariate Binomial Logistic Regression Determines the Association between Cer/SM Species and COVID-19 Clinical Severity and Mortality
3.6. Correlation of Values of SM Species with Immunological, Clinical, and Laboratory Markers in COVID-19
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Gandhi, R.T.; Lynch, J.B.; del Rio, C. Mild or Moderate COVID-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.; Gomersall, C.D.; Fowler, R.A. Care for Critically Ill Patients with COVID-19. J. Am. Med. Assoc. 2020, 323, 1499–1500. [Google Scholar] [CrossRef] [PubMed]
- Erdinc, B.; Sahni, S.; Gotlieb, V. Hematological manifestations and complications of COVID-19. Adv. Clin. Exp. Med. 2021, 30, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; de Giglio, M.; Roviello, G. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr. Med. Chem. 2020, 27, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- Carpinteiro, A.; Edwards, M.J.; Hoffmann, M.; Kochs, G.; Gripp, B.; Weigang, S.; Adams, C.; Carpinteiro, E.; Gulbins, A.; Keitsch, S.; et al. Pharmacological Inhibition of Acid Sphingomyelinase Prevents Uptake of SARS-CoV-2 by Epithelial Cells. Cell Rep. Med. 2020, 1, 100142. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J.; Schneider-Schaulies, S. Sphingolipids in viral infection. Biol. Chem. 2015, 396, 585–595. [Google Scholar] [CrossRef]
- Simonis, A.; Schubert-Unkmeir, A. The role of acid sphingomyelinase and modulation of sphingolipid metabolism in bacterial infection. Biol. Chem. 2018, 399, 1135–1146. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Gomez-Larrauri, A.; Presa, N.; Dominguez-Herrera, A.; Ouro, A.; Trueba, M.; Gomez-Muñoz, A. Role of bioactive sphingolipids in physiology and pathology. Essays Biochem. 2020, 64, 579–589. [Google Scholar] [CrossRef]
- Hussain, G.; Wang, J.; Rasul, A.; Anwar, H.; Imran, A.; Qasim, M.; Zafar, S.; Kamran, S.K.S.; Razzaq, A.; Aziz, N.; et al. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis. 2019, 18, 26. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Vitner, E.B.; Achdout, H.; Avraham, R.; Politi, B.; Cherry, L.; Tamir, H.; Yahalom-Ronen, Y.; Paran, N.; Melamed, S.; Erez, N.; et al. Glucosylceramide synthase inhibitors prevent replication of SARS-CoV-2 and influenza virus. J. Biol. Chem. 2021, 296, 100470. [Google Scholar] [CrossRef]
- Bezgovsek, J.; Gulbins, E.; Friedrich, S.-K.; Lang, K.S.; Duhan, V. Sphingolipids in early viral replication and innate immune activation. Biol. Chem. 2018, 399, 1115–1123. [Google Scholar] [CrossRef]
- Wigger, D.; Gulbins, E.; Kleuser, B.; Schumacher, F. Monitoring the Sphingolipid de novo Synthesis by Stable-Isotope Labeling and Liquid Chromatography-Mass Spectrometry. Front. Cell Dev. Biol. 2019, 7, 210. [Google Scholar] [CrossRef]
- Kurz, J.; Parnham, M.J.; Geisslinger, G.; Schiffmann, S. Ceramides as Novel Disease Biomarkers. Trends Mol. Med. 2019, 25, 20–32. [Google Scholar] [CrossRef]
- Gandy, K.A.O.; Obeid, L.M. Regulation of the Sphingosine Kinase/Sphingosine 1-Phosphate Pathway BT—Sphingolipids in Disease; Gulbins, E., Petrache, I., Eds.; Springer: Vienna, Austria, 2013; pp. 275–303. [Google Scholar] [CrossRef]
- Pyne, S.; Adams, D.R.; Pyne, N.J. Sphingosine 1-phosphate and sphingosine kinases in health and disease: Recent advances. Prog. Lipid Res. 2016, 62, 93–106. [Google Scholar] [CrossRef]
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854. [Google Scholar] [CrossRef]
- Vitner, E.B.; Avraham, R.; Politi, B.; Melamed, S.; Israely, T. Elevation in sphingolipid upon SARS-CoV-2 infection: Possible implications for COVID-19 pathology. Life Sci. Alliance 2022, 5, e202101168. [Google Scholar] [CrossRef]
- Marín-Corral, J.; Rodríguez-Morató, J.; Gomez-Gomez, A.; Pascual-Guardia, S.; Muñoz-Bermúdez, R.; Salazar-Degracia, A.; Pérez-Terán, P.; Restrepo, M.I.; Khymenets, O.; Haro, N.; et al. Metabolic Signatures Associated with Severity in Hospitalized COVID-19 Patients. Int. J. Mol. Sci. 2021, 22, 4794. [Google Scholar] [CrossRef]
- Lee, J.W.; Su, Y.; Baloni, P.; Chen, D.; Pavlovitch-Bedzyk, A.J.; Yuan, D.; Duvvuri, V.R.; Ng, R.H.; Choi, J.; Xie, J.; et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol. 2022, 40, 110–120. [Google Scholar] [CrossRef]
- Torretta, E.; Garziano, M.; Poliseno, M.; Capitanio, D.; Biasin, M.; Santantonio, T.A.; Clerici, M.; Lo Caputo, S.; Trabattoni, D.; Gelfi, C. Severity of COVID-19 Patients Predicted by Serum Sphingolipids Signature. Int. J. Mol. Sci. 2021, 22, 10198. [Google Scholar] [CrossRef] [PubMed]
- Janneh, A.H.; Kassir, M.F.; Dwyer, C.J.; Chakraborty, P.; Pierce, J.S.; Flume, P.A.; Li, H.; Nadig, S.N.; Mehrotra, S.; Ogretmen, B. Alterations of lipid metabolism provide serologic biomarkers for the detection of asymptomatic versus symptomatic COVID-19 patients. Sci. Rep. 2021, 11, 14232. [Google Scholar] [CrossRef] [PubMed]
- Diagnosis and treatment protocol for novel coronavirus pneumonia (Trial version 7). Chin. Med. J. 2020, 133, 1087–1095. [CrossRef] [PubMed]
- Wan, S.; Xiang, Y.; Fang, W.; Zheng, Y.; Li, B.; Hu, Y.; Lang, C.; Huang, D.; Sun, Q.; Xiong, Y.; et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J. Med. Virol. 2020, 92, 797–806. [Google Scholar] [CrossRef]
- Reimann, C.-M.; Gräler, M. Extraction and Quantification of Sphingosine 1-Phosphate (S1P). Bio-Protocol 2016, 6, e1817. [Google Scholar] [CrossRef]
- Andréani, P.; Gräler, M.H. Comparative quantification of sphingolipids and analogs in biological samples by high-performance liquid chromatography after chloroform extraction. Anal. Biochem. 2006, 358, 239–246. [Google Scholar] [CrossRef]
- Shaner, R.L.; Allegood, J.C.; Park, H.; Wang, E.; Kelly, S.; Haynes, C.A.; Sullards, M.C.; Merrill, A.H. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009, 50, 1692–1707. [Google Scholar] [CrossRef]
- Fedorova, M.; Lange, M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584. [Google Scholar] [CrossRef]
- R Development Core Team. R a Language and Environment for Statistical Computing: Reference Index; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- RStudio Team. RStudio: Integrated development environment for R. In RStudio: Integrated Development Environment for R; RStudio Team: Boston, MA, USA, 2019. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Benjaminit, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008, 8, R19. [Google Scholar] [CrossRef]
- Epskamp, S.; Cramer, A.O.J.; Waldorp, L.J.; Schmittmann, V.D.; Borsboom, D. qgraph: Network Visualizations of Relationships in Psychometric Data. J. Stat. Softw. 2012, 48, 1–18. Available online: http://www.jstatsoft.org/ (accessed on 19 May 2023). [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Mühle, C.; Bilbao Canalejas, R.D.; Kornhuber, J. Sphingomyelin Synthases in Neuropsychiatric Health and Disease. Neurosignals 2019, 27, 54–76. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, J.S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef]
- Sassa, T.; Hirayama, T.; Kihara, A. Enzyme Activities of the Ceramide Synthases CERS2–6 Are Regulated by Phosphorylation in the C-terminal Region. J. Biol. Chem. 2016, 291, 7477–7487. [Google Scholar] [CrossRef]
- Coant, N.; Sakamoto, W.; Mao, C.; Hannun, Y.A. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv. Biol. Regul. 2017, 63, 122–131. [Google Scholar] [CrossRef]
- Prakash, H.; Upadhyay, D.; Bandapalli, O.R.; Jain, A.; Kleuser, B. Host sphingolipids: Perspective immune adjuvant for controlling SARS-CoV-2 infection for managing COVID-19 disease. Prostaglandins Other Lipid Mediat. 2021, 152, 106504. [Google Scholar] [CrossRef]
- Harvald, E.B.; Olsen, A.S.B.; Færgeman, N.J. Autophagy in the light of sphingolipid metabolism. Apoptosis 2015, 20, 658–670. [Google Scholar] [CrossRef]
- Ghidoni, R.; Caretti, A.; Signorelli, P. Role of Sphingolipids in the Pathobiology of Lung Inflammation. Mediat. Inflamm. 2015, 2015, 487508. [Google Scholar] [CrossRef]
- van Blitterswijk, W.J.; van der Luit, A.H.; Veldman, R.J.; Verheij, M.; Borst, J. Ceramide: Second messenger or modulator of membrane structure and dynamics? Biochem. J. 2003, 369, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, F.; Cecchetti, S.; Fantuzzi, L. Macrophages and Phospholipases at the Intersection between Inflammation and the Pathogenesis of HIV-1 Infection. Int. J. Mol. Sci. 2017, 18, 1390. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Smyth, M.J.; Martinet, L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 2014, 21, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Bai, A.; Guo, Y. Acid sphingomyelinase mediates human CD4+ T-cell signaling: Potential roles in T-cell responses and diseases. Cell Death Dis. 2017, 8, e2963. [Google Scholar] [CrossRef]
- Peng, H.; Li, C.; Kadow, S.; Henry, B.D.; Steinmann, J.; Becker, K.A.; Riehle, A.; Beckmann, N.; Wilker, B.; Li, P.-L.; et al. Acid sphingomyelinase inhibition protects mice from lung edema and lethal Staphylococcus aureus sepsis. J. Mol. Med. 2015, 93, 675–689. [Google Scholar] [CrossRef]
- Chung, H.-Y.; Witt, C.J.; Jbeily, N.; Hurtado-Oliveros, J.; Giszas, B.; Lupp, A.; Gräler, M.H.; Bruns, T.; Stallmach, A.; Gonnert, F.A.; et al. Acid Sphingomyelinase Inhibition Prevents Development of Sepsis Sequelae in the Murine Liver. Sci. Rep. 2017, 7, 12348. [Google Scholar] [CrossRef]
- Wigger, D.; Schumacher, F.; Schneider-Schaulies, S.; Kleuser, B. Sphingosine 1-phosphate metabolism and insulin signaling. Cell Signal 2021, 82, 109959. [Google Scholar] [CrossRef]
- Chaurasia, B.; Summers, S.A. Ceramides in Metabolism: Key Lipotoxic Players. Annu. Rev. Physiol. 2021, 83, 303–330. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Noriyuki, O.; Masafumi, S.; Kyoko, S.; Kiyoko, O.; Yoshio, M.; Kentaro, H.; Makoto, T. Both Sphingomyelin and Cholesterol in the Host Cell Membrane Are Essential for Rubella Virus Entry. J. Virol. 2017, 92, 10-1128. [Google Scholar] [CrossRef]
- Radenkovic, D.; Chawla, S.; Pirro, M.; Sahebkar, A.; Banach, M. Cholesterol in Relation to COVID-19: Should We Care about It? J. Clin. Med. 2020, 9, 1909. [Google Scholar] [CrossRef]
- Audi, A.; Soudani, N.; Dbaibo, G.; Zaraket, H. Depletion of Host and Viral Sphingomyelin Impairs Influenza Virus Infection. Front. Microbiol. 2020, 11, 612. [Google Scholar] [CrossRef]
- Abusukhun, M.; Winkler, M.S.; Pöhlmann, S.; Moerer, O.; Meissner, K.; Tampe, B.; Hofmann-Winkler, H.; Bauer, M.; Gräler, M.H.; Claus, R.A. Activation of Sphingomyelinase-Ceramide-Pathway in COVID-19 Purposes Its Inhibition for Therapeutic Strategies. Front. Immunol. 2021, 12, 784989. [Google Scholar] [CrossRef]
- Creeden, J.F.; Imami, A.S.; Eby, H.M.; Gillman, C.; Becker, K.N.; Reigle, J.; Andari, E.; Pan, Z.K.; O’Donovan, S.M.; McCullumsmith, R.E.; et al. Fluoxetine as an anti-inflammatory therapy in SARS-CoV-2 infection. Biomed. Pharmacother. 2021, 138, 111437. [Google Scholar] [CrossRef]
- Sukhatme, V.P.; Reiersen, A.M.; Vayttaden, S.J.; Sukhatme, V.V. Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19. Front. Pharmacol. 2021, 12, 652688. [Google Scholar] [CrossRef]
- Whitley, R. Molnupiravir—A Step toward Orally Bioavailable Therapies for COVID-19. N. Engl. J. Med. 2021, 386, 592–593. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L. In the age of Omicron variant: Paxlovid raises new hopes of COVID-19 recovery. J. Med. Virol. 2022, 94, 1766–1767. [Google Scholar] [CrossRef]

| Variables | Healthy Controls n = 55 | COVID-19 Patients n = 204 | COVID-19 Patients | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| Asy-to-Mild n = 36 | Moderate n = 60 | Severe n = 67 | Critical n = 41 | Convalescent n = 77 | ||||
| Demographic characteristics | ||||||||
| Age, M ± SD, and (IQR) | 35 ± 12.9 (19–69) | 55 ± 19 (20–96) | 37.5 ± 11.4 (21–67) | 49 ± 18 (24–92) | 63 ± 15.9 (30–96) | 71 ± 17.1 (20–94) | 46 ± 9.7 (30–66) | a,d,e < 0.0001; c 0.0002 f 0.0041 |
| Age < 50, n (%) | 45 (81.8) | 79 (38.7) | 28 (77.8) | 31 (51.7) | 17 (19.4) | 7 (17.1) | 42 (54.5) | a,d,e < 0.0001, c 0.0008 f 0.0014 |
| Age ≥ 50, n (%) | 10 (18.2) | 125 (61.3) | 8 (22.2) | 29 (48.3) | 54 (80.6) | 34 (82.9) | 35 (45.4) | |
| Sex, n (%) | ||||||||
| Man | 24 (43.6) | 116 (56.9) | 15 (41.7) | 36 (60) | 40 (59.7) | 25 (61) | 9 (11.7) | f < 0.0001 |
| Woman | 31 (56.4) | 88 (43.1) | 21 (58.3) | 24 (40) | 27 (40.3) | 16 (39) | 68 (88.3) | |
| BMI (kg/m3) | 25.4 ± 4.2 (15.4–34.9) | 28.4 ± 5.9 (15.8–50.3) | 27.8 ± 5.3 (15.8–43.8) | 28.3 ± 5.7 (17.4–42.1) | 28.1 ± 6.1 (20.2–47.7) | 29.4 ± 6.1 (21.7–50.3) | 29 ± 5.1 (20.7–45.5) | a 0.0002; c 0.0240 d 0.0007 e 0.0003 f 0.0041 |
| Comorbidities, n (%) | ||||||||
| Hypertension | 6 (10.9) | 90 (44.1) | 2 (5.5) | 19 (31.7) | 46 (68.6) | 23 (56.1) | 18 (23.4) | a,d,e < 0.0001; c 0.0118 |
| Cardiovascular disorder | 7 (12.7) | 21 (10.3) | 4 (11.1) | 9 (15) | 6 (8.9) | 2 (4.9) | - | |
| Diabetes mellitus | 3 (5.4) | 62 (30.4) | 3 (8.3) | 16 (32) | 29 (43.3) | 14 (34.1) | 13 (16.9) | a,d < 0.0001; c 0.0006 e 0.0004 |
| History of smoking | 6 (10.9) | 39 (19.1) | 4 (11.1) | 9 (15) | 15 (22.4) | 11 (26.8) | 2 (2.6) | |
| Neurological disorder | - | 34 (16.7) | 9 (25) | 10 (16.7) | 10 (14.9) | 5 (12.2) | 14 (18.2) | |
| Presenting symptoms, n (%) | ||||||||
| Dyspnea | - | 127 (62.2) | - | 45 (75) | 47 (70.1) | 35 (85.4) | 60 (77.9) | f 0.0157 |
| Fever | - | 64 (31.4) | 2 (5.5) | 14 (23.3) | 33 (49.2) | 15 (36.6) | 53 (68.8) | f < 0.0001 |
| Myalgia | - | 45 (22.1) | - | 7 (11.7) | 23 (34.3) | 15 (36.6) | 68 (88.3) | f < 0.0001 |
| Diarrhea | - | 52 (25.5) | 12 (33.3) | 21 (35) | 14 (20.9) | 5 (12.2) | 47 (61.1) | f < 0.0001 |
| Cough | - | 145 (71.1) | 26 (72.2) | 42 (70) | 51 (76.1) | 26 (63.4) | 53 (93) | f 0.0004 |
| Hyperactive delirium | - | 12 (5.9) | - | 5 (8.3) | - | 7 (17.1) | - | |
| Dysgeusia | - | 53 (26) | 21 (58.3) | 22 (36.7) | 8 (12) | 2 (4.9) | 62 (80.5) | f < 0.0001 |
| Anosmia | - | 58 (28.4) | 22 (61.1) | 23 (38.3) | 11 (16.4) | 2 (4.9) | 58 (75.3) | f < 0.0001 |
| Laboratory findings, M ± SD, and (IQR) | ||||||||
| Erythrocytes × 109/L | 4.7 ± 0.5 (3.6–5.8) | 4.5 ± 0.7 (2.2–5.9) | 4.8 ± 0.5 (3.9–5.8) | 4.5 ± 0.6 (3.0–5.9) | 4.3 ± 0.8 (2.2–5.8) | 4.0 ± 0.8 (2.3–5.7) | 4.6 ± 0.4 (3.7–5.4) | a 0.0076; d 0.0026; e < 0.0001 |
| Hemoglobin (g/dL) | 14.5 ± 1.5 (10.5–17.4) | 13.3 ± 2.4 (6.6–18.2) | 15 ± 1.2 (12–16.9) | 13.6 ± 2.2 (8.1–18.2) | 12.6 ± 2.3 (6.8–16.5) | 12.4 ± 2.6 (6.6–18.2) | 13.8 ± 1.4 (9.4–16.5) | a,d,e < 0.0001; c 0.0142 |
| Leukocytes × 109/L | 7.4 ± 1.8 (4.1–11.3) | 8.4 ± 4.4 (1.6–26.1) | 7.3 ± 2.3 (3.2–13.6) | 7.4 ± 2.7 (2.6–15.7) | 8.6 ± 4.1 (1.6–21.9) | 11.1 ± 6.0 (4.6–26.1) | 5.9 ± 1.8 (2.1–12.3) | e < 0.0001; f 0.0098 |
| Neutrophils × 109/L | 4.3 ± 1.3 (2.3–7.4) | 6.0 ± 4.1 (1.6–23.8) | 4.1 ± 1.7 (1.6–9.9) | 5.0 ± 2.6 (1.6–13.4) | 7.2 ± 3.5 (2.9–18.8) | 9.5 ± 5.2 (3.2–23.7) | 3.1 ± 1.3 (1.1–8.6) | a,d,e < 0.0001; f 0.0299 |
| Lymphocytes × 109/L | 2.3 ± 0.6 (1.0–3.9) | 1.3 ± 0.9 (0.1–4.3) | 2.3 ± 0.7 (1.1–4.3) | 1.5 ± 0.8 (0.3–3.8) | 1.0 ± 0.6 (0.1–2.8) | 1.0 ± 0.5 (0.2–2.2) | 2.1 ± 0.5 (1.0–3.6) | a,d,e < 0.0001; c 0.0004 |
| Neutrophil–lymphocyte ratio | 1.9 ± 0.6 (1.0–3.3) | 4.9 ± 5.6 (0.2–28.7) | 1.7 ± 0.6 (0.7–3.6) | 3.3 ± 3.1 (0.6–15.2) | 6.8 ± 4.3 (1.0–23) | 9.1 ± 6.7 (2.3–26.7) | 1.5 ± 0.7 (0.5–4.3) | a,d,e < 0.0001; c 0.0145 |
| Monocytes × 109/L | 0.5 ± 0.1 (0.3–0.9) | 0.5 ± 0.3 (0.1–1.6) | 0.5 ± 0.1 (0.2–0.9) | 0.4 ± 0.2 (0.1–1.1) | 0.5 ± 0.3 (0.1–1.3) | 0.5 ± 0.4 (0.1–1.6) | 0.4 ± 0.1 (0.2–1.0) | |
| Platelets × 109/L | 212 ± 43.8 (129–363) | 235 ± 89.5 (50–515) | 233 ± 63.1 (135–365) | 228 ± 93.8 (117–515) | 257 ± 102 (85–506) | 212 ±67 (50–370) | 213 ± 54.6 (116–386) | |
| Glycemia (mg/dL) | 89 ± 14.6 (63–146) | 114.5 ± 69 (65–409) | 87 ± 13.4 (71–127) | 101 ± 33 (65–2003) | 132 ± 78.4 (89–409) | 143 ± 81 (79–384) | 98.5± 18.6 (67–168) | a,d,e < 0.0001; c 0.0109 |
| Hospital support, n (%) | ||||||||
| Infirmary | - | 100 (49.0) | - | 34 (56.7) | 63 (94) | 3 (7.3) | - | |
| Intensive care unit | - | 44 (21.6) | - | 2 (3.3) | 4 (6.0) | 38 (92.7) | - | |
| Hospitalization data, n | ||||||||
| Days in hospital | - | 9 ± 4.1 (1–19) | 12 ± 4.9 (2–18) | 9 ± 4.0 (1–19) | 7 ± 3.2 (1–17) | 9 ± 3.8 (4–19) | - | |
| Days from symptom onset to recruitment | - | 4 ± 4.2 (1–17) | 9 ± 3.7 (2–17) | 4 ± 3.9 (1–15) | 3 ± 3.5 (1–16) | 3 ± 4.6 (1–16) | - | |
| Days recovery until recruitment | - | - | - | - | - | - | 30 ± 17.4 (15–90) | |
| Respiratory support received (%) | ||||||||
| High flow nasal cannula | - | 65 (31.9) | - | 24 (40) | 39 (58.2) | 2 (4.8) | - | |
| Oxygen masks/noninvasive | - | 35 (17.1) | - | 3 (5) | 26 (38.8) | 6 (14.6) | - | |
| Invasive ventilation | - | 33 (16.2) | - | - | 1 (1.5) | 32 (78) | - | |
| Oxygen saturation, M ± SD (IQR) | 99 ± 1.8 (90–99) | 94 ± 8.1 (54–99) | 97.5 ± 1.7 (94–99) | 96 ± 3.9 (80–99) | 91 ± 8.6 (54–99) | 89 ± 9.1 (60–96) | - | a,d,e,f < 0.0001; c 0.0008 |
| Medications, n (%) | ||||||||
| Glucocorticoid | 2 (3.6) | 125 (61.3) | 5 (13.9) | 30 (50) | 55 (82.1) | 35 (85.4) | - | a < 0.0001 |
| Azithromycin | - | 121 (59.3) | 8 (22.2) | 39 (65) | 46 (68.6) | 28 (68.3) | - | |
| Ceftriaxone | - | 93 (45.6) | - | 23 (38.3) | 46 (68.7) | 24 (58.5) | - | |
| Oseltamivir | - | 60 (29.4) | 4 (11.1) | 10 (16.7) | 34 (50.7) | 12 (29.3) | - | |
| Colchicine | - | 6 (2.9) | - | 1 (1.7) | - | 5 (12.2) | - | |
| CQ/HCQs | - | 27 (13.2) | - | 4 (6.7) | 13 (19.4) | 10 (24.4) | - | |
| Anticoagulant | - | 18 (8.8) | 1 (2.8) | 7 (11.7) | 1 (1.5) | 9 (21.9) | - | |
| Ivermectin | - | 11 (5.4) | 5 (13.9) | 6 (10) | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toro, D.M.; da Silva-Neto, P.V.; de Carvalho, J.C.S.; Fuzo, C.A.; Pérez, M.M.; Pimentel, V.E.; Fraga-Silva, T.F.C.; Oliveira, C.N.S.; Caruso, G.R.; Vilela, A.F.L.; et al. Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells 2023, 12, 1938. https://doi.org/10.3390/cells12151938
Toro DM, da Silva-Neto PV, de Carvalho JCS, Fuzo CA, Pérez MM, Pimentel VE, Fraga-Silva TFC, Oliveira CNS, Caruso GR, Vilela AFL, et al. Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells. 2023; 12(15):1938. https://doi.org/10.3390/cells12151938
Chicago/Turabian StyleToro, Diana Mota, Pedro V. da Silva-Neto, Jonatan C. S. de Carvalho, Carlos A. Fuzo, Malena M. Pérez, Vinícius E. Pimentel, Thais F. C. Fraga-Silva, Camilla N. S. Oliveira, Glaucia R. Caruso, Adriana F. L. Vilela, and et al. 2023. "Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19" Cells 12, no. 15: 1938. https://doi.org/10.3390/cells12151938
APA StyleToro, D. M., da Silva-Neto, P. V., de Carvalho, J. C. S., Fuzo, C. A., Pérez, M. M., Pimentel, V. E., Fraga-Silva, T. F. C., Oliveira, C. N. S., Caruso, G. R., Vilela, A. F. L., Nobre-Azevedo, P., Defelippo-Felippe, T. V., Argolo, J. G. M., Degiovani, A. M., Ostini, F. M., Feitosa, M. R., Parra, R. S., Vilar, F. C., Gaspar, G. G., ... ImmunoCovid Consortium Group. (2023). Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells, 12(15), 1938. https://doi.org/10.3390/cells12151938