Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives
Abstract
1. The Current State-of-the-Art Research of ALS
1.1. Risk Factors for ALS Onset and Progression
1.2. ALS Genetics and Associated Mechanisms
2. Extracellular Vesicles and Their Role in ALS Onset and Development
2.1. EVs Overview
2.2. The Role of EVs in ALS
2.2.1. EVs in ALS Disease Progression and Pathological Mechanisms
2.2.2. miRNAs and Misfolded Proteins EVs Cargo in ALS: Potential ALS Biomarkers
3. Therapeutic Perspectives with EVs in ALS
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; van den Berg, L.H. Amyotrophic Lateral Sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef] [PubMed]
- Cividini, C.; Basaia, S.; Spinelli, E.G.; Canu, E.; Castelnovo, V.; Riva, N.; Cecchetti, G.; Caso, F.; Magnani, G.; Falini, A.; et al. Amyotrophic Lateral Sclerosis-Frontotemporal Dementia: Shared and Divergent Neural Correlates Across the Clinical Spectrum. Neurology 2021, 98, e402–e415. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.J.; Abrahams, S.; Goldstein, L.H.; Woolley, S.; Mclaughlin, P.; Snowden, J.; Mioshi, E.; Roberts-South, A.; Benatar, M.; HortobáGyi, T.; et al. Amyotrophic Lateral Sclerosis—Frontotemporal Spectrum Disorder (ALS-FTSD): Revised Diagnostic Criteria. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 153–174. [Google Scholar] [CrossRef] [PubMed]
- Saxon, J.A.; Thompson, J.C.; Jones, M.; Harris, J.M.; Richardson, A.M.; Langheinrich, T.; Neary, D.; Mann, D.M.; Snowden, J.S. Examining the Language and Behavioural Profile in FTD and ALS-FTD. J. Neurol. Neurosurg. Psychiatry 2017, 88, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Longinetti, E.; Fang, F. Epidemiology of Amyotrophic Lateral Sclerosis: An Update of Recent Literature. Curr. Opin. Neurol. 2019, 32, 771–776. [Google Scholar] [CrossRef]
- Renton, A.E.; Chiò, A.; Traynor, B.J. State of Play in Amyotrophic Lateral Sclerosis Genetics. Nat. Neurosci. 2014, 17, 17–23. [Google Scholar] [CrossRef]
- McCann, E.P.; Henden, L.; Fifita, J.A.; Zhang, K.Y.; Grima, N.; Bauer, D.C.; Chan Moi Fat, S.; Twine, N.A.; Pamphlett, R.; Kiernan, M.C.; et al. Evidence for Polygenic and Oligogenic Basis of Australian Sporadic Amyotrophic Lateral Sclerosis. J. Med. Genet. 2020, 58, 87–95. [Google Scholar] [CrossRef]
- Jaiswal, M.K. Riluzole and Edaravone: A Tale of Two Amyotrophic Lateral Sclerosis Drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Oliveira Santos, M.; Gromicho, M.; Pinto, S.; De Carvalho, M. Clinical Characteristics in Young-Adult ALS—Results from a Portuguese Cohort Study. Amyotroph. Lateral Scler. Front. Degener. 2020, 21, 620–623. [Google Scholar] [CrossRef]
- Cunha-Oliveira, T.; Montezinho, L.; Mendes, C.; Firuzi, O.; Saso, L.; Oliveira, P.J.; Silva, F.S.G. Oxidative Stress in Amyotrophic Lateral Sclerosis: Pathophysiology and Opportunities for Pharmacological Intervention. Oxid. Med. Cell. Longev. 2020, 2020, 5021694. [Google Scholar] [CrossRef]
- McCombe, P.A.; Henderson, R.D. Effects of Gender in Amyotrophic Lateral Sclerosis. Gend. Med. 2010, 7, 557–570. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, E.; Factor-Litvak, P.; Santella, R.M.; Mitsumoto, H. Clinical Perspective on Oxidative Stress in Sporadic Amyotrophic Lateral Sclerosis. Free Radic. Biol. Med. 2013, 65, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Weisskopf, M.G.; McCullough, M.L.; Calle, E.E.; Thun, M.J.; Cudkowicz, M.; Ascherio, A. Prospective Study of Cigarette Smoking and Amyotrophic Lateral Sclerosis. Am. J. Epidemiol. 2004, 160, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Zhou, X. Causal Effects of Blood Lipids on Amyotrophic Lateral Sclerosis: A Mendelian Randomization Study. Hum. Mol. Genet. 2019, 28, 688–697. [Google Scholar] [CrossRef]
- Seelen, M.; Toro Campos, R.A.; Veldink, J.H.; Visser, A.E.; Hoek, G.; Brunekreef, B.; van der Kooi, A.J.; de Visser, M.; Raaphorst, J.; van den Berg, L.H.; et al. Long-Term Air Pollution Exposure and Amyotrophic Lateral Sclerosis in Netherlands: A Population-Based Case-Control Study. Environ. Health Perspect. 2017, 125, 097023. [Google Scholar] [CrossRef]
- Fang, F.; Kwee, L.C.; Allen, K.D.; Umbach, D.M.; Ye, W.; Watson, M.; Keller, J.; Oddone, E.Z.; Sandler, D.P.; Schmidt, S.; et al. Association between Blood Lead and the Risk of Amyotrophic Lateral Sclerosis. Am. J. Epidemiol. 2010, 171, 1126–1133. [Google Scholar] [CrossRef]
- Park, R.M.; Schulte, P.A.; Bowman, J.D.; Walker, J.T.; Bondy, S.C.; Yost, M.G.; Touchstone, J.A.; Dosemeci, M. Potential Occupational Risks for Neurodegenerative Diseases. Am. J. Ind. Med. 2005, 48, 63–77. [Google Scholar] [CrossRef]
- Jafari, H.; Couratier, P.; Camu, W. Motor Neuron Disease after Electric Injury. J. Neurol. Neurosurg. Psychiatry 2001, 71, 265–267. [Google Scholar] [CrossRef]
- Gallagher, J.P.; Sanders, M. Trauma and Amyotrophic Lateral Sclerosis: A Report of 78 Patients. Acta Neurol. Scand. 1987, 75, 145–150. [Google Scholar] [CrossRef]
- Coffman, C.J.; Horner, R.D.; Grambow, S.C.; Lindquist, J.; VA Cooperative Studies Program Project #500. Estimating the Occurrence of Amyotrophic Lateral Sclerosis among Gulf War (1990–1991) Veterans Using Capture-Recapture Methods. Neuroepidemiology 2005, 24, 141–150. [Google Scholar] [CrossRef]
- Chiò, A.; Benzi, G.; Dossena, M.; Mutani, R.; Mora, G. Severely Increased Risk of Amyotrophic Lateral Sclerosis among Italian Professional Football Players. Brain 2005, 128, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Scarmeas, N.; Shih, T.; Stern, Y.; Ottman, R.; Rowland, L.P. Premorbid Weight, Body Mass, and Varsity Athletics in ALS. Neurology 2002, 59, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Julian, T.H.; Glascow, N.; Barry, A.D.F.; Moll, T.; Harvey, C.; Klimentidis, Y.C.; Newell, M.; Zhang, S.; Snyder, M.P.; Cooper-Knock, J.; et al. Physical Exercise Is a Risk Factor for Amyotrophic Lateral Sclerosis: Convergent Evidence from Mendelian Randomisation, Transcriptomics and Risk Genotypes. EBioMedicine 2021, 68, 103397. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, B.; Horton, D.K.; Mitsumoto, H. Potential Environmental Factors in Amyotrophic Lateral Sclerosis. Neurol. Clin. 2015, 33, 877–888. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, L.; Tang, L.; Xia, K.; Huang, T.; Fan, D. Physical Activity and Amyotrophic Lateral Sclerosis: A Mendelian Randomization Study. Neurobiol. Aging 2021, 105, 374.e1–374.e4. [Google Scholar] [CrossRef] [PubMed]
- Savage, A.L.; Lopez, A.I.; Iacoangeli, A.; Bubb, V.J.; Smith, B.; Troakes, C.; Alahmady, N.; Koks, S.; Schumann, G.G.; Al-Chalabi, A.; et al. Frequency and Methylation Status of Selected Retrotransposition Competent L1 Loci in Amyotrophic Lateral Sclerosis. Mol. Brain 2020, 13, 154. [Google Scholar] [CrossRef]
- Morahan, J.M.; Yu, B.; Trent, R.J.; Pamphlett, R. A Genome-Wide Analysis of Brain DNA Methylation Identifies New Candidate Genes for Sporadic Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. 2009, 10, 418–429. [Google Scholar] [CrossRef]
- Figueroa-Romero, C.; Hur, J.; Bender, D.E.; Delaney, C.E.; Cataldo, M.D.; Smith, A.L.; Yung, R.; Ruden, D.M.; Callaghan, B.C.; Feldman, E.L. Identification of Epigenetically Altered Genes in Sporadic Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e52672. [Google Scholar] [CrossRef]
- Cai, Z.; Jia, X.; Liu, M.; Yang, X.; Cui, L. Epigenome-Wide DNA Methylation Study of Whole Blood in Patients with Sporadic Amyotrophic Lateral Sclerosis. Chin. Med. J. 2022, 135, 1466–1473. [Google Scholar] [CrossRef]
- Chen, L.-X.; Xu, H.-F.; Wang, P.-S.; Yang, X.-X.; Wu, Z.-Y.; Li, H.-F. Mutation Spectrum and Natural History of ALS Patients in a 15-Year Cohort in Southeastern China. Front. Genet. 2021, 12, 746060. [Google Scholar] [CrossRef]
- Dupuis, L.; Gonzalez de Aguilar, J.-L.; Oudart, H.; de Tapia, M.; Barbeito, L.; Loeffler, J.-P. Mitochondria in Amyotrophic Lateral Sclerosis: A Trigger and a Target. Neurodegener. Dis. 2004, 1, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Otomo, A.; Pan, L.; Hadano, S. Dysregulation of the Autophagy-Endolysosomal System in Amyotrophic Lateral Sclerosis and Related Motor Neuron Diseases. Neurol. Res. Int. 2012, 2012, 498428. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Moreau, C.; Lassalle, P.; Perez, T.; De Seze, J.; Brunaud-Danel, V.; Destée, A.; Tonnel, A.B.; Just, N. Low Levels of the Vascular Endothelial Growth Factor in CSF from Early ALS Patients. Neurology 2004, 62, 2127–2129. [Google Scholar] [CrossRef]
- Suzuki, M.; Watanabe, T.; Mikami, H.; Nomura, M.; Yamazaki, T.; Irie, T.; Ishikawa, H.; Yasui, K.; Ono, S. Immunohistochemical Studies of Vascular Endothelial Growth Factor in Skin of Patients with Amyotrophic Lateral Sclerosis. J. Neurol. Sci. 2009, 285, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Thakur, K.; Gupta, P.K. ALS and Oxidative Stress: The Neurovascular Scenario. Oxid. Med. Cell. Longev. 2013, 2013, 635831. [Google Scholar] [CrossRef]
- Mejzini, R.; Flynn, L.L.; Pitout, I.L.; Fletcher, S.; Wilton, S.D.; Akkari, P.A. ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now? Front. Neurosci. 2019, 13, 1310. [Google Scholar] [CrossRef]
- Nagano, S.; Araki, T. Axonal Transport and Local Translation of mRNA in Amyotrophic Lateral Sclerosis. In Amyotrophic Lateral Sclerosis; Araki, T., Ed.; Exon Publications: Brisbane, Australia; ISBN 9780645001778.
- Pehar, M.; Vargas, M.R.; Robinson, K.M.; Cassina, P.; Díaz-Amarilla, P.J.; Hagen, T.M.; Radi, R.; Barbeito, L.; Beckman, J.S. Mitochondrial Superoxide Production and Nuclear Factor Erythroid 2-Related Factor 2 Activation in p75 Neurotrophin Receptor-Induced Motor Neuron Apoptosis. J. Neurosci. 2007, 27, 7777–7785. [Google Scholar] [CrossRef]
- Juliani, J.; Vassileff, N.; Spiers, J.G. Inflammatory-Mediated Neuron-Glia Communication Modulates ALS Pathophysiology. J. Neurosci. 2021, 41, 1142–1144. [Google Scholar] [CrossRef]
- Radford, R.A.W.; Vidal-Itriago, A.; Scherer, N.M.; Lee, A.; Graeber, M.; Chung, R.S.; Morsch, M. Evidence for a Growing Involvement of Glia in Amyotrophic Lateral Sclerosis. In Spectrums of Amyotrophic Lateral Sclerosis; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 123–142. [Google Scholar]
- Laferriere, F.; Polymenidou, M. Advances and Challenges in Understanding the Multifaceted Pathogenesis of Amyotrophic Lateral Sclerosis. Swiss Med. Wkly. 2015, 145, w14054. [Google Scholar] [CrossRef]
- Scarrott, J.M.; Herranz-Martín, S.; Alrafiah, A.R.; Shaw, P.J.; Azzouz, M. Current Developments in Gene Therapy for Amyotrophic Lateral Sclerosis. Expert Opin. Biol. Ther. 2015, 15, 935–947. [Google Scholar] [CrossRef]
- Yun, Y.; Ha, Y. CRISPR/Cas9-Mediated Gene Correction to Understand ALS. Int. J. Mol. Sci. 2020, 21, 3801. [Google Scholar] [CrossRef] [PubMed]
- Cudkowicz, M.E.; Brown, R.H., Jr. An Update on Superoxide Dismutase 1 in Familial Amyotrophic Lateral Sclerosis. J. Neurol. Sci. 1996, 139, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Patterson, D.; Warner, H.R.; Fox, L.M.; Rahmani, Z. Superoxide Dismutase, Oxygen Radical Metabolism, and Amyotrophic Lateral Sclerosis. Mol. Genet. Med. 1994, 4, 79–118. [Google Scholar] [PubMed]
- Andrus, P.K.; Fleck, T.J.; Gurney, M.E.; Hall, E.D. Protein Oxidative Damage in a Transgenic Mouse Model of Familial Amyotrophic Lateral Sclerosis. J. Neurochem. 1998, 71, 2041–2048. [Google Scholar] [CrossRef]
- Culik, R.M.; Sekhar, A.; Nagesh, J.; Deol, H.; Rumfeldt, J.A.O.; Meiering, E.M.; Kay, L.E. Effects of Maturation on the Conformational Free-Energy Landscape of SOD1. Proc. Natl. Acad. Sci. USA 2018, 115, E2546–E2555. [Google Scholar] [CrossRef]
- Soon, C.P.W.; Crouch, P.J.; Turner, B.J.; McLean, C.A.; Laughton, K.M.; Atkin, J.D.; Masters, C.L.; White, A.R.; Li, Q.-X. Serum Matrix Metalloproteinase-9 Activity Is Dysregulated with Disease Progression in the Mutant SOD1 Transgenic Mice. Neuromuscul. Disord. 2010, 20, 260–266. [Google Scholar] [CrossRef]
- Haidet-Phillips, A.M.; Hester, M.E.; Miranda, C.J.; Meyer, K.; Braun, L.; Frakes, A.; Song, S.; Likhite, S.; Murtha, M.J.; Foust, K.D.; et al. Astrocytes from Familial and Sporadic ALS Patients Are Toxic to Motor Neurons. Nat. Biotechnol. 2011, 29, 824–828. [Google Scholar] [CrossRef]
- Lee, J.; Hyeon, S.J.; Im, H.; Ryu, H.; Kim, Y.; Ryu, H. Astrocytes and Microglia as Non-Cell Autonomous Players in the Pathogenesis of ALS. Exp. Neurobiol. 2016, 25, 233–240. [Google Scholar] [CrossRef]
- Zhao, W.; Beers, D.R.; Appel, S.H. Immune-Mediated Mechanisms in the Pathoprogression of Amyotrophic Lateral Sclerosis. J. Neuroimmune Pharmacol. 2013, 8, 888–899. [Google Scholar] [CrossRef]
- Meissner, F.; Molawi, K.; Zychlinsky, A. Mutant Superoxide Dismutase 1-Induced IL-1beta Accelerates ALS Pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 13046–13050. [Google Scholar] [CrossRef]
- Luís, J.P.; Simões, C.J.V.; Brito, R.M.M. The Therapeutic Prospects of Targeting IL-1R1 for the Modulation of Neuroinflammation in Central Nervous System Disorders. Int. J. Mol. Sci. 2022, 23, 1731. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic Reactive Astrocytes Are Induced by Activated Microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Genc, B.; Gozutok, O.; Kocak, N.; Ozdinler, P.H. The Timing and Extent of Motor Neuron Vulnerability in ALS Correlates with Accumulation of Misfolded SOD1 Protein in the Cortex and in the Spinal Cord. Cells 2020, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Smeyers, J.; Banchi, E.-G.; Latouche, M. C9ORF72: What It Is, What It Does, and Why It Matters. Front. Cell. Neurosci. 2021, 15, 661447. [Google Scholar] [CrossRef]
- Pang, W.; Hu, F. Cellular and Physiological Functions of C9ORF72 and Implications for ALS/FTD. J. Neurochem. 2021, 157, 334–350. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.G.; Bogdanik, L.; Yáñez, A.; Lall, D.; Wolf, A.J.; Muhammad, A.K.M.G.; Ho, R.; Carmona, S.; Vit, J.P.; Zarrow, J.; et al. C9orf72 Is Required for Proper Macrophage and Microglial Function in Mice. Science 2016, 351, 1324–1329. [Google Scholar] [CrossRef]
- Sullivan, P.M.; Zhou, X.; Robins, A.M.; Paushter, D.H.; Kim, D.; Smolka, M.B.; Hu, F. The ALS/FTLD Associated Protein C9orf72 Associates with SMCR8 and WDR41 to Regulate the Autophagy-Lysosome Pathway. Acta Neuropathol. Commun. 2016, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Sheng, J.; Xu, L.; Zhan, X.; Liu, J.; Jiang, H.; Shu, X.; Liu, X.; Zhang, T.; Jiang, L.; et al. Cryo-EM Structure of C9ORF72-SMCR8-WDR41 Reveals the Role as a GAP for Rab8a and Rab11a. Proc. Natl. Acad. Sci. USA 2020, 117, 9876–9883. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Weng, S.-M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzschmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9ORF72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Yokogawa, M.; Nakagawa, I.; Onodera, K.; Okano, H.; Inoue, H.; Hattori, M.; Okada, Y.; Tsuiji, H. C9ORF72 Dipeptide Repeat Proteins Disrupt Formation of GEM Bodies and Induce Aberrant Accumulation of Survival of Motor Neuron Protein. bioRxiv 2021. [Google Scholar] [CrossRef]
- Westergard, T.; Jensen, B.K.; Wen, X.; Cai, J.; Kropf, E.; Iacovitti, L.; Pasinelli, P.; Trotti, D. Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72-ALS/FTD. Cell Rep. 2016, 17, 645–652. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, R.; Lu, Y.; Gendron, T.F.; Karydas, A.; Tran, H.; Yang, D.; Petrucelli, L.; Miller, B.L.; Almeida, S.; Gao, F.B. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron 2016, 92, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, B.; Hartmann, H.; May, S.; Möhl, C.; Ederle, H.; Michaelsen, M.; Schludi, M.H.; Dormann, D.; Edbauer, D. Cytoplasmic Poly-GA Aggregates Impair Nuclear Import of TDP-43 in C9orf72 ALS/FTLD. Hum. Mol. Genet. 2016, 26, 790–800. [Google Scholar] [CrossRef]
- Prudencio, M.; Humphrey, J.; Pickles, S.; Brown, A.-L.; Hill, S.E.; Kachergus, J.M.; Shi, J.; Heckman, M.G.; Spiegel, M.R.; Cook, C.; et al. Truncated Stathmin-2 Is a Marker of TDP-43 Pathology in Frontotemporal Dementia. J. Clin. Investig. 2020, 130, 6080–6092. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Lin, Z.; Kim, E.; Henstridge, C.M.; Pena Altamira, E.; Hunt, C.K.; Burchill, E.; Callaghan, I.; Loreto, A.; Brown-Wright, H.; et al. Sarm1 Deletion Suppresses TDP-43-Linked Motor Neuron Degeneration and Cortical Spine Loss. Acta Neuropathol. Commun. 2019, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Andrade, N.S.; Ramic, M.; Esanov, R.; Liu, W.; Rybin, M.J.; Gaidosh, G.; Abdallah, A.; Del’Olio, S.; Huff, T.C.; Chee, N.T.; et al. Dipeptide Repeat Proteins Inhibit Homology-Directed DNA Double Strand Break Repair in C9ORF72 ALS/FTD. Mol. Neurodegener. 2020, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, D.; Pasetto, L.; Bonetto, V.; Basso, M. Role of Extracellular Vesicles in Amyotrophic Lateral Sclerosis. Front. Neurosci. 2018, 12, 574. [Google Scholar] [CrossRef]
- Jo, M.; Lee, S.; Jeon, Y.-M.; Kim, S.; Kwon, Y.; Kim, H.-J. The Role of TDP-43 Propagation in Neurodegenerative Diseases: Integrating Insights from Clinical and Experimental Studies. Exp. Mol. Med. 2020, 52, 1652–1662. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Arai, T.; Nonaka, T.; Kametani, F.; Yoshida, M.; Hashizume, Y.; Beach, T.G.; Buratti, E.; Baralle, F.; Morita, M.; et al. Phosphorylated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Ann. Neurol. 2008, 64, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Suk, T.R.; Rousseaux, M.W.C. The Role of TDP-43 Mislocalization in Amyotrophic Lateral Sclerosis. Mol. Neurodegener. 2020, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Fifita, J.A.; Hogan, A.; Grima, N.; Smith, B.; Troakes, C.; Vance, C.; Shaw, C.; Al-Sarraj, S.; Blair, I.P.; et al. Co-Deposition of SOD1, TDP-43 and p62 Proteinopathies in ALS: Evidence for Multifaceted Pathways Underlying Neurodegeneration. Acta Neuropathol. Commun. 2022, 10, 122. [Google Scholar] [CrossRef]
- Pokrishevsky, E.; Grad, L.I.; Yousefi, M.; Wang, J.; Mackenzie, I.R.; Cashman, N.R. Aberrant Localization of FUS and TDP43 Is Associated with Misfolding of SOD1 in Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e35050. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.; Scotter, E.L.; Nishimura, A.L.; Troakes, C.; Mitchell, J.C.; Kathe, C.; Urwin, H.; Manser, C.; Miller, C.C.; Hortobágyi, T.; et al. ALS Mutant FUS Disrupts Nuclear Localization and Sequesters Wild-Type FUS within Cytoplasmic Stress Granules. Hum. Mol. Genet. 2013, 22, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Clatterbuck-Soper, S.F.; Jackrel, M.E.; Shorter, J.; Mili, S. FUS Inclusions Disrupt RNA Localization by Sequestering Kinesin-1 and Inhibiting Microtubule Detyrosination. J. Cell Biol. 2017, 216, 1015–1034. [Google Scholar] [CrossRef]
- Baldwin, K.R.; Godena, V.K.; Hewitt, V.L.; Whitworth, A.J. Axonal Transport Defects Are a Common Phenotype in Drosophila Models of ALS. Hum. Mol. Genet. 2016, 25, 2378–2392. [Google Scholar] [CrossRef]
- Tsai, Y.-L.; Coady, T.H.; Lu, L.; Zheng, D.; Alland, I.; Tian, B.; Shneider, N.A.; Manley, J.L. ALS/FTD-Associated Protein FUS Induces Mitochondrial Dysfunction by Preferentially Sequestering Respiratory Chain Complex mRNAs. Genes Dev. 2020, 34, 785–805. [Google Scholar] [CrossRef]
- Ishigaki, S.; Sobue, G. Importance of Functional Loss of FUS in FTLD/ALS. Front. Mol. Biosci. 2018, 5, 44. [Google Scholar] [CrossRef]
- Sproviero, D.; La Salvia, S.; Giannini, M.; Crippa, V.; Gagliardi, S.; Bernuzzi, S.; Diamanti, L.; Ceroni, M.; Pansarasa, O.; Poletti, A.; et al. Pathological Proteins Are Transported by Extracellular Vesicles of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Neurosci. 2018, 12, 487. [Google Scholar] [CrossRef]
- Gagliardi, D.; Bresolin, N.; Comi, G.P.; Corti, S. Extracellular Vesicles and Amyotrophic Lateral Sclerosis: From Misfolded Protein Vehicles to Promising Clinical Biomarkers. Cell. Mol. Life Sci. 2021, 78, 561–572. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-like Vesicles, and Apoptotic Bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- D’Acunzo, P.; Pérez-González, R.; Kim, Y.; Hargash, T.; Miller, C.; Alldred, M.J.; Erdjument-Bromage, H.; Penikalapati, S.C.; Pawlik, M.; Saito, M.; et al. Mitovesicles Are a Novel Population of Extracellular Vesicles of Mitochondrial Origin Altered in Down Syndrome. Sci. Adv. 2021, 7, eabe5085. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Zhao, S.; Sheng, S.; Wang, Y.; Ding, L.; Xu, X.; Xia, X.; Zheng, J.C. Astrocyte-Derived Extracellular Vesicles: A Double-Edged Sword in Central Nervous System Disorders. Neurosci. Biobehav. Rev. 2021, 125, 148–159. [Google Scholar] [CrossRef]
- Bağcı, C.; Sever-Bahcekapili, M.; Belder, N.; Bennett, A.P.S.; Erdener, Ş.E.; Dalkara, T. Overview of Extracellular Vesicle Characterization Techniques and Introduction to Combined Reflectance and Fluorescence Confocal Microscopy to Distinguish Extracellular Vesicle Subpopulations. Neurophotonics 2022, 9, 021903. [Google Scholar] [CrossRef]
- Deng, F.; Miller, J. A Review on Protein Markers of Exosome from Different Bio-Resources and the Antibodies Used for Characterization. J. Histotechnol. 2019, 42, 226–239. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology Function and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-Derived Microvesicles: Important and Underappreciated Mediators of Cell-to-Cell Communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel Biomarkers for Clinical Diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Rota, R.; Ciarapica, R.; Giordano, A.; Miele, L.; Locatelli, F. MicroRNAs in Rhabdomyosarcoma: Pathogenetic Implications and Translational Potentiality. Mol. Cancer 2011, 10, 120. [Google Scholar] [CrossRef]
- Camussi, G.; Deregibus, M.-C.; Bruno, S.; Grange, C.; Fonsato, V.; Tetta, C. Exosome/microvesicle-Mediated Epigenetic Reprogramming of Cells. Am. J. Cancer Res. 2011, 1, 98–110. [Google Scholar] [PubMed]
- Sharma, A. Bioinformatic Analysis Revealing Association of Exosomal mRNAs and Proteins in Epigenetic Inheritance. J. Theor. Biol. 2014, 357, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; van Niel, G.; Stahl, P.D. Extracellular Vesicles and Homeostasis-An Emerging Field in Bioscience Research. FASEB Bioadv. 2021, 3, 456–458. [Google Scholar] [CrossRef]
- Smalheiser, N.R. Exosomal Transfer of Proteins and RNAs at Synapses in the Nervous System. Biol. Direct 2007, 2, 35. [Google Scholar] [CrossRef]
- Zappulli, V.; Friis, K.P.; Fitzpatrick, Z.; Maguire, C.A.; Breakefield, X.O. Extracellular Vesicles and Intercellular Communication within the Nervous System. J. Clin. Investig. 2016, 126, 1198–1207. [Google Scholar] [CrossRef]
- Lee, Y.; El Andaloussi, S.; Wood, M.J.A. Exosomes and Microvesicles: Extracellular Vesicles for Genetic Information Transfer and Gene Therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef]
- Cantaluppi, V.; Figliolini, F.; Deregibus, M.C.; Camussi, G. Membrane-Derived Extracellular Vesicles from Endothelial Progenitor Cells Activate Angiogenesis. In Tumor Dormancy, Quiescence, and Senescence; Springer: Dordrecht, The Netherlands, 2014; Volume 2, pp. 17–25. [Google Scholar]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Lo Iacono, M.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial Progenitor Cell Derived Microvesicles Activate an Angiogenic Program in Endothelial Cells by a Horizontal Transfer of mRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef]
- Cruz, L.; Romero, J.A.A.; Iglesia, R.P.; Lopes, M.H. Extracellular Vesicles: Decoding a New Language for Cellular Communication in Early Embryonic Development. Front. Cell Dev. Biol. 2018, 6, 94. [Google Scholar] [CrossRef]
- Draebing, T.; Heigwer, J.; Juergensen, L.; Katus, H.A.; Hassel, D. Extracellular Vesicle-Delivered Bone Morphogenetic Proteins: A Novel Paracrine Mechanism during Embryonic Development. bioRxiv 2018. [Google Scholar] [CrossRef]
- Xiao, T.; Zhang, W.; Jiao, B.; Pan, C.-Z.; Liu, X.; Shen, L. The Role of Exosomes in the Pathogenesis of Alzheimer’ Disease. Transl. Neurodegener. 2017, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, R.; Zingg, W.; Shetty, S.; Shetty, A.K. Astrocyte-Derived Extracellular Vesicles: Neuroreparative Properties and Role in the Pathogenesis of Neurodegenerative Disorders. J. Control. Release 2020, 323, 225–239. [Google Scholar] [CrossRef]
- Prada, I.; Furlan, R.; Matteoli, M.; Verderio, C. Classical and Unconventional Pathways of Vesicular Release in Microglia. Glia 2013, 61, 1003–1017. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Racchetti, G.; Rupnik, M.; Meldolesi, J. The Regulated Exocytosis of Enlargeosomes Is Mediated by a SNARE Machinery That Includes VAMP4. J. Cell Sci. 2008, 121, 2983–2991. [Google Scholar] [CrossRef] [PubMed]
- Turola, E.; Furlan, R.; Bianco, F.; Matteoli, M.; Verderio, C. Microglial Microvesicle Secretion and Intercellular Signaling. Front. Physiol. 2012, 3, 149. [Google Scholar] [CrossRef] [PubMed]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.-J. Apoptotic Cell-Derived Exosomes: Messages from Dying Cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef]
- Busatto, S.; Morad, G.; Guo, P.; Moses, M.A. The Role of Extracellular Vesicles in the Physiological and Pathological Regulation of the Blood-Brain Barrier. FASEB Bioadv. 2021, 3, 665–675. [Google Scholar] [CrossRef]
- Fauré, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes Are Released by Cultured Cortical Neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef]
- Potolicchio, I.; Carven, G.J.; Xu, X.; Stipp, C.; Riese, R.J.; Stern, L.J.; Santambrogio, L. Proteomic Analysis of Microglia-Derived Exosomes: Metabolic Role of the Aminopeptidase CD13 in Neuropeptide Catabolism. J. Immunol. 2005, 175, 2237–2243. [Google Scholar] [CrossRef]
- Taylor, A.R.; Robinson, M.B.; Gifondorwa, D.J.; Tytell, M.; Milligan, C.E. Regulation of Heat Shock Protein 70 Release in Astrocytes: Role of Signaling Kinases. Dev. Neurobiol. 2007, 67, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Krämer-Albers, E.-M.; Bretz, N.; Tenzer, S.; Winterstein, C.; Möbius, W.; Berger, H.; Nave, K.-A.; Schild, H.; Trotter, J. Oligodendrocytes Secrete Exosomes Containing Major Myelin and Stress-Protective Proteins: Trophic Support for Axons? Proteom. Clin. Appl. 2007, 1, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Zuñiga-Garcia, P.; Sanchez-Mosquera, P.; Egia, A.; Perez, A.; Loizaga, A.; Arceo, R.; Lacasa, I.; Rabade, A.; Arrieta, E.; et al. Different EV Enrichment Methods Suitable for Clinical Settings Yield Different Subpopulations of Urinary Extracellular Vesicles from Human Samples. J. Extracell. Vesicles 2016, 5, 29497. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, M.I.; Brisson, A.R.; van Herwijnen, M.J.C.; Tan, S.; van de Lest, C.H.A.; Redegeld, F.A.; Garssen, J.; Wauben, M.H.M.; Nolte-’t Hoen, E.N.M. Recovery of Extracellular Vesicles from Human Breast Milk Is Influenced by Sample Collection and Vesicle Isolation Procedures. J. Extracell. Vesicles 2014, 3, 24215. [Google Scholar] [CrossRef]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and Proteomic Profiling of Exosomes in Human Cerebrospinal Fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef]
- Morad, G.; Moses, M.A. Brainwashed by Extracellular Vesicles: The Role of Extracellular Vesicles in Primary and Metastatic Brain Tumour Microenvironment. J. Extracell. Vesicles 2019, 8, 1627164. [Google Scholar] [CrossRef]
- Brites, D.; Vaz, A.R. Microglia Centered Pathogenesis in ALS: Insights in Cell Interconnectivity. Front. Cell. Neurosci. 2014, 8, 117. [Google Scholar] [CrossRef]
- Brites, D.; Fernandes, A. Neuroinflammation and Depression: Microglia Activation, Extracellular Microvesicles and microRNA Dysregulation. Front. Cell. Neurosci. 2015, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Pulliam, L. Exosomes as Mediators of Neuroinflammation. J. Neuroinflamm. 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Simons, M. Exosomes: Vesicular Carriers for Intercellular Communication in Neurodegenerative Disorders. Cell Tissue Res. 2013, 352, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, T.; Masuda-Suzukake, M.; Arai, T.; Hasegawa, Y.; Akatsu, H.; Obi, T.; Yoshida, M.; Murayama, S.; Mann, D.M.A.; Akiyama, H.; et al. Prion-like Properties of Pathological TDP-43 Aggregates from Diseased Brains. Cell Rep. 2013, 4, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Grad, L.I.; Yerbury, J.J.; Turner, B.J.; Guest, W.C.; Pokrishevsky, E.; O’Neill, M.A.; Yanai, A.; Silverman, J.M.; Zeineddine, R.; Corcoran, L.; et al. Intercellular Propagated Misfolding of Wild-Type Cu/Zn Superoxide Dismutase Occurs via Exosome-Dependent and -Independent Mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 3620–3625. [Google Scholar] [CrossRef]
- Cicardi, M.E.; Marrone, L.; Azzouz, M.; Trotti, D. Proteostatic Imbalance and Protein Spreading in Amyotrophic Lateral Sclerosis. EMBO J. 2021, 40, e106389. [Google Scholar] [CrossRef]
- Salvany, S.; Casanovas, A.; Piedrafita, L.; Gras, S.; Calderó, J.; Esquerda, J.E. Accumulation of Misfolded SOD1 Outlines Distinct Patterns of Motor Neuron Pathology and Death during Disease Progression in a SOD1 Mouse Model of Amyotrophic Lateral Sclerosis. Brain Pathol. 2022, 32, e13078. [Google Scholar] [CrossRef]
- Dobrowolny, G.; Martini, M.; Scicchitano, B.M.; Romanello, V.; Boncompagni, S.; Nicoletti, C.; Pietrangelo, L.; De Panfilis, S.; Catizone, A.; Bouchè, M.; et al. Muscle Expression of SOD1 Triggers the Dismantlement of Neuromuscular Junction via PKC-Theta. Antioxid. Redox Signal. 2018, 28, 1105–1119. [Google Scholar] [CrossRef]
- Peggion, C.; Scalcon, V.; Massimino, M.L.; Nies, K.; Lopreiato, R.; Rigobello, M.P.; Bertoli, A. SOD1 in ALS: Taking Stock in Pathogenic Mechanisms and the Role of Glial and Muscle Cells. Antioxidants 2022, 11, 614. [Google Scholar] [CrossRef]
- Thome, A.D.; Thonhoff, J.R.; Zhao, W.; Faridar, A.; Wang, J.; Beers, D.R.; Appel, S.H. Extracellular Vesicles Derived from Expanded Regulatory T Cells Modulate and Inflammation. Front. Immunol. 2022, 13, 875825. [Google Scholar] [CrossRef]
- Chen, P.-C.; Wu, D.; Hu, C.-J.; Chen, H.-Y.; Hsieh, Y.-C.; Huang, C.-C. Exosomal TAR DNA-Binding Protein-43 and Neurofilaments in Plasma of Amyotrophic Lateral Sclerosis Patients: A Longitudinal Follow-up Study. J. Neurol. Sci. 2020, 418, 117070. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Ma, M.; Teng, J.; Teng, R.K.F.; Zhou, S.; Yin, J.; Fonkem, E.; Huang, J.H.; Wu, E.; Wang, X. Exposure to ALS-FTD-CSF Generates TDP-43 Aggregates in Glioblastoma Cells through Exosomes and TNTs-like Structure. Oncotarget 2015, 6, 24178–24191. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.; Cunha, C.; Barbosa, M.; Vaz, A.R.; Brites, D. Exosomes from NSC-34 Cells Transfected with hSOD1-G93A Are Enriched in miR-124 and Drive Alterations in Microglia Phenotype. Front. Neurosci. 2017, 11, 273. [Google Scholar] [CrossRef]
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant Copper-Zinc Superoxide Dismutase (SOD1) Induces Protein Secretion Pathway Alterations and Exosome Release in Astrocytes: Implications for Disease Spreading and Motor Neuron Pathology in Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2013, 288, 15699–15711. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xia, K.; Chen, L.; Fan, D. Increased Interleukin-6 Levels in the Astrocyte-Derived Exosomes of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Neurosci. 2019, 13, 574. [Google Scholar] [CrossRef]
- Le Gall, L.; Duddy, W.J.; Martinat, C.; Mariot, V.; Connolly, O.; Milla, V.; Anakor, E.; Ouandaogo, Z.G.; Millecamps, S.; Lainé, J.; et al. Muscle Cells of Sporadic Amyotrophic Lateral Sclerosis Patients Secrete Neurotoxic Vesicles. J. Cachexia Sarcopenia Muscle 2022, 13, 1385–1402. [Google Scholar] [CrossRef] [PubMed]
- Anakor, E.; Milla, V.; Connolly, O.; Martinat, C.; Pradat, P.F.; Dumonceaux, J.; Duddy, W.; Duguez, S. The Neurotoxicity of Vesicles Secreted by ALS Patient Myotubes Is Specific to Exosome-like and Not Larger Subtypes. Cells 2022, 11, 845. [Google Scholar] [CrossRef]
- Laneve, P.; Tollis, P.; Caffarelli, E. RNA Deregulation in Amyotrophic Lateral Sclerosis: The Noncoding Perspective. Int. J. Mol. Sci. 2021, 22, 10285. [Google Scholar] [CrossRef]
- Barbo, M.; Ravnik-Glavač, M. Extracellular Vesicles as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Genes 2023, 14, 325. [Google Scholar] [CrossRef]
- Yin, H.; He, H.; Shen, X.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; Xia, L.; et al. miR-9-5p Inhibits Skeletal Muscle Satellite Cell Proliferation and Differentiation by Targeting IGF2BP3 through the IGF2-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 1655. [Google Scholar] [CrossRef]
- Wang, M.; Gao, Q.; Chen, Y.; Li, Z.; Yue, L.; Cao, Y. PAK4, a Target of miR-9-5p, Promotes Cell Proliferation and Inhibits Apoptosis in Colorectal Cancer. Cell. Mol. Biol. Lett. 2019, 24, 58. [Google Scholar] [CrossRef] [PubMed]
- Saucier, D.; Wajnberg, G.; Roy, J.; Beauregard, A.-P.; Chacko, S.; Crapoulet, N.; Fournier, S.; Ghosh, A.; Lewis, S.M.; Marrero, A.; et al. Identification of a Circulating miRNA Signature in Extracellular Vesicles Collected from Amyotrophic Lateral Sclerosis Patients. Brain Res. 2019, 1708, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ha, S.E.; Wei, L.; Jin, B.; Zogg, H.; Poudrier, S.M.; Jorgensen, B.G.; Park, C.; Ronkon, C.F.; Bartlett, A.; et al. miR-10b-5p Rescues Diabetes and Gastrointestinal Dysmotility. Gastroenterology 2021, 160, 1662–1678.e18. [Google Scholar] [CrossRef]
- Ge, G.; Yang, D.; Tan, Y.; Chen, Y.; Jiang, D.; Jiang, A.; Li, Q.; Liu, Y.; Zhong, Z.; Li, X.; et al. miR-10b-5p Regulates C2C12 Myoblasts Proliferation and Differentiation. Biosci. Biotechnol. Biochem. 2019, 83, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Faur, C.I.; Roman, R.C.; Jurj, A.; Raduly, L.; Almășan, O.; Rotaru, H.; Chirilă, M.; Moldovan, M.A.; Hedeșiu, M.; Dinu, C. Salivary Exosomal MicroRNA-486-5p and MicroRNA-10b-5p in Oral and Oropharyngeal Squamous Cell Carcinoma. Medicina 2022, 58, 1478. [Google Scholar] [CrossRef]
- Yelick, J.; Men, Y.; Jin, S.; Seo, S.; Espejo-Porras, F.; Yang, Y. Elevated Exosomal Secretion of miR-124-3p from Spinal Neurons Positively Associates with Disease Severity in ALS. Exp. Neurol. 2020, 333, 113414. [Google Scholar] [CrossRef] [PubMed]
- Banack, S.A.; Dunlop, R.A.; Stommel, E.W.; Mehta, P.; Cox, P.A. miRNA Extracted from Extracellular Vesicles Is a Robust Biomarker of Amyotrophic Lateral Sclerosis. J. Neurol. Sci. 2022, 442, 120396. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Q.; Li, J.; Chen, W.; Jin, X.; Wang, Y. MicroRNA-15a-5p Inhibits Endometrial Carcinoma Proliferation, Invasion and Migration via Downregulation of VEGFA and Inhibition of the Wnt/β-Catenin Signaling Pathway. Oncol. Lett. 2021, 21, 310. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, H.; Wang, X.; Zhou, L.; Li, H.; Deng, T.; Qu, Y.; Duan, J.; Bai, M.; Ge, S.; et al. The miR-24-Bim Pathway Promotes Tumor Growth and Angiogenesis in Pancreatic Carcinoma. Oncotarget 2015, 6, 43831–43842. [Google Scholar] [CrossRef]
- Katsu, M.; Hama, Y.; Utsumi, J.; Takashina, K.; Yasumatsu, H.; Mori, F.; Wakabayashi, K.; Shoji, M.; Sasaki, H. MicroRNA Expression Profiles of Neuron-Derived Extracellular Vesicles in Plasma from Patients with Amyotrophic Lateral Sclerosis. Neurosci. Lett. 2019, 708, 134176. [Google Scholar] [CrossRef]
- Li, S.; Hu, C.; Li, J.; Liu, L.; Jing, W.; Tang, W.; Tian, W.; Long, J. Effect of miR-26a-5p on the Wnt/Ca(2+) Pathway and Osteogenic Differentiation of Mouse Adipose-Derived Mesenchymal Stem Cells. Calcif. Tissue Int. 2016, 99, 174–186. [Google Scholar] [CrossRef]
- Lo, T.-W.; Figueroa-Romero, C.; Hur, J.; Pacut, C.; Stoll, E.; Spring, C.; Lewis, R.; Nair, A.; Goutman, S.A.; Sakowski, S.A.; et al. Extracellular Vesicles in Serum and Central Nervous System Tissues Contain microRNA Signatures in Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2021, 14, 739016. [Google Scholar] [CrossRef]
- Zhou, L.; Liang, X.; Zhang, L.; Yang, L.; Nagao, N.; Wu, H.; Liu, C.; Lin, S.; Cai, G.; Liu, J. MiR-27a-3p Functions as an Oncogene in Gastric Cancer by Targeting BTG2. Oncotarget 2016, 7, 51943–51954. [Google Scholar] [CrossRef]
- Xi, T.; Jin, F.; Zhu, Y.; Wang, J.; Tang, L.; Wang, Y.; Liebeskind, D.S.; Scalzo, F.; He, Z. miR-27a-3p Protects against Blood-Brain Barrier Disruption and Brain Injury after Intracerebral Hemorrhage by Targeting Endothelial Aquaporin-11. J. Biol. Chem. 2018, 293, 20041–20050. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the Extraction and Determination of Serum Exosome and miRNA in Serum and the Detection of miR-27a-3p in Serum Exosome of ALS Patients. Intractable Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Ye, L.; Chen, C.; She, J.; Song, Y. MiR-29b-3p Promotes Particulate Matter-Induced Inflammatory Responses by Regulating the C1QTNF6/AMPK Pathway. Aging 2020, 12, 1141–1158. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Gao, M.; Wang, Z.; Wang, W.; Zhan, L.; Wei, B. Upregulation of MicroRNA-34a Sensitizes Ovarian Cancer Cells to Resveratrol by Targeting Bcl-2. Yonsei Med. J. 2021, 62, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative Mechanisms of miR-34a Regulation in Cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, M.; Melzi, V.; Gagliardi, D.; Resnati, D.; Meneri, M.; Dioni, L.; Masrori, P.; Hersmus, N.; Poesen, K.; Locatelli, M.; et al. Insights into the Identification of a Molecular Signature for Amyotrophic Lateral Sclerosis Exploiting Integrated microRNA Profiling of iPSC-Derived Motor Neurons and Exosomes. Cell. Mol. Life Sci. 2022, 79, 189. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Zhu, T.; Liu, H.; Zhu, Y.; Peng, Y.; Tang, T.; Zhou, S.; Hu, C.; Chen, H.; Guo, A.; et al. The miR-100-5p Targets SMARCA5 to Regulate the Apoptosis and Intracellular Survival of BCG in Infected THP-1 Cells. Cells 2023, 12, 476. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, Y.; Huang, S.; Yan, M.; Li, J.; Jin, T.; Bao, S. MiR-100-5p, miR-199a-3p and miR-199b-5p Induce Autophagic Death of Endometrial Carcinoma Cell through Targeting mTOR. Int. J. Clin. Exp. Pathol. 2017, 10, 9262–9272. [Google Scholar] [PubMed]
- Zeng, J.; Wang, L.; Zhao, J.; Zheng, Z.; Peng, J.; Zhang, W.; Wen, T.; Nie, J.; Ding, L.; Yi, D. MiR-100-5p Regulates Cardiac Hypertrophy through Activation of Autophagy by Targeting mTOR. Hum. Cell 2021, 34, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Dubes, S.; Soula, A.; Benquet, S.; Tessier, B.; Poujol, C.; Favereaux, A.; Thoumine, O.; Letellier, M. miR-124-Dependent Tagging of Synapses by Synaptopodin Enables Input-Specific Homeostatic Plasticity. EMBO J. 2022, 41, e109012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-B.; Xiong, L.-L.; Xue, L.-L.; Deng, Y.-P.; Du, R.-L.; Hu, Q.; Xu, Y.; Yang, S.-J.; Wang, T.-H. MiR-127-3p Targeting CISD1 Regulates Autophagy in Hypoxic-Ischemic Cortex. Cell Death Dis. 2021, 12, 279. [Google Scholar] [CrossRef]
- Shen, L.; Li, Q.; Wang, J.; Zhao, Y.; Niu, L.; Bai, L.; Shuai, S.; Li, X.; Zhang, S.; Zhu, L. miR-144-3p Promotes Adipogenesis Through Releasing C/EBPα From Klf3 and CtBP2. Front. Genet. 2018, 9, 677. [Google Scholar] [CrossRef]
- Pan, J.; Du, M.; Cao, Z.; Zhang, C.; Hao, Y.; Zhu, J.; He, H. miR-146a-5p Attenuates IL-1β-Induced IL-6 and IL-1β Expression in a Cementoblast-Derived Cell Line. Oral Dis. 2020, 26, 1308–1317. [Google Scholar] [CrossRef]
- Banack, S.A.; Dunlop, R.A.; Cox, P.A. An miRNA Fingerprint Using Neural-Enriched Extracellular Vesicles from Blood Plasma: Towards a Biomarker for Amyotrophic Lateral Sclerosis/motor Neuron Disease. Open Biol. 2020, 10, 200116. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhu, H.; Lei, Q.; Chen, L.; Yang, D.; Sui, W. MicroRNA-149-3p Inhibits Cell Proliferation by Targeting AKT2 in Oral Squamous Cell Carcinoma. Mol. Med. Rep. 2021, 23, 172. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ye, G.; Liu, Y.; Huang, D.; Luo, Q.; Chen, W.; Qi, Z. miR-150-3p Enhances Neuroprotective Effects of Neural Stem Cell Exosomes after Hypoxic-Ischemic Brain Injury by Targeting CASP2. Neurosci. Lett. 2022, 779, 136635. [Google Scholar] [CrossRef]
- Wang, H.; Shu, J.; Zhang, C.; Wang, Y.; Shi, R.; Yang, F.; Tang, X. Extracellular Vesicle-Mediated miR-150-3p Delivery in Joint Homeostasis: A Potential Treatment for Osteoarthritis? Cells 2022, 11, 2766. [Google Scholar] [CrossRef]
- Zhang, Y.; Hao, T.; Zhang, H.; Wei, P.; Li, X. Over-expression of miR-151a-3p inhibits proliferation and migration of PC-3 prostate cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2018, 34, 247–252. [Google Scholar] [PubMed]
- Zhou, R.; Wang, R.; Qin, Y.; Ji, J.; Xu, M.; Wu, W.; Chen, M.; Wu, D.; Song, L.; Shen, H.; et al. Mitochondria-Related miR-151a-5p Reduces Cellular ATP Production by Targeting CYTB in Asthenozoospermia. Sci. Rep. 2015, 5, 17743. [Google Scholar] [CrossRef]
- Ma, Z.; Qiu, X.; Wang, D.; Li, Y.; Zhang, B.; Yuan, T.; Wei, J.; Zhao, B.; Zhao, X.; Lou, J.; et al. MiR-181a-5p Inhibits Cell Proliferation and Migration by Targeting Kras in Non-Small Cell Lung Cancer A549 Cells. Acta Biochim. Biophys. Sin. 2015, 47, 630–638. [Google Scholar] [CrossRef]
- Cao, Y.; Zhao, D.; Li, P.; Wang, L.; Qiao, B.; Qin, X.; Li, L.; Wang, Y. MicroRNA-181a-5p Impedes IL-17-Induced Nonsmall Cell Lung Cancer Proliferation and Migration through Targeting VCAM-1. Cell. Physiol. Biochem. 2017, 42, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, M.; Liu, G.; Xiong, C.; Rao, J. microRNA-181a-5p Impedes the Proliferation, Migration, and Invasion of Retinoblastoma Cells by Targeting the NRAS Proto-Oncogene. Clinics 2022, 77, 100026. [Google Scholar] [CrossRef] [PubMed]
- Assmann, J.L.J.C.; Leon, L.G.; Stavast, C.J.; van den Bogaerdt, S.E.; Schilperoord-Vermeulen, J.; Sandberg, Y.; Bellido, M.; Erkeland, S.J.; Feith, D.J.; Loughran, T.P., Jr.; et al. miR-181a Is a Novel Player in the STAT3-Mediated Survival Network of TCRαβ+ CD8+ T Large Granular Lymphocyte Leukemia. Leukemia 2022, 36, 983–993. [Google Scholar] [CrossRef]
- Li, X.; Han, J.; Zhu, H.; Peng, L.; Chen, Z. miR-181b-5p Mediates TGF-β1-Induced Epithelial-to-Mesenchymal Transition in Non-Small Cell Lung Cancer Stem-like Cells Derived from Lung Adenocarcinoma A549 Cells. Int. J. Oncol. 2017, 51, 158–168. [Google Scholar] [CrossRef]
- Liu, B.; Guo, Z.; Gao, W. miR-181b-5p Promotes Proliferation and Inhibits Apoptosis of Hypertrophic Scar Fibroblasts through Regulating the MEK/ERK/p21 Pathway. Exp. Ther. Med. 2019, 17, 1537–1544. [Google Scholar] [CrossRef]
- Yeon, M.; Kim, Y.; Pathak, D.; Kwon, E.; Kim, D.Y.; Jeong, M.S.; Jung, H.S.; Jeoung, D. The CAGE-MiR-181b-5p-S1PR1 Axis Regulates Anticancer Drug Resistance and Autophagy in Gastric Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 666387. [Google Scholar] [CrossRef]
- Graham, A.; Holbert, J.; Nothnick, W.B. miR-181b-5p Modulates Cell Migratory Proteins, Tissue Inhibitor of Metalloproteinase 3, and Annexin A2 During In Vitro Decidualization in a Human Endometrial Stromal Cell Line. Reprod. Sci. 2017, 24, 1264–1274. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, J.; Zhao, C.; Ren, H.; Yuan, Z.; Zhang, B.; Zhuang, J.; Wang, J.; Feng, B. MiR-181b-5p Modulates Chemosensitivity of Glioma Cells to Temozolomide by Targeting Bcl-2. Biomed. Pharmacother. 2019, 109, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Ou, R.; Gu, X.; Cao, B.; Shang, H. MicroRNA-183-5p Is Stress-Inducible and Protects Neurons against Cell Death in Amyotrophic Lateral Sclerosis. J. Cell. Mol. Med. 2020, 24, 8614–8622. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Sheng, L.; Qu, W.; Xue, X.; Chen, H.; Zheng, G.; Chen, W. MiR-194-5p Enhances the Sensitivity of Nonsmall-Cell Lung Cancer to Doxorubicin through Targeted Inhibition of Hypoxia-Inducible Factor-1. World J. Surg. Oncol. 2021, 19, 174. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Gong, A. The Expression of microRNA-197-3p Regulates the Proliferation of Ovarian Granulosa Cells through CUL3 in Polycystic Ovarian Syndrome. Acta Biochim. Pol. 2022, 69, 599–604. [Google Scholar] [CrossRef]
- Jiang, J.; Tang, Q.; Gong, J.; Jiang, W.; Chen, Y.; Zhou, Q.; Aldeen, A.; Wang, S.; Li, C.; Lv, W.; et al. Radiosensitizer EXO-miR-197-3p Inhibits Nasopharyngeal Carcinoma Progression and Radioresistance by Regulating the AKT/mTOR Axis and HSPA5-Mediated Autophagy. Int. J. Biol. Sci. 2022, 18, 1878–1895. [Google Scholar] [CrossRef]
- Chang, R.-M.; Fu, Y.; Zeng, J.; Zhu, X.-Y.; Gao, Y. Cancer-Derived Exosomal miR-197-3p Confers Angiogenesis via Targeting TIMP2/3 in Lung Adenocarcinoma Metastasis. Cell Death Dis. 2022, 13, 1032. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, J.; Dong, J.; Bai, R.; Song, D.; Ma, X.; Zhao, L.; Yao, Y.; Zhang, H.; Liu, T. Association of miR-197-5p, a Circulating Biomarker for Heart Failure, with Myocardial Fibrosis and Adverse Cardiovascular Events among Patients with Stage C or D Heart Failure. Cardiology 2018, 141, 212–225. [Google Scholar] [CrossRef]
- Ghosh, A.; Dasgupta, D.; Ghosh, A.; Roychoudhury, S.; Kumar, D.; Gorain, M.; Butti, R.; Datta, S.; Agarwal, S.; Gupta, S.; et al. MiRNA199a-3p Suppresses Tumor Growth, Migration, Invasion and Angiogenesis in Hepatocellular Carcinoma by Targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017, 8, e2706. [Google Scholar] [CrossRef]
- Ren, K.; Li, T.; Zhang, W.; Ren, J.; Li, Z.; Wu, G. miR-199a-3p Inhibits Cell Proliferation and Induces Apoptosis by Targeting YAP1, Suppressing Jagged1-Notch Signaling in Human Hepatocellular Carcinoma. J. Biomed. Sci. 2016, 23, 79. [Google Scholar] [CrossRef]
- Liu, J.; Quan, Z.; Gao, Y.; Wu, X.; Zheng, Y. MicroRNA-199b-3p Suppresses Malignant Proliferation by Targeting Phospholipase Cε and Correlated with Poor Prognosis in Prostate Cancer. Biochem. Biophys. Res. Commun. 2021, 576, 73–79. [Google Scholar] [CrossRef]
- Huang, G.-H.; Shan, H.; Li, D.; Zhou, B.; Pang, P.-F. MiR-199a-5p Suppresses Tumorigenesis by Targeting Clathrin Heavy Chain in Hepatocellular Carcinoma. Cell Biochem. Funct. 2017, 35, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Y.-Z.; Zhao, K.-X. MiR-298 Suppresses the Malignant Progression of Osteosarcoma by Targeting JMJD6. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 2250–2258. [Google Scholar] [PubMed]
- Ye, L.; Wang, F.; Wu, H.; Yang, H.; Yang, Y.; Ma, Y.; Xue, A.; Zhu, J.; Chen, M.; Wang, J.; et al. Functions and Targets of miR-335 in Cancer. Onco Targets Ther. 2021, 14, 3335–3349. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Moro, A.; Wang, J.; Meng, D.; Zhan, X.; Wei, Q. MicroRNA-338-3p as a Novel Therapeutic Target for Intervertebral Disc Degeneration. Exp. Mol. Med. 2021, 53, 1356–1365. [Google Scholar] [CrossRef]
- Zhang, C.; Li, H.; Wang, J.; Zhang, J.; Hou, X. MicroRNA-338-3p Suppresses Cell Proliferation, Migration and Invasion in Human Malignant Melanoma by Targeting MACC1. Exp. Ther. Med. 2019, 18, 997–1004. [Google Scholar] [CrossRef]
- Fu, Y.; Hu, X.; Zheng, C.; Sun, G.; Xu, J.; Luo, S.; Cao, P. Intrahippocampal miR-342-3p Inhibition Reduces β-Amyloid Plaques and Ameliorates Learning and Memory in Alzheimer’s Disease. Metab. Brain Dis. 2019, 34, 1355–1363. [Google Scholar] [CrossRef]
- Montag, J.; Hitt, R.; Opitz, L.; Schulz-Schaeffer, W.J.; Hunsmann, G.; Motzkus, D. Upregulation of miRNA Hsa-miR-342-3p in Experimental and Idiopathic Prion Disease. Mol. Neurodegener. 2009, 4, 36. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Calin, G.A.; Yuen, K.S.; Jin, D.Y.; Chim, C.S. Epigenetic Silencing of miR-342-3p in B Cell Lymphoma and Its Impact on Autophagy. Clin. Epigenet. 2020, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Luo, R.; Yang, W.; Zhou, Z.; Li, C. miR-363-3p Is Activated by MYB and Regulates Osteoporosis Pathogenesis via PTEN/PI3K/AKT Signaling Pathway. Vitr. Cell. Dev. Biol. Anim. 2019, 55, 376–386. [Google Scholar] [CrossRef]
- Lv, Z.; Qiu, X.; Jin, P.; Li, Z.; Zhang, Y.; Lv, L.; Song, F. MiR-371a-5p Positively Associates with Hepatocellular Carcinoma Malignancy but Sensitizes Cancer Cells to Oxaliplatin by Suppressing BECN1-Dependent Autophagy. Life 2022, 12, 1651. [Google Scholar] [CrossRef]
- Muys, B.R.; Sousa, J.F.; Plaça, J.R.; de Araújo, L.F.; Sarshad, A.A.; Anastasakis, D.G.; Wang, X.; Li, X.L.; de Molfetta, G.A.; Ramão, A.; et al. miR-450a Acts as a Tumor Suppressor in Ovarian Cancer by Regulating Energy Metabolism. Cancer Res. 2019, 79, 3294–3305. [Google Scholar] [CrossRef] [PubMed]
- Varcianna, A.; Myszczynska, M.A.; Castelli, L.M.; O’Neill, B.; Kim, Y.; Talbot, J.; Nyberg, S.; Nyamali, I.; Heath, P.R.; Stopford, M.J.; et al. Micro-RNAs Secreted through Astrocyte-Derived Extracellular Vesicles Cause Neuronal Network Degeneration in C9orf72 ALS. EBioMedicine 2019, 40, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, M.; Liu, W.; Guo, C.; Zhan, L.; Zhan, X. miR-502-5p Inhibits the Proliferation, Migration and Invasion of Gastric Cancer Cells by Targeting SP1. Oncol. Lett. 2020, 20, 2757–2762. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Tan, S.; Tang, H.; Chen, Y.; Shu, P. miR-512-5p Suppresses Proliferation, Migration and Invasion, and Induces Apoptosis in Non-small Cell Lung Cancer Cells by Targeting ETS1. Mol. Med. Rep. 2019, 19, 3604–3614. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Q.; Huang, Z.-P.; Li, H.-F.; Ou, Y.-L.; Huo, F.; Hu, L.-K. MicroRNA-520f-3p Inhibits Proliferation of Gastric Cancer Cells via Targeting SOX9 and Thereby Inactivating Wnt Signaling. Sci. Rep. 2020, 10, 6197. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Q.; Dai, Y.; Jiang, T.; Zhou, Y. miR-532-3p Inhibits Proliferation and Promotes Apoptosis of Lymphoma Cells by Targeting β-Catenin. J. Cancer 2020, 11, 4762–4770. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Gao, L.; Yu, Z. Tumor-Suppressive microRNA-551b-3p Targets H6PD to Inhibit Gallbladder Cancer Progression. Cancer Gene Ther. 2021, 28, 693–705. [Google Scholar] [CrossRef]
- Xuan, Z.; Chen, C.; Tang, W.; Ye, S.; Zheng, J.; Zhao, Y.; Shi, Z.; Zhang, L.; Sun, H.; Shao, C. TKI-Resistant Renal Cancer Secretes Low-Level Exosomal miR-549a to Induce Vascular Permeability and Angiogenesis to Promote Tumor Metastasis. Front. Cell Dev. Biol. 2021, 9, 689947. [Google Scholar] [CrossRef]
- Chen, M.; Wang, D.; Liu, J.; Zhou, Z.; Ding, Z.; Liu, L.; Su, D.; Li, H. Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting Ribosomal Protein SA. Biomed. Res. Int. 2020, 2020, 3280530. [Google Scholar] [CrossRef]
- Zheng, H.; Ma, R.; Wang, Q.; Zhang, P.; Li, D.; Wang, Q.; Wang, J.; Li, H.; Liu, H.; Wang, Z. MiR-625-3p Promotes Cell Migration and Invasion via Inhibition of SCAI in Colorectal Carcinoma Cells. Oncotarget 2015, 6, 27805–27815. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, S.; Wang, J.; Zhu, Z.; Luo, L.; Xiang, Q.; Zhou, M.; Ma, Y.; Wang, Z.; Zhao, Z. MiR-629-5p Promotes Prostate Cancer Development and Metastasis by Targeting AKAP13. Front. Oncol. 2021, 11, 754353. [Google Scholar] [CrossRef]
- Cong, J.; Liu, R.; Wang, X.; Jiang, H.; Zhang, Y. MiR-634 Decreases Cell Proliferation and Induces Apoptosis by Targeting mTOR Signaling Pathway in Cervical Cancer Cells. Artif. Cells Nanomed. Biotechnol. 2015, 44, 1694–1701. [Google Scholar] [CrossRef]
- Chen, D.; Wu, X.; Zhao, J.; Zhao, X. MicroRNA-634 Functions as a Tumor Suppressor in Pancreatic Cancer via Directly Targeting Heat Shock-Related 70-kDa Protein 2. Exp. Ther. Med. 2019, 17, 3949–3956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Wu, M.; Wang, H.; Wu, L.; Xu, B.; Zhou, W.; Fan, X.; Shao, J.; Yang, T. MicroRNA-664a-5p Promotes Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Directly Downregulating HMGA2. Biochem. Biophys. Res. Commun. 2020, 521, 9–14. [Google Scholar] [CrossRef]
- Watanabe, K.; Yamaji, R.; Ohtsuki, T. MicroRNA-664a-5p Promotes Neuronal Differentiation of SH-SY5Y Cells. Genes Cells 2018, 23, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. MicroRNA-766-3p Contributes to Anti-Inflammatory Responses through the Indirect Inhibition of NF-κB Signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef]
- Gao, J.; Fei, L.; Wu, X.; Li, H. MiR-766-3p Suppresses Malignant Behaviors and Stimulates Apoptosis of Colon Cancer Cells via Targeting TGFBI. Can. J. Gastroenterol. Hepatol. 2022, 2022, 7234704. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Diao, H.; Wang, P.; Guan, F.; Liu, H. microRNA-877-5p Exerts Tumor-Suppressive Functions in Prostate Cancer through Repressing Transcription of Forkhead Box M1. Bioengineered 2021, 12, 9094–9102. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, G.; Gao, H.; Li, Y.; Zhuang, L.; Meng, Z.; Liu, L. miR-939-5p Contributes to the Migration and Invasion of Pancreatic Cancer by Targeting ARHGAP4. Onco Targets Ther. 2020, 13, 389–399. [Google Scholar] [CrossRef]
- Bertolazzi, G.; Cipollina, C.; Benos, P.V.; Tumminello, M.; Coronnello, C. miR-1207-5p Can Contribute to Dysregulation of Inflammatory Response in COVID-19 Targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020, 10, 586592. [Google Scholar] [CrossRef]
- Song, M.; Xie, D.; Gao, S.; Bai, C.-J.; Zhu, M.-X.; Guan, H.; Zhou, P.-K. A Biomarker Panel of Radiation-Upregulated miRNA as Signature for Ionizing Radiation Exposure. Life 2020, 10, 361. [Google Scholar] [CrossRef]
- Mo, L.-J.; Song, M.; Huang, Q.-H.; Guan, H.; Liu, X.-D.; Xie, D.-F.; Huang, B.; Huang, R.-X.; Zhou, P.-K. Exosome-Packaged miR-1246 Contributes to Bystander DNA Damage by Targeting LIG4. Br. J. Cancer 2018, 119, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Shi, L.; Yang, C.; Ge, Y.; Lin, L.; Fan, H.; He, Y.; Zhang, D.; Miao, Y.; Yang, L. miR-1254 Inhibits Cell Proliferation, Migration, and Invasion by down-Regulating Smurf1 in Gastric Cancer. Cell Death Dis. 2019, 10, 32. [Google Scholar] [CrossRef]
- Xin, Y.; Wang, X.; Meng, K.; Ni, C.; Lv, Z.; Guan, D. Identification of Exosomal miR-455-5p and miR-1255a as Therapeutic Targets for Breast Cancer. Biosci. Rep. 2020, 40, BSR20190303. [Google Scholar] [CrossRef]
- Huang, Z.; Zhen, S.; Jin, L.; Chen, J.; Han, Y.; Lei, W.; Zhang, F. miRNA-1260b Promotes Breast Cancer Cell Migration and Invasion by Downregulating CCDC134. Curr. Gene Ther. 2023, 23, 60–71. [Google Scholar]
- Lei, T.; Zhang, L.; Song, Y.; Wang, B.; Shen, Y.; Zhang, N.; Yang, M. Transcriptionally Modulated by an Enhancer Genetic Variant Improves Efficiency of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors in Advanced Lung Adenocarcinoma. DNA Cell Biol. 2020, 39, 1111–1118. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Ren, J.; Deng, S.; Yi, G.; Guo, M.; Shu, S.; Zhao, L.; Peng, Y.; Qi, S. miR-1268a Regulates ABCC1 Expression to Mediate Temozolomide Resistance in Glioblastoma. J. Neurooncol. 2018, 138, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.-J.; Chen, X.; Wang, Y.-W.; Liu, H.-T.; Ma, R.-R.; Gao, P. MiR-1268b Confers Chemosensitivity in Breast Cancer by Targeting ERBB2-Mediated PI3K-AKT Pathway. Oncotarget 2017, 8, 89631–89642. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, Z.; Zheng, P.; Zhao, W.; Han, N. MicroRNA-1285-5p Influences the Proliferation and Metastasis of Non-Small-Cell Lung Carcinoma Cells via Downregulating CDH1 and Smad4. Tumour Biol. 2017, 39, 1010428317705513. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, X.-Y.; Zhang, Z.-M.; Li, S.; Ren, L.-H.; Cao, R.-S.; Feng, Y.-D.; Ji, Y.-L.; Zhao, Y.; Shi, R.-H. MicroRNA-1290 Promotes Esophageal Squamous Cell Carcinoma Cell Proliferation and Metastasis. World J. Gastroenterol. 2015, 21, 3245–3255. [Google Scholar] [CrossRef]
- miRDB—MicroRNA Target Prediction Database. Available online: https://mirdb.org/ (accessed on 21 June 2023).
- Byun, Y.J.; Piao, X.M.; Jeong, P.; Kang, H.W.; Seo, S.P.; Moon, S.K.; Lee, J.Y.; Choi, Y.H.; Lee, H.Y.; Kim, W.T.; et al. Urinary microRNA-1913 to microRNA-3659 Expression Ratio as a Non-Invasive Diagnostic Biomarker for Prostate Cancer. Investig. Clin. Urol. 2021, 62, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wan, X.; Chen, X.; Fang, Y.; Cheng, X.; Xie, X.; Lu, W. miR-2861 Acts as a Tumor Suppressor via Targeting EGFR/AKT2/CCND1 Pathway in Cervical Cancer Induced by Human Papillomavirus Virus 16 E6. Sci. Rep. 2016, 6, 28968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Deng, L.; Ji, Y.; Cheng, G.; Su, D.; Qiu, B. Novel Long Noncoding RNA (lncRNA) Panel as Biomarkers for Prognosis in Lung Squamous Cell Carcinoma via Competitive Endogenous RNA (ceRNA) Network Analysis. Transl. Cancer Res. 2021, 10, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, Y.; Feng, Z.; Sheng, J.; Dong, A.; Zhang, M.; Cao, L. miR-345-5p Regulates Adipogenesis via Targeting VEGF-B. Aging 2020, 12, 17114–17121. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cao, S.; Hong, S.; Lin, X.; Guan, H.; Chen, S.; Zhang, Q.; Lv, W.; Li, Y.; Xiao, H. miR-3619-3p Promotes Papillary Thyroid Carcinoma Progression via Wnt/β-Catenin Pathway. Ann. Transl. Med. 2019, 7, 643. [Google Scholar] [CrossRef]
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jonsson, G.; Naya, H.; Hoglund, M.; et al. Identification of New microRNAs in Paired Normal and Tumor Breast Tissue Suggests a Dual Role for the ERBB2/Her2 Gene. Cancer Res. 2011, 71, 78–86. [Google Scholar] [CrossRef]
- Komina, A.; Palkina, N.; Aksenenko, M.; Tsyrenzhapova, S.; Ruksha, T. Antiproliferative and Pro-Apoptotic Effects of MiR-4286 Inhibition in Melanoma Cells. PLoS ONE 2016, 11, e0168229. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, S.; Wang, H.; Zhang, B.; Zhang, P. miR-4286 Promotes Prostate Cancer Progression via Targeting the Expression of SALL1. J. Gene Med. 2019; Early View. [Google Scholar]
- He, Z.; Xue, H.; Liu, P.; Han, D.; Xu, L.; Zeng, X.; Wang, J.; Yang, B.; Luo, B. miR-4286/TGF-β1/Smad3-Negative Feedback Loop Ameliorated Vascular Endothelial Cell Damage by Attenuating Apoptosis and Inflammatory Response. J. Cardiovasc. Pharmacol. 2020, 75, 446–454. [Google Scholar] [CrossRef]
- Ling, C.; Wang, X.; Zhu, J.; Tang, H.; Du, W.; Zeng, Y.; Sun, L.; Huang, J.-A.; Liu, Z. MicroRNA-4286 Promotes Cell Proliferation, Migration, and Invasion via PTEN Regulation of the PI3K/Akt Pathway in Non-Small Cell Lung Cancer. Cancer Med. 2019, 8, 3520–3531. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, W.; Luan, Q.; Liu, Y. miR-4284 Promotes Cell Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells and Is Associated with Postoperative Prognosis. Cancer Manag. Res. 2021, 13, 5865–5872. [Google Scholar] [CrossRef]
- Zuo, X.-M.; Sun, H.-W.; Fang, H.; Wu, Y.; Shi, Q.; Yu, Y.-F. miR-4443 Targets TRIM14 to Suppress Metastasis and Energy Metabolism of Papillary Thyroid Carcinoma (PTC) in Vitro. Cell Biol. Int. 2021, 45, 1917–1925. [Google Scholar] [CrossRef]
- Herrero-Aguayo, V.; Jiménez-Vacas, J.M.; Sáez-Martínez, P.; Gómez-Gómez, E.; López-Cánovas, J.L.; Garrido-Sánchez, L.; Herrera-Martínez, A.D.; García-Bermejo, L.; Macías-González, M.; López-Miranda, J.; et al. Influence of Obesity in the miRNome: miR-4454, a Key Regulator of Insulin Response via Splicing Modulation in Prostate. J. Clin. Endocrinol. Metab. 2021, 106, e469–e484. [Google Scholar] [CrossRef]
- Dasari, S.; Pandhiri, T.; Grassi, T.; Visscher, D.W.; Multinu, F.; Agarwal, K.; Mariani, A.; Shridhar, V.; Mitra, A.K. Signals from the Metastatic Niche Regulate Early and Advanced Ovarian Cancer Metastasis through miR-4454 Downregulation. Mol. Cancer Res. 2020, 18, 1202–1217. [Google Scholar] [CrossRef]
- Wang, H.; Hu, H.; Luo, Z.; Liu, S.; Wu, W.; Zhu, M.; Wang, J.; Liu, Y.; Lu, Z. miR-4454 up-Regulated by HPV16 E6/E7 Promotes Invasion and Migration by Targeting ABHD2/NUDT21 in Cervical Cancer. Biosci. Rep. 2020, 40, BSR20200796. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, Z.; Patil, V.; Thinagararjan, S.; Rao, S.A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Impact of Somatic Copy Number Alterations on the Glioblastoma miRNome: miR-4484 Is a Genomically Deleted Tumour Suppressor. Mol. Oncol. 2017, 11, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Kannathasan, T.; Kuo, W.-W.; Chen, M.-C.; Viswanadha, V.P.; Shen, C.-Y.; Tu, C.-C.; Yeh, Y.-L.; Bharath, M.; Shibu, M.A.; Huang, C.-Y. Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway. Cancers 2020, 12, 1231. [Google Scholar] [CrossRef]
- Morishita, A.; Iwama, H.; Fujihara, S.; Sakamoto, T.; Fujita, K.; Tani, J.; Miyoshi, H.; Yoneyama, H.; Himoto, T.; Masaki, T. MicroRNA Profiles in Various Hepatocellular Carcinoma Cell Lines. Oncol. Lett. 2016, 12, 1687–1692. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Wang, L.; Kang, Q.; Yu, G.; Wan, X.; Wang, J.; Zhu, K. MiR-4505 Aggravates Lipopolysaccharide-Induced Vascular Endothelial Injury by Targeting Heat Shock Protein A12B. Mol. Med. Rep. 2018, 17, 1389–1395. [Google Scholar] [CrossRef]
- Zhao, M.; Tang, Z.; Wang, Y.; Ding, J.; Guo, Y.; Zhang, N.; Gao, T. MIR-4507 Targets to Facilitate the Malignant Progression of Non-Small-Cell Lung Cancer. J. Cancer 2021, 12, 6600–6609. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Wu, S.; Wang, W.; Mao, L.; Yu, Y.; Jiang, L.; Yuan, W.; Zhang, M.; Sang, L.; Huang, Q.; et al. miRNA Sequencing Reveals miRNA-4508 from Peripheral Blood Lymphocytes as Potential Diagnostic Biomarker for Silica-Related Pulmonary Fibrosis: A Multistage Study. Respirology 2020, 25, 511–517. [Google Scholar] [CrossRef]
- Yang, L.; Hou, Y.; Du, Y.-E.; Li, Q.; Zhou, F.; Li, Y.; Zeng, H.; Jin, T.; Wan, X.; Guan, S.; et al. Mirtronic miR-4646-5p Promotes Gastric Cancer Metastasis by Regulating ABHD16A and Metabolite Lysophosphatidylserines. Cell Death Differ. 2021, 28, 2708–2727. [Google Scholar] [CrossRef]
- Jia, J.; Ouyang, Z.; Wang, M.; Ma, W.; Liu, M.; Zhang, M.; Yu, M. MicroRNA-361-5p Slows down Gliomas Development through Regulating UBR5 to Elevate ATMIN Protein Expression. Cell Death Dis. 2021, 12, 746. [Google Scholar] [CrossRef] [PubMed]
- Icli, B.; Li, H.; Pérez-Cremades, D.; Wu, W.; Ozdemir, D.; Haemmig, S.; Guimaraes, R.B.; Manica, A.; Marchini, J.F.; Orgill, D.P.; et al. MiR-4674 Regulates Angiogenesis in Tissue Injury by Targeting p38K Signaling in Endothelial Cells. Am. J. Physiol. Cell Physiol. 2020, 318, C524–C535. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ji, J.; Li, J.; Ren, Q.; Gu, J.; Zhao, Y.; Hong, D.; Guo, Q.; Tan, Y. Several Critical Genes and microRNAs Associated with the Development of Polycystic Ovary Syndrome. Ann. Endocrinol. 2020, 81, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, C.; Liu, Y.; Hu, Y.; Wu, H. Circular RNA hsa_circ_0001649 Inhibits Hepatocellular Carcinoma Progression Multiple miRNAs Sponge. Aging 2019, 11, 3362–3375. [Google Scholar] [CrossRef]
- Deng, G.; Wang, R.; Sun, Y.; Huang, C.-P.; Yeh, S.; You, B.; Feng, C.; Li, G.; Ma, S.; Chang, C. Targeting Androgen Receptor (AR) with Antiandrogen Enzalutamide Increases Prostate Cancer Cell Invasion yet Decreases Bladder Cancer Cell Invasion via Differentially Altering the AR/circRNA-ARC1/miR-125b-2-3p or miR-4736/PPARγ/MMP-9 Signals. Cell Death Differ. 2021, 28, 2145–2159. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Meng, Z.; Zhang, S.; Li, N.; Hu, W.; Li, H. miR-4739/ITGA10/PI3K Signaling Regulates Differentiation and Apoptosis of Osteoblast. Regen. Ther. 2022, 21, 342–350. [Google Scholar] [CrossRef]
- Ma, H.-P.; Fu, M.; Masula, M.; Xing, C.-S.; Zhou, Q.; Tan, J.-T.; Wang, J. miR-3064-5p and miR-4745-5p Affect Heparin Sensitivity in Patients Undergoing Cardiac Surgery by Regulating AT-III and Factor X mRNA Levels. Front. Physiol. 2022, 13, 914333. [Google Scholar] [CrossRef]
- Mercuri, R.L.; Conceição, H.B.; Guardia, G.D.A.; Goldstein, G.; Vibranovski, M.D.; Hinske, L.C.; Galante, P.A.F. Retro-miRs: Novel and Functional miRNAs Originated from mRNA Retrotransposition. bioRxiv 2023. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.-J.; Yuan, Y.-G.; Das, J.; Yasuda, H.; Kim, J.-H. MicroRNA-7641 Is a Regulator of Ribosomal Proteins and a Promising Targeting Factor to Improve the Efficacy of Cancer Therapy. Sci. Rep. 2017, 7, 8365. [Google Scholar] [CrossRef] [PubMed]
- Karere, G.M.; Glenn, J.P.; Li, G.; Konar, A.; VandeBerg, J.L.; Cox, L.A. Potential miRNA Biomarkers and Therapeutic Targets for Early Atherosclerotic Lesions. Sci. Rep. 2023, 13, 3467. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Yang, G.; He, H.; Gao, P.; Jiang, T.; Chen, Y.; Zhao, G. Identification of Cerebrospinal Fluid MicroRNAs Associated with Leptomeningeal Metastasis from Lung Adenocarcinoma. Front. Oncol. 2020, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Gohir, W.; Klement, W.; Singer, L.G.; Palmer, S.M.; Mazzulli, T.; Keshavjee, S.; Husain, S. Identifying Host microRNAs in Bronchoalveolar Lavage Samples from Lung Transplant Recipients Infected with Aspergillus. J. Heart Lung Transplant. 2020, 39, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Horiguchi, H.; Kikuchi, S.; Iyama, S.; Ikeda, H.; Goto, A.; Kawano, Y.; Murase, K.; Takada, K.; Miyanishi, K.; et al. miR-7977 Inhibits the Hippo-YAP Signaling Pathway in Bone Marrow Mesenchymal Stromal Cells. PLoS ONE 2019, 14, e0213220. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Z.; Zhu, H.; Yuan, X.; Sun, M.; Wang, J.; Zuo, M.; Cui, X.; Han, Y.; Zhang, Y.; et al. Hyperinsulinemia Contributes to Impaired-Glucose-Tolerance-Induced Renal Injury Mir-7977/SIRT3 Signaling. Ther. Adv. Chronic Dis. 2020, 11, 2040622320916008. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wang, X.; Chen, S.; Liu, H.; Wang, Y.; Xu, X.; Cheng, J.; Jia, J.; Zhen, X. MicroRNA Let-7c-5p Protects against Cerebral Ischemia Injury via Mechanisms Involving the Inhibition of Microglia Activation. Brain Behav. Immun. 2015, 49, 75–85. [Google Scholar] [CrossRef]
- Pregnolato, F.; Cova, L.; Doretti, A.; Bardelli, D.; Silani, V.; Bossolasco, P. Exosome microRNAs in Amyotrophic Lateral Sclerosis: A Pilot Study. Biomolecules 2021, 11, 1220. [Google Scholar] [CrossRef]
- Rizzuti, M.; Filosa, G.; Melzi, V.; Calandriello, L.; Dioni, L.; Bollati, V.; Bresolin, N.; Comi, G.P.; Barabino, S.; Nizzardo, M.; et al. MicroRNA Expression Analysis Identifies a Subset of Downregulated miRNAs in ALS Motor Neuron Progenitors. Sci. Rep. 2018, 8, 10105. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Steinert, J.R.; Shalom-Feuerstein, R.; Rouleau, M.; Aberdam, D.; Forsythe, I.D.; Young, K.W.; Ventura, A.; Concepcion, C.P.; et al. microRNA-34a Regulates Neurite Outgrowth, Spinal Morphology, and Function. Proc. Natl. Acad. Sci. USA 2011, 108, 21099–21104. [Google Scholar] [CrossRef]
- Sproviero, D.; Gagliardi, S.; Zucca, S.; Arigoni, M.; Giannini, M.; Garofalo, M.; Olivero, M.; Dell’Orco, M.; Pansarasa, O.; Bernuzzi, S.; et al. Different miRNA Profiles in Plasma Derived Small and Large Extracellular Vesicles from Patients with Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 2737. [Google Scholar] [CrossRef]
- Deng, D.; Wang, L.; Chen, Y.; Li, B.; Xue, L.; Shao, N.; Wang, Q.; Xia, X.; Yang, Y.; Zhi, F. MicroRNA-124-3p Regulates Cell Proliferation, Invasion, Apoptosis, and Bioenergetics by Targeting PIM1 in Astrocytoma. Cancer Sci. 2016, 107, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, S.; Yan, J.; Sun, M.-Z.; Greenaway, F.T. The Potential Role of miR-124-3p in Tumorigenesis and Other Related Diseases. Mol. Biol. Rep. 2021, 48, 3579–3591. [Google Scholar] [CrossRef] [PubMed]
- Long, H.-D.; Ma, Y.-S.; Yang, H.-Q.; Xue, S.-B.; Liu, J.-B.; Yu, F.; Lv, Z.-W.; Li, J.-Y.; Xie, R.-T.; Chang, Z.-Y.; et al. Reduced Hsa-miR-124-3p Levels Are Associated with the Poor Survival of Patients with Hepatocellular Carcinoma. Mol. Biol. Rep. 2018, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Majid, A.; Wang, J.; Nawaz, M.; Abdul, S.; Ayesha, M.; Guo, C.; Liu, Q.; Liu, S.; Sun, M.-Z. miR-124-3p Suppresses the Invasiveness and Metastasis of Hepatocarcinoma Cells via Targeting CRKL. Front. Mol. Biosci. 2020, 7, 223. [Google Scholar] [CrossRef]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased Microglial Exosomal miR-124-3p Alleviates Neurodegeneration and Improves Cognitive Outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef]
- Vuokila, N.; Lukasiuk, K.; Bot, A.M.; van Vliet, E.A.; Aronica, E.; Pitkänen, A.; Puhakka, N. miR-124-3p Is a Chronic Regulator of Gene Expression after Brain Injury. Cell. Mol. Life Sci. 2018, 75, 4557–4581. [Google Scholar] [CrossRef]
- Iacona, J.R.; Lutz, C.S. miR-146a-5p: Expression, Regulation, and Functions in Cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, R.; Wu, W.; Lei, L. miR-4454 Promotes Hepatic Carcinoma Progression by Targeting Vps4A and Rab27A. Oxid. Med. Cell. Longev. 2021, 2021, 9230435. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.-J.; Yang, H.-Q.; Wang, R.; Wu, H.-J. MicroRNA-10b Expression Predicts Long-Term Survival in Patients with Solid Tumor. J. Cell. Physiol. 2019, 234, 1248–1256. [Google Scholar] [CrossRef]
- Li, Z.; Yi, N.; Chen, R.; Meng, Y.; Wang, Y.; Liu, H.; Cao, W.; Hu, Y.; Gu, Y.; Tong, C.; et al. miR-29b-3p Protects Cardiomyocytes against Endotoxin-Induced Apoptosis and Inflammatory Response through Targeting FOXO3A. Cell Signal. 2020, 74, 109716. [Google Scholar] [CrossRef]
- Foggin, S.; Mesquita-Ribeiro, R.; Dajas-Bailador, F.; Layfield, R. Biological Significance of microRNA Biomarkers in ALS-Innocent Bystanders or Disease Culprits? Front. Neurol. 2019, 10, 578. [Google Scholar] [CrossRef]
- Toivonen, J.M.; Manzano, R.; Oliván, S.; Zaragoza, P.; García-Redondo, A.; Osta, R. MicroRNA-206: A Potential Circulating Biomarker Candidate for Amyotrophic Lateral Sclerosis. PLoS ONE 2014, 9, e89065. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Lee, Y.S.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-Specific microRNA miR-206 Promotes Muscle Differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.J. MicroRNA-206: The Skeletal Muscle-Specific myomiR. Biochim. Biophys. Acta 2008, 1779, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 Delays ALS Progression and Promotes Regeneration of Neuromuscular Synapses in Mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Przanowska, R.K.; Sobierajska, E.; Su, Z.; Jensen, K.; Przanowski, P.; Nagdas, S.; Kashatus, J.A.; Kashatus, D.F.; Bhatnagar, S.; Lukens, J.R.; et al. miR-206 Family Is Important for Mitochondrial and Muscle Function, but Not Essential for Myogenesis in Vitro. FASEB J. 2020, 34, 7687–7702. [Google Scholar] [CrossRef]
- Casola, I.; Scicchitano, B.M.; Lepore, E.; Mandillo, S.; Golini, E.; Nicoletti, C.; Barberi, L.; Dobrowolny, G.; Musarò, A. Circulating myomiRs in Muscle Denervation: From Surgical to ALS Pathological Condition. Cells 2021, 10, 2043. [Google Scholar] [CrossRef]
- Waller, R.; Goodall, E.F.; Milo, M.; Cooper-Knock, J.; Da Costa, M.; Hobson, E.; Kazoka, M.; Wollff, H.; Heath, P.R.; Shaw, P.J.; et al. Serum miRNAs miR-206, 143–3p and 374b-5p as Potential Biomarkers for Amyotrophic Lateral Sclerosis (ALS). Neurobiol. Aging 2017, 55, 123–131. [Google Scholar] [CrossRef]
- de Andrade, H.M.T.; de Albuquerque, M.; Avansini, S.H.; de S. Rocha, C.; Dogini, D.B.; Nucci, A.; Carvalho, B.; Lopes-Cendes, I.; França, M.C., Jr. MicroRNAs-424 and 206 Are Potential Prognostic Markers in Spinal Onset Amyotrophic Lateral Sclerosis. J. Neurol. Sci. 2016, 368, 19–24. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of microRNA-1 and microRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Vassileff, N.; Vella, L.J.; Rajapaksha, H.; Shambrook, M.; Kenari, A.N.; McLean, C.; Hill, A.F.; Cheng, L. Revealing the Proteome of Motor Cortex Derived Extracellular Vesicles Isolated from Amyotrophic Lateral Sclerosis Human Postmortem Tissues. Cells 2020, 9, 1709. [Google Scholar] [CrossRef] [PubMed]
- Pasetto, L.; Callegaro, S.; Corbelli, A.; Fiordaliso, F.; Ferrara, D.; Brunelli, L.; Sestito, G.; Pastorelli, R.; Bianchi, E.; Cretich, M.; et al. Decoding Distinctive Features of Plasma Extracellular Vesicles in Amyotrophic Lateral Sclerosis. Mol. Neurodegener. 2021, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Sjoqvist, S.; Otake, K. A Pilot Study Using Proximity Extension Assay of Cerebrospinal Fluid and Its Extracellular Vesicles Identifies Novel Amyotrophic Lateral Sclerosis Biomarker Candidates. Biochem. Biophys. Res. Commun. 2022, 613, 166–173. [Google Scholar] [CrossRef]
- Hayes, A.J.; Farrugia, B.L.; Biose, I.J.; Bix, G.J.; Melrose, J.; Perlecan, A. Multi-Functional, Cell-Instructive, Matrix-Stabilizing Proteoglycan with Roles in Tissue Development Has Relevance to Connective Tissue Repair and Regeneration. Front. Cell Dev. Biol. 2022, 10, 856261. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.G.; Gray, E.; Mäger, I.; Thézénas, M.-L.; Charles, P.D.; Talbot, K.; Fischer, R.; Kessler, B.M.; Wood, M.; Turner, M.R. CSF Extracellular Vesicle Proteomics Demonstrates Altered Protein Homeostasis in Amyotrophic Lateral Sclerosis. Clin. Proteomics 2020, 17, 31. [Google Scholar] [CrossRef]
- Riise, R.; Odqvist, L.; Mattsson, J.; Monkley, S.; Abdillahi, S.M.; Tyrchan, C.; Muthas, D.; Yrlid, L.F. Bleomycin Hydrolase Regulates the Release of Chemokines Important for Inflammation and Wound Healing by Keratinocytes. Sci. Rep. 2019, 9, 20407. [Google Scholar] [CrossRef]
- Skytthe, M.K.; Graversen, J.H.; Moestrup, S.K. Targeting of CD163+ Macrophages in Inflammatory and Malignant Diseases. Int. J. Mol. Sci. 2020, 21, 5497. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Dela Cruz, C.S. Chitotriosidase: A Marker and Modulator of Lung Disease. Eur. Respir. Rev. 2020, 29, 190143. [Google Scholar] [CrossRef]
- Man, J.; Zhang, X. CUEDC2: An Emerging Key Player in Inflammation and Tumorigenesis. Protein Cell 2011, 2, 699–703. [Google Scholar] [CrossRef]
- Otake, K.; Kamiguchi, H.; Hirozane, Y. Identification of Biomarkers for Amyotrophic Lateral Sclerosis by Comprehensive Analysis of Exosomal mRNAs in Human Cerebrospinal Fluid. BMC Med. Genom. 2019, 12, 7. [Google Scholar] [CrossRef]
- Kamelgarn, M.; Chen, J.; Kuang, L.; Arenas, A.; Zhai, J.; Zhu, H.; Gal, J. Proteomic Analysis of FUS Interacting Proteins Provides Insights into FUS Function and Its Role in ALS. Biochim. Biophys. Acta 2016, 1862, 2004–2014. [Google Scholar] [CrossRef]
- Zuehlke, A.D.; Moses, M.A.; Neckers, L. Heat Shock Protein 90: Its Inhibition and Function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160527. [Google Scholar] [CrossRef]
- Sugano, Y.; Takeuchi, M.; Hirata, A.; Matsushita, H.; Kitamura, T.; Tanaka, M.; Miyajima, A. Junctional Adhesion Molecule-A, JAM-A, Is a Novel Cell-Surface Marker for Long-Term Repopulating Hematopoietic Stem Cells. Blood 2008, 111, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.J.; Updegraff, B.L.; Bellich, R.M.; Goff, M.R.; Lu, D.; Minkin, S.C., Jr.; Karlstad, M.D.; Collier, J.J. Regulation of iNOS Gene Transcription by IL-1β and IFN-γ Requires a Coactivator Exchange Mechanism. Mol. Endocrinol. 2013, 27, 1724–1742. [Google Scholar] [CrossRef]
- Ordway, G.A.; Garry, D.J. Myoglobin: An Essential Hemoprotein in Striated Muscle. J. Exp. Biol. 2004, 207, 3441–3446. [Google Scholar] [CrossRef] [PubMed]
- Garry, D.J.; Mammen, P.P.A. Molecular Insights into the Functional Role of Myoglobin. Adv. Exp. Med. Biol. 2007, 618, 181–193. [Google Scholar]
- Bunton-Stasyshyn, R.K.A.; Saccon, R.A.; Fratta, P.; Fisher, E.M.C. SOD1 Function and Its Implications for Amyotrophic Lateral Sclerosis Pathology: New and Renascent Themes. Neuroscientist 2015, 21, 519–529. [Google Scholar] [CrossRef]
- Heyne, K.; Förster, J.; Schüle, R.; Roemer, K. Transcriptional Repressor NIR Interacts with the p53-Inhibiting Ubiquitin Ligase MDM2. Nucleic Acids Res. 2014, 42, 3565–3579. [Google Scholar] [CrossRef]
- Hayashi, N.; Doi, H.; Kurata, Y.; Kagawa, H.; Atobe, Y.; Funakoshi, K.; Tada, M.; Katsumoto, A.; Tanaka, K.; Kunii, M.; et al. Proteomic Analysis of Exosome-Enriched Fractions Derived from Cerebrospinal Fluid of Amyotrophic Lateral Sclerosis Patients. Neurosci. Res. 2020, 160, 43–49. [Google Scholar] [CrossRef]
- Vomhof-Dekrey, E.E.; Picklo, M.J., Sr. The Nrf2-Antioxidant Response Element Pathway: A Target for Regulating Energy Metabolism. J. Nutr. Biochem. 2012, 23, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, F.; Nyberg, S.; Giunti, D.; Torazza, C.; Parodi, B.; Bonifacino, T.; Usai, C.; Kerlero de Rosbo, N.; Milanese, M.; Uccelli, A.; et al. Micro-RNAs Shuttled by Extracellular Vesicles Secreted from Mesenchymal Stem Cells Dampen Astrocyte Pathological Activation and Support Neuroprotection in In-Vitro Models of ALS. Cells 2022, 11, 3923. [Google Scholar] [CrossRef]
- Bourtchuladze, R.; Frenguelli, B.; Blendy, J.; Cioffi, D.; Schutz, G.; Silva, A.J. Deficient Long-Term Memory in Mice with a Targeted Mutation of the cAMP-Responsive Element-Binding Protein. Cell 1994, 79, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Ban, J.-J.; Kim, K.Y.; Jeon, G.S.; Im, W.; Sung, J.-J.; Kim, M. Adipose-Derived Stem Cell Exosomes Alleviate Pathology of Amyotrophic Lateral Sclerosis in Vitro. Biochem. Biophys. Res. Commun. 2016, 479, 434–439. [Google Scholar] [CrossRef]
- Liang, H.; Ward, W.F. PGC-1alpha: A Key Regulator of Energy Metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.E. An Overview of Phenylalanine and Tyrosine Kinetics in Humans. J. Nutr. 2007, 137, 1549S–1555S; discussion 1573S–1575S. [Google Scholar] [CrossRef]
- Morasso, C.F.; Sproviero, D.; Mimmi, M.C.; Giannini, M.; Gagliardi, S.; Vanna, R.; Diamanti, L.; Bernuzzi, S.; Piccotti, F.; Truffi, M.; et al. Raman Spectroscopy Reveals Biochemical Differences in Plasma Derived Extracellular Vesicles from Sporadic Amyotrophic Lateral Sclerosis Patients. Nanomedicine 2020, 29, 102249. [Google Scholar] [CrossRef]
- Butler, R.E.; Krishnan, N.; Garcia-Jimenez, W.; Francis, R.; Martyn, A.; Mendum, T.; Felemban, S.; Locker, N.; Salguero, F.J.; Robertson, B.; et al. Susceptibility of Mycobacterium Tuberculosis-Infected Host Cells to Phospho-MLKL Driven Necroptosis Is Dependent on Cell Type and Presence of TNFα. Virulence 2017, 8, 1820–1832. [Google Scholar] [CrossRef]
- Ren, L.-Q.; Liu, W.; Li, W.-B.; Liu, W.-J.; Sun, L. Peptidylprolyl Cis/trans Isomerase Activity and Molecular Evolution of Vertebrate Cyclophilin A. Yi Chuan 2016, 38, 736–745. [Google Scholar] [PubMed]
- Boylan, K.B.; Glass, J.D.; Crook, J.E.; Yang, C.; Thomas, C.S.; Desaro, P.; Johnston, A.; Overstreet, K.; Kelly, C.; Polak, M.; et al. Phosphorylated Neurofilament Heavy Subunit (pNF-H) in Peripheral Blood and CSF as a Potential Prognostic Biomarker in Amyotrophic Lateral Sclerosis. J. Neurol. Neurosurg. Psychiatry 2013, 84, 467–472. [Google Scholar] [CrossRef]
- Scotter, E.L.; Chen, H.-J.; Shaw, C.E. TDP-43 Proteinopathy and ALS: Insights into Disease Mechanisms and Therapeutic Targets. Neurotherapeutics 2015, 12, 352–363. [Google Scholar] [CrossRef]
- Feneberg, E.; Steinacker, P.; Lehnert, S.; Schneider, A.; Walther, P.; Thal, D.R.; Linsenmeier, M.; Ludolph, A.C.; Otto, M. Limited Role of Free TDP-43 as a Diagnostic Tool in Neurodegenerative Diseases. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 351–356. [Google Scholar] [CrossRef]
- Medler, J.; Wajant, H. Tumor Necrosis Factor Receptor-2 (TNFR2): An Overview of an Emerging Drug Target. Expert Opin. Ther. Targets 2019, 23, 295–307. [Google Scholar] [CrossRef]
- Groen, E.J.N.; Gillingwater, T.H. UBA1: At the Crossroads of Ubiquitin Homeostasis and Neurodegeneration. Trends Mol. Med. 2015, 21, 622–632. [Google Scholar] [CrossRef]
- Bobis-Wozowicz, S.; Marbán, E. Editorial: Extracellular Vesicles as Next Generation Therapeutics. Front. Cell Dev. Biol. 2022, 10, 919426. [Google Scholar] [CrossRef]
- Cataldi, M.; Vigliotti, C.; Mosca, T.; Cammarota, M.; Capone, D. Emerging Role of the Spleen in the Pharmacokinetics of Monoclonal Antibodies, Nanoparticles and Exosomes. Int. J. Mol. Sci. 2017, 18, 1249. [Google Scholar] [CrossRef]
- Liu, S.; Wu, X.; Chandra, S.; Lyon, C.; Ning, B.; Jiang, L.; Fan, J.; Hu, T.Y. Extracellular Vesicles: Emerging Tools as Therapeutic Agent Carriers. Acta Pharm. Sin. B 2022, 12, 3822–3842. [Google Scholar] [CrossRef]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular Vesicles as Drug Delivery Systems: Why and How? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Bonafede, R.; Mariotti, R. ALS Pathogenesis and Therapeutic Approaches: The Role of Mesenchymal Stem Cells and Extracellular Vesicles. Front. Cell. Neurosci. 2017, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Belkozhayev, A.M.; Al-Yozbaki, M.; George, A.; Ye Niyazova, R.; Sharipov, K.O.; Byrne, L.J.; Wilson, C.M. Extracellular Vesicles, Stem Cells and the Role of miRNAs in Neurodegeneration. Curr. Neuropharmacol. 2022, 20, 1450–1478. [Google Scholar]
- Sadanandan, N.; Lee, J.-Y.; Garbuzova-Davis, S. Extracellular Vesicle-Based Therapy for Amyotrophic Lateral Sclerosis. Brain Circ. 2021, 7, 23–28. [Google Scholar] [PubMed]
- Bonafede, R.; Scambi, I.; Peroni, D.; Potrich, V.; Boschi, F.; Benati, D.; Bonetti, B.; Mariotti, R. Exosome Derived from Murine Adipose-Derived Stromal Cells: Neuroprotective Effect on In Vitro Model of Amyotrophic Lateral Sclerosis. Exp. Cell Res. 2016, 340, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Jin, T.; Botchway, B.O.A.; Fan, R.; Wang, L.; Liu, X. Adipose-Derived Mesenchymal Stem Cells Combined With Extracellular Vesicles May Improve Amyotrophic Lateral Sclerosis. Front. Aging Neurosci. 2022, 14, 830346. [Google Scholar] [CrossRef] [PubMed]
- Bonafede, R.; Brandi, J.; Manfredi, M.; Scambi, I.; Schiaffino, L.; Merigo, F.; Turano, E.; Bonetti, B.; Marengo, E.; Cecconi, D.; et al. The Anti-Apoptotic Effect of ASC-Exosomes in an In Vitro ALS Model and Their Proteomic Analysis. Cells 2019, 8, 1087. [Google Scholar] [CrossRef] [PubMed]
- Bonafede, R.; Turano, E.; Scambi, I.; Busato, A.; Bontempi, P.; Virla, F.; Schiaffino, L.; Marzola, P.; Bonetti, B.; Mariotti, R. ASC-Exosomes Ameliorate the Disease Progression in SOD1(G93A) Murine Model Underlining Their Potential Therapeutic Use in Human ALS. Int. J. Mol. Sci. 2020, 21, 3651. [Google Scholar] [CrossRef]
- Calabria, E.; Scambi, I.; Bonafede, R.; Schiaffino, L.; Peroni, D.; Potrich, V.; Capelli, C.; Schena, F.; Mariotti, R. ASCs-Exosomes Recover Coupling Efficiency and Mitochondrial Membrane Potential in an Model of ALS. Front. Neurosci. 2019, 13, 1070. [Google Scholar] [CrossRef]
- Garbuzova-Davis, S.; Willing, A.E.; Ehrhart, J.; Wang, L.; Sanberg, P.R.; Borlongan, C.V. Cell-Free Extracellular Vesicles Derived from Human Bone Marrow Endothelial Progenitor Cells as Potential Therapeutics for Microvascular Endothelium Restoration in ALS. Neuromolecular Med. 2020, 22, 503–516. [Google Scholar] [CrossRef]
- Niu, X.; Chen, J.; Gao, J. Nanocarriers as a Powerful Vehicle to Overcome Blood-Brain Barrier in Treating Neurodegenerative Diseases: Focus on Recent Advances. Asian J. Pharm. Sci. 2019, 14, 480–496. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Ren, C.; Wang, Y.; He, W.; Jiang, Y. Extracellular Vesicles as Innovative Treatment Strategy for Amyotrophic Lateral Sclerosis. Front. Cell Dev. Biol. 2021, 9, 754630. [Google Scholar] [CrossRef]
- Thakur, A.; Sidu, R.K.; Zou, H.; Alam, M.K.; Yang, M.; Lee, Y. Inhibition of Glioma Cells’ Proliferation by Doxorubicin-Loaded Exosomes via Microfluidics. Int. J. Nanomed. 2020, 15, 8331–8343. [Google Scholar] [CrossRef]
- Kiernan, M.C.; Vucic, S.; Talbot, K.; McDermott, C.J.; Hardiman, O.; Shefner, J.M.; Al-Chalabi, A.; Huynh, W.; Cudkowicz, M.; Talman, P.; et al. Improving Clinical Trial Outcomes in Amyotrophic Lateral Sclerosis. Nat. Rev. Neurol. 2021, 17, 104–118. [Google Scholar] [CrossRef]
- Zhu, X.; Gao, M.; Yang, Y.; Li, W.; Bao, J.; Li, Y. The CRISPR/Cas9 System Delivered by Extracellular Vesicles. Pharmaceutics 2023, 15, 984. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular Nanovesicles for Packaging of CRISPR-Cas9 Protein and sgRNA to Induce Therapeutic Exon Skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
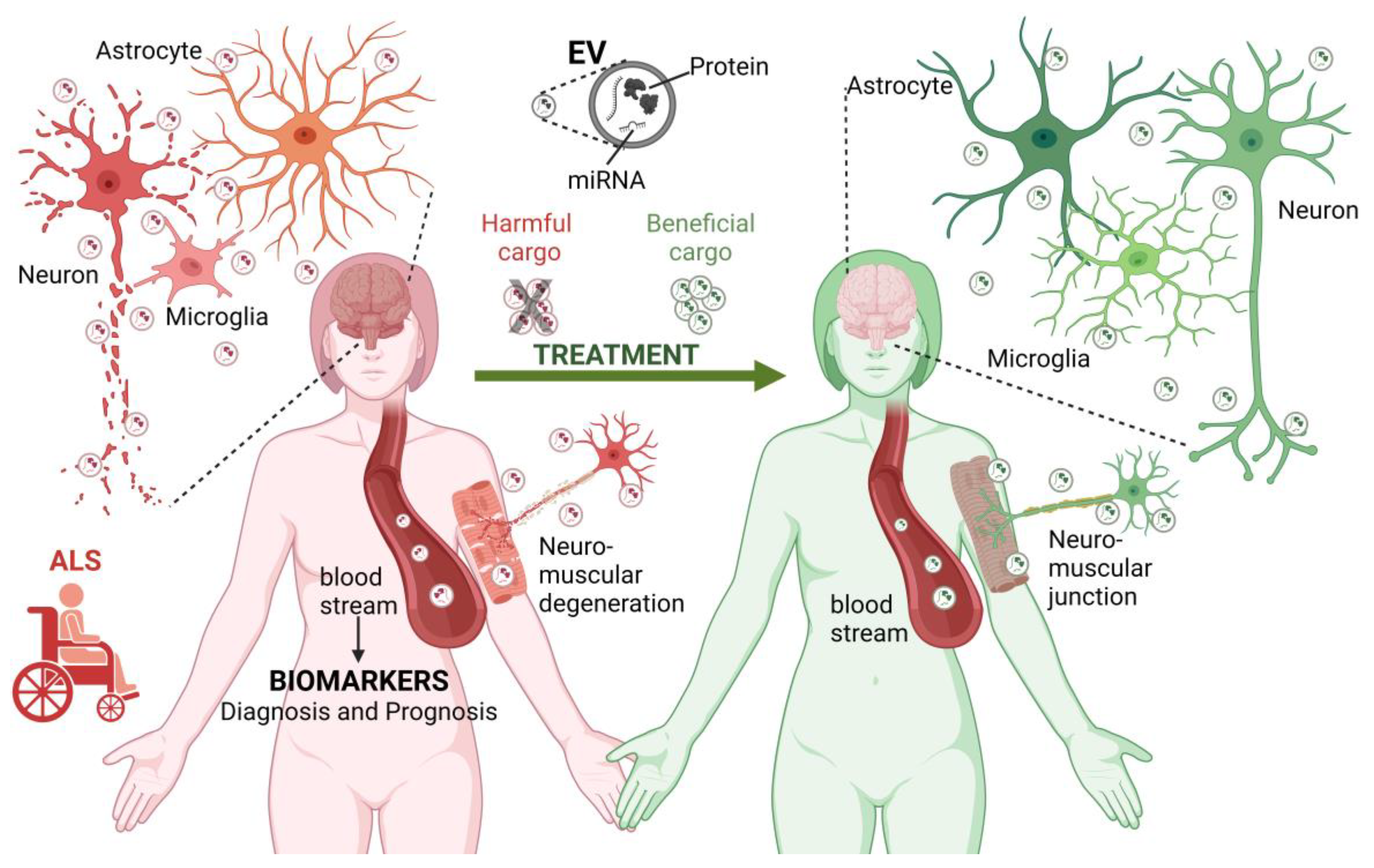
| miRNA | Main Targets | Biological Role | Expression |
|---|---|---|---|
| miR-9-1-5p, miR-9-2-5p, miR-9-3-5p | PAK4, CoREST, CPEB3, ECAD, elavl3, FoxG1, Hes1, IGF2BP3, Nr2e1/TLX, REST, Sirt1, Zic5; IGF2-PI3K/Akt signaling | Cell differentiation regulation, neuronal function, synaptic plasticity neurotransmitter release; skeletal muscle cell proliferation, and differentiation inhibition regulation [143]; apoptosis inhibition [144] | downregulated: blood plasma-derived EVs [145] |
| miR-10b-5p | NFAT5; KLF11-KIT signaling | Regulation of insulin production, lipid metabolism and gastrointestinal motility [146], and myoblasts differentiation [147]. Tumorigenic inhibitor [148] | downregulated: CSF exosomes and blood plasma-derived EVs [149,150] |
| miR-15a-5p | BCL2, Cyclin D1, FEAT, PD-1, ROR1, CXCL10-ERK-LIN28a-let-7 axis, NF-κB signaling, Wtn/β-catenin signaling | Tumor progression inhibition [151] | upregulated: blood plasma-derived EVs [145] |
| miR-24-3p | eNOS, GATA2, PAK4 | Tumor suppression, angiogenesis regulation, and cell protection against apoptosis [152] | upregulated: blood plasma-derived EVs [153] |
| miR-26a-5p | ADAM17, Bid, FAF1, SERBP1, Wnt5; TGF-β signaling | Osteogenic differentiation and cell proliferation regulation [154] | upregulated: serum [155] |
| miR-27a-3p | AQP11, BTG2 | Tumor suppression [156] and protection against the blood–brain barrier and brain injury after brain hemorrhage [157] | downregulated: serum-derived exosomes [158] |
| miR-29b-3p | C1QTNF6/AMPK signaling | Modulation of inflammatory response [159] | downregulated: CSF exosomes and blood plasma-derived EVs [149,150] |
| miR-34a | AXIN2, BCL2, BIRC5, CD44, DGKζ, E2F3, MDMX, MET; MYCN, NOTCH1, NANOG, PD-L1, SIRT1, SNAI1, SOX2; cyclins, cyclin-dependent kinases, TGF-β1/Smad signaling | Cell proliferation, apoptosis, autophagy and cellular senescence regulation [160,161], and matrix proteins deposition [161] | downregulated: ALS iMNs-derived exosomes and CSF [162] |
| miR-100-5p | ANKAR, AP1AR, EPDR1, ICK, NR1I3, SMARCA5, ST6GALNAC4, TMPRSS13, TTC39A; mTOR signaling | Cell survival regulation (e.g., apoptosis) [163] and autophagy [164,165] | downregulated: blood plasma-derived EVs [145] |
| miR-124–3p | CDK6, EfnB1, PTBP1, REST, SCP1, Sox9; NeuroD1; | Synaptic connectivity and plasticity regulation [166] | upregulated: CSF exosomes [149] |
| miR-127-3p | BCCIP, BOLA1, FAM27D1, KCNK2, LOC100134822, MTCP1, PSD95, RBPMS, SIRT3, SLC25A2, TPTE2, ZNF3, NeuroD1, NR2A-subunit | Neurogenesis, synapse formation and motor neuron integrity maintenance, mitophagy, ROS, and misfolded proteins accumulation [167] | upregulated: serum [155]; downregulated: blood plasma [145,155] |
| miR-144-3p | ABCA1, CCNT2, FoxO1, FST, GABRA1, HGF, IGIP, NFE2L2, ST3GAL6, UBE2D1, UBR3 | Adipogenesis regulation, metastasis, and cell proliferation inhibition [168] | upregulated: blood plasma-derived EVs [145] |
| miR-146a-5p | IRAK-1; NF-κB signaling | Immune cell activity, hematopoiesis, and malignant transformation regulation [169] | upregulated: blood plasma-derived EVs [150,170] |
| miR-149-3p | AKT2 | Cell proliferation inhibition in cancer [171] | upregulated in blood plasma-derived EVs [153] |
| miR-150-3p | CASP2, SP1 | Neuroprotection of neural stem cells exosomes after ischemic insult and cell proliferation inhibition [172,173] | downregulated: blood plasma-derived EVs [153] |
| miR-151a-3p | SOCS5, SP3, YTHDF3; JAK2/STAT3 signaling | Tumorigenic inhibitor [174] | upregulated: blood plasma-derived EVs [150,170] |
| miR-151a-5p | AGMAT, CYTB, SMARCA5 | Cellular ATP production regulation [175] | upregulated: blood plasma-derived EVs [150,170] |
| miR-181a-1-5p | Kras, NRAS, VCAM-1, ZNF780A, ZNF780B, ZNF204P, ZNF439, ZNF527, ZNF559, ZNF594, ZNF781, ZNF844 | Tumorigenic suppressor, immune response regulation, and cell proliferation [176,177,178] | downregulated: blood plasma-derived EVs [145] |
| miR-181a-2-5p | STAT3, TGFβR3 | Tumorigenic suppressor [179] | downregulated: blood plasma-derived EVs [145] |
| miR-181b-1-5p | BAZ2B, NOVA1, TGFβ1, ZNF780A, ZNF780B, ZNF439, ZNF527, ZNF559, ZNF594, ZNF781, ZNF844; MEK/ERK/p21 pathway | Cell proliferation, invasion and metastasis in cancer [180], apoptosis inhibition [181], and autophagy [182] | downregulated: blood plasma-derived EVs [145] |
| miR-181b-2-5p | BCL2, TIMP3; annexin A2 | Cell migratory proteins modulation [183] and chemosensitivity in cancer cells [184] | downregulated: blood plasma-derived EVs [145] |
| miR-183-5p | AKAP12, CCDC121, DHRSX, FKSG83, GNG5, NUDT4, PFN2, PDCD4, PSEN2, RIPK3, SLAIN1, XPNPEP3, | Neuron protection against motor cell death in ALS (under stress conditions) [185] | upregulated: blood plasma-derived EVs [145] |
| miR-194-5p | HIF-1, NR2F2, NR2F6, PAK2; MAPK1/PTEN/AKT signaling | Tumorigenic inhibition [186] | upregulated: ALS iMNs-derived exosomes [145,162] |
| miR-197-3p | HSPA5, KIAA0101, TIMP2/3; AKT/mTOR axis signaling | Tumor suppression, cell proliferation [187], autophagy regulation [188], and angiogenesis promotion [189]. Recognized biomarker for myocardial fibrosis and heart failure [190] | downregulated: postmortem frontal cortex and spinal cord [155] |
| miR-199a-1-3p | BCAR3, CDNF, DNMT3a, FABP12, HVCN1, KLHL3, RAP2a; SERPINE2SRR, TMEM161B, TSGA10, WFDC8 | Growth and angiogenesis inhibition in tumors [191] | downregulated: blood plasma-derived EVs [145] |
| miR-199a-2-3p | caveolin-2, Ppargc1a, Sirt1 | Regulation of cell proliferation and survival [192] | downregulated: blood plasma-derived EVs [145] |
| miR-199a-3p | CCND1, CD44, c-MYC, DNMT3a, EGFR, ETNK1, YAP1; mTOR | Cell proliferation regulation and apoptosis induction [192] | upregulated: blood plasma-derived EVs [150,170] |
| miR-199b-3p | CDNF, BCAR3, FABP12, HVCN1, KLHL3, SERPINE2 TSGA10, SRR, TMEM161B, WFDC8; Phospholipase Cε | Tumor suppression [193] | downregulated: blood plasma-derived EVs [145] |
| miR-199a-5p | DDK1, ITGA3, WTN2; CREB/BDNF signaling, NF-κB signaling | Tumorigenic inhibitor [194] | upregulated: blood plasma-derived EVs [150,170] |
| miR-298 | JMJD6 | Tumor suppression, cell proliferation, and metastasis inhibition [195] | downregulated: postmortem frontal cortex, spinal cord and serum [155] |
| miR-335 | ROCK1, survivin | Tumor suppression [196] | downregulated: ALS iMNs-derived exosomes [162] |
| miR-338-3p | C5ORF47, C6ORF141, DGKB, IDNK PREX2, IZUMO3, PIM1, ROBO1, SP3, TAX1BP3, ZNF141, ZNF208 | Tumor suppression; cell proliferation, migration, and invasion inhibition [197,198] | downregulated: blood plasma-derived EVs [145] |
| miR-342-3p | ATF3, FOXQ1, RAP2B, MAP1LC3B; HDAC7/PTEN axis signaling, RhoC GTPase | Prion-based neurodegeneration and intracellular motor proteins, axon guidance, cell proliferation and apoptosis regulation [199,200], tumor suppression, autophagy, and reduction of cell stemness [201] | upregulated: postmortem frontal cortex, spinal cord and serum-derived EVs [155] |
| miR-363-3p | CD69, DCAF6, FAM24A, FBXW7, FNIP1, MAN2A1, FBXW7, KLF4, PTEN; PI3K/AKT signaling | Osteogenic differentiation [202] | upregulated: blood plasma-derived EVs [145] |
| miR-371a-5p | BCL2; BECN1, SOX2 | Tumor suppression; cell proliferation, migration, and autophagy [203] | upregulated in blood plasma-derived EVs [153] |
| miR-450a-2-3p | FOXP3, IGF2, MAPK1, KSR2 | Tumorigenic inhibition [204] | upregulated: postmortem spinal cord and serum [155] |
| miR-494-3p | SEMA3A | Axonal maintenance (negative regulation of semaphorin 3A (SEM3A)) [205] | downregulated: astrocyte-derived EVs and in cortico-spinal tract tissue [205] |
| miR-502-5p | SP1 | Tumor suppression, regulation of cell proliferation, and migration [206] | downregulated: postmortem frontal cortex and spinal cord [155] |
| miR-512-5p | ETS1, hTERT, SOD2; Wnt/β-catenin signaling | Tumor suppression and apoptosis induction [207] | upregulated: postmortem frontal cortex [155] |
| miR-520f-3p | C2orf69, NDST4, SOX9, Wnt signaling | Tumor suppression [208] | upregulated: serum [155] |
| miR-532-3p | C13orf34, C22orf46, DNAL4, ENSA, FOXP3, KLHL12; OPHN1, RPRML, RPS3, ZNF514; β-catenin | Cell proliferation, metastasis inhibition, and apoptosis enhancing [209] | upregulated: blood plasma-derived EVs [145] |
| miR-551b-3p | H6PD, Cyclin D1, TRIM31/Akt signaling | Tumor inhibition [210] | upregulated: serum [155] |
| miR-549a | yet to be studied | Angiogenesis and metastasis induction [211] | downregulated: postmortem frontal cortex and spinal cord [155] |
| miR-587 | RPSA | Tumor suppression [212] | downregulated: serum-derived EVs [155] |
| miR-625-3p | GABBR2, SCAI | Cancer cells migration and invasion inhibition [213] | downregulated: ALS iMNs-derived exosomes and CSF [162] |
| miR-629-5p | AKAP13, CAV1, SFRP2 | Tumor cell growth regulation [214] | upregulated: ALS iMNs- derived exosomes [145,162] |
| miR-634 | HSPA2; mTOR signaling | Tumor suppression and apoptosis enhancing [215,216] | downregulated: blood plasma-derived EVs [153] |
| miR-664a-5p | AC093802.1, ANKRD36; CCNDBP1, DNASE2, FBXO17, HMGA2, IDH2, PTCD3, SEPT7, ZNF256, ZNF772 | Osteogenic differentiation, controlled apoptosis [217], and neuronal differentiation [218] | downregulated: blood plasma-derived EVs [145] |
| miR-766-3p | NF-κB signaling, TGFBI signaling | Inhibition of inflammatory responses [219] and apoptosis promotion in cancer [220] | downregulated: serum-derived EVs [155] |
| miR-877-5p | FOXM1 | Tumor suppression, cell proliferation, migration, and invasion reduction [221] | downregulated: serum-derived EVs [155] |
| miR-939-5p | ARHGAP4, HIF-1 alpha, IGF-1R; PI3K/Akt signaling | Cell migration and invasion in certain types of cancer [222] | upregulated: blood plasma-derived EVs [153] |
| miR-1207-5p | CX3CR1; NF-κB signaling, SARS-CoV-2 RNA | inflammatory response regulation [223] | upregulated: blood plasma-derived EVs [153] |
| miR-1246 | CDR1as, DNAH, FAM53C, FAM169B, GSG1L, KIAA1370, LIG4; NFE2L3, NR2F2, SGOL1, WDR77, ZNF23, ZNF267; NHEJ signaling | Modulation of DNA damage following ionizing radiation exposure [224,225] | upregulated: blood plasma-derived EVs [145] |
| miR-1254 | Smurf1; PIK/Akt signaling, | Cell proliferation, migration, and invasion inhibition [226] | downregulated: postmortem frontal cortex, spinal cord, and serum [155] |
| miR-1255a | SMAD4; TGF-β signaling | Related with breast cancer malignant phenotype and downstream effector of TGF-β [227] | upregulated in serum [155] |
| miR-1260b | C2orf48, CASP8, CTAGE1, GOLGA8A, MED13L, PABPN1, USP48, ZNF256, ZNF594, ZNF788; MAPK pathway | Tumorigenesis promotion [228] | downregulated: blood plasma-derived EVs [145] |
| miR-1262 | SCL2A1, ULK1 | Tumor suppression [229] | upregulated in serum [155] |
| miR-1268a | ABCC1 | Mediation of temozolomide resistance in glioblastoma [230] | downregulated: blood plasma-derived EVs [153] |
| miR-1268b | AKT, BCL2, ERBB2, PI3KCA, PI3K-AKT signaling | Apoptosis inhibition [231] | Upregulated: serum [155] |
| miR-1285-5p | CDH1, Smad4, TMEM194A | Cell proliferation regulation [232] | upregulated: postmortem frontal cortex [155] |
| miR-1290 | AKAP7, CDR1as, FAM19A5, HIGD2A, OGN MYO10, OSBPL6, RP11-1167A19.2, SGOL1, WDR77 | Cell proliferation, migration, and invasion regulation in cancer [233] | downregulated: blood plasma-derived EVs [145] |
| miR-1913 | not yet studied, but 732 predicted targets in [234] | Potential noninvasive biomarker for prostate cancer [235] | downregulated: blood plasma-derived EVs [153] |
| miR-2861 | STAT3, MMP2, EGFR/AKT2/CCND1 signaling | Tumor suppression, cell proliferation regulation, and apoptosis [236] | downregulated: blood plasma-derived EVs [153] |
| miR-3176 | AR, PTEN | Promotion of tumorigenesis and tumor progression [237] | downregulated: blood plasma-derived EVs [153] |
| miR-3177-3p | not yet studied, but 65 predicted targets in [234] | to be studied | downregulated: blood plasma-derived EVs [153] |
| miR-3605-5p | SCABR2 | Adipocyte lipolysis regulation [238] | downregulated: blood plasma-derived EVs [153] |
| miR-3619-3p | Wnt/β-catenin signaling | Cell migration and invasion promotion [239] | upregulated: blood plasma-derived EVs [153] |
| miR-3911 | not yet studied | Possible sALS biomarker [142] | downregulated: blood plasma-derived EVs [153] |
| miR-3940-3p | BIRC5, IL-2Ry, KCNA5, Integrin α6 | Regulation of maternal insulin resistance, T-cell activity promotion, and metastasis inhibition in cancers [240] | downregulated: blood plasma-derived EVs [153] |
| miR-4286 | APLN, C15orf34, CBX2, FAM222B, HKDC1, INPP4A, ZFP36L1, PARVG, PRX PTEN, RNF43, SALL1; TMSB4X, JAK2/STAT3 signaling, PI3K/Akt signaling, TGF-ß/TGF-ß1/Smad signaling | Cell proliferation, apoptosis, and inflammatory response modulation [241,242,243,244] | downregulated: blood plasma-derived EVs [145] |
| miR-4298 | SOD2, TGIF2 | Cell proliferation, migration, and invasion of cancer cells [245] | upregulated: blood plasma-derived EVs [153] |
| miR-4443 | INPP4A, METLL3, TRIM14; JAK2/STAT3 signaling, NF-κB signaling, Ras signaling, TGF-β1 signaling | Metastasis and energy metabolism suppression [246] | downregulated: postmortem frontal cortex and spinal cord [155] |
| miR-4454 | ABHD2/NUDT21, Vps4a, Rab27A; GNL3L/NF-κB signaling; TGF-β/MAPK pathway | Insulin signaling [247], metastasis progression in cancer [248,249,250] and apoptosis regulation [251] | downregulated: CSF exosomes and blood plasma-derived EVs [145,149,150]; upregulated: serum [155] |
| miR-4505 | HSPA12B | Tumorigenesis [252] vascular function [253] | upregulated: blood plasma-derived EVs [153] |
| miR-4507 | TP53; PI3K-AKT signaling | Cell proliferation and migration in lung cancer [254] | downregulated: blood plasma-derived EVs [153] |
| miR-4508 | 125 predicted in miRDB [234] | Potential involvement in pulmonary fibrosis through an unknown mechanism [255] | downregulated: blood plasma-derived EVs [153] |
| miR-4646-5p | ABHD16A; PHD3; PHD3/HIF1A signaling | Ubiquitination and cell proliferation and invasion regulation [256,257] | downregulated: blood plasma-derived EVs [153] |
| miR-4674 | p38k | Angiogenesis regulation [258] | downregulated: blood plasma-derived EVs [153] |
| miR-4687-5p | ATP10D, THRSP | Involved in polycystic ovary syndrome [259] | downregulated: blood plasma-derived EVs [153] |
| miR-4688 | not yet studied | miRNA sponge and cancer progression [260] | upregulated blood plasma-derived EVs [153] |
| miR-4700-5p | not yet studied | not yet studied | upregulated blood plasma-derived EVs [153] |
| miR-4736 | AR | Inflammatory response regulation [261] | upregulated blood plasma-derived EVs [153] |
| miR-4739 | BMP-7; ITGA10/PI3K signaling | Apoptosis and differentiation regulation [262] | upregulated blood plasma-derived EVs [153] |
| miR-4745-5p | SIRT6/PCSk9 signaling | sensibility to anesthetics regulation [263] | upregulated blood plasma-derived EVs [153] |
| miR-4788 | not yet studied, but 29 predicted targets in miRDB [234] | Nervous system development, neurotransmitter levels regulation and transport, and synapsis [264] | downregulated: blood plasma-derived EVs [153] |
| miR-7641-1 | BCL2, CAS9, C9orf153, PARP, RAB7L1, RPS16, TMEM156, TMPRSS11BNL, TNFSF10 | Apoptotic signaling in cancer [265] | downregulated: blood plasma-derived EVs [145] |
| miR-7975 | MTDH | Possible involvement in atherosclerosis [266], in lung inflammation, and cancer [267,268] | upregulated: serum of sALS patients [155] |
| miR-7977 | CD84, MRPS12, MRPL27, MUC19, TRAPPC2, SIRT3, Hyppo-YAP signaling | Hematopoiesis regulation [269], oxidative stress, and insulin resistance [270] | downregulated: blood plasma-derived EVs [145] |
| let-7c-5p | ARID3B, C14orf28, DNA2, FIGN, HMGA2, LIN28B, TRIM71 SMARCAD1; CTHRC1/AKT/ERK signaling | Microglia activation inhibition. Cell migration and proliferation. Inhibition and apoptosis enhancement [271] | downregulated: blood plasma-derived EVs [145] |
| Protein Content | Biological Function | Vesicle/Sample Type | Main Results |
|---|---|---|---|
| BLMH | Enzyme with proteolytic activity. Involved in release of inflammatory chemokines and in wound healing [301] | EVs from CSF of ALS patients (C9orf72 mutation) | Downregulation in ALS patients CSF-derived EVs |
| CD163, FOXP3, IL2RA, MRC1 | Anti-inflammatory transcripts [302] | Treg-derived EVs from spinal cord from SOD1G93A mice model and iPSC-derived from myeloid cells | - Upregulation in spleen-derived myeloid cells after Treg- derived EVs treatment [133]; - Intranasal administration of enriched Treg EVs slowed disease progression, increased survival, and modulated inflammation within the SOD1G93A mice spinal cord [133] |
| CD177, CHMP4B, CSPG5, DYNC1I2, IGHV3-43, LBP, RPS29, S100A9, SAA1, SCAMP4, SCN2B, SLC16A1, STAU1, FXYD6, DYNC1I1, DHX30 | Involvement in stress granule dynamics [296] | MCEVs from ALS patients’ postmortem motor cortex tissue | - 12 RNA-binding proteins only found in MCEVs from ALS patients (mainly downregulated) [296]; - 4 proteins significantly upregulated in MCEVs from ALS patients (DYNC1I1, DHX30, FXYD6, STAU1) [296] |
| CHIT1 | Cleavage of chitin (protein found in cell walls of various pathogens). Expressed during the later stages of macrophage differentiation. Important in inflammation and tissue remodeling. In the ALS context, plays a role in the feed-forward loop that maintains inflammation [303] | EVs from ALS patients’ CSF | Upregulation in ALS patients CSF-derived EVs [298] |
| CUEDC2 | Regulates ubiquitin-proteasome pathways and inflammatory response [304] | Exos from sALS patients’ CSF | Only expressed in ALS group [305] |
| FUS and pFUS | DNA repair, RNA splicing, dendritic RNA transport, miRNA function and biogenesis [306] | MVs and Exos from sALS patients’ plasma | Protein levels are higher in ALS patients’ plasma-derived MVs than Exos [81] |
| HSP90 | Chaperone protein involved in protein folding [307] | EVs from ALS patients and symptomatic SOD1G93A and TDP-43Q331K ALS mice models plasma | Downregulation in EVs from sALS patients [297] |
| JAM-A | Regulation of several processes including paracellular permeability, platelet activation, angiogenesis and the modulation of junctional tightness in the blood brain barrier (BBB) [308] | EVs from ALS patients CSF | Downregulation in ALS patients CSF-derived EVs [298] |
| IL-6, iNOS, IL-1b, IFN-y | Proinflammatory cytokines (IL-1b, IFN-y, IL-6) and enzyme (iNOS) produced in response to cytokines [309] | Treg-derived EVs from spinal cord from SOD1G93A mice model and iPSC-derived from myeloid cells | Downregulation in spleen-derived myeloid cells after Treg-derived EVs treatment [133] |
| MB | Oxygen-binding molecule expressed in skeletal and cardiac muscle tissue [310,311] | EVs from ALS patients CSF | Downregulation in ALS patients CSF-derived EVs [298] |
| mfSOD1 | Antioxidant enzyme, protects cells from ROS [312] | Vacuoles and EVs from spinal cord samples of SOD1G93A mice model | Accumulation of mfSOD1-vacuoles in degenerating MNs, released into the extracellular space in the form of extracellular vesicles [130] |
| NIR | Translocates from the nucleolus to the nucleoplasm in response to the nucleolar stress [313] | Exos from sALS patients CSF and anterior horn postmortem tissue sections | Upregulation in sALS patients CSF-derived exos [314] |
| Nrf2 | Antioxidant factor [315] | EVs from spinal cord tissue of SOD1G93A mice model | Upregulation after exposure to MSCs-derived EVs, with consequent reduction of ROS [316] |
| pCREB | Involved in the synthesis of proteins required for LTP [317] | Exos from SOD1G93A mice model SVZ-derived NSCs, differentiated into G93A neuronal cells | Downregulation in G93A cells, normalized with ADSC-exos treatment [318] |
| PGC-1α | Involved in the regulation cell metabolism [319] | Exos from SOD1G93A mice model SVZ-derived NSCs, differentiated into G93A neuronal cells | Downregulation levels in G93A cells, normalized with ADSC-exos treatment [318] |
| Phenylalanine | Precursor for tyrosine [320], the monoamine neurotransmitters dopamine, norepinephrine, and epinephrine | lEVs and sEVs from sALS patients’ plasma | Downregulation in EVs from sALS patients [321] |
| pMLKL | Effector of necroptotic pathways [322] | Vacuoles and EVs from spinal cord samples of SOD1G93A mice model | Upregulation in vacuoles of degenerating MNs (necroptotic pathway activation/phenotype 3) [130] |
| PPIA | Ubiquitous protein involved in protein folding, transport and signaling (e.g apoptosis, inflammation, etc) [323] | EVs from ALS patients and symptomatic SOD1G93A and TDP-43Q331K ALS mice models plasma | Protein levels and EV size distribution distinguish fast and slow ALS disease progression [297] |
| pNFH | Chaperone involved in TDP-43 trafficking and function [324] | EVs from ALS patients and symptomatic SOD1G93A and TDP-43Q331K ALS mice models plasma | Upregulation in EVs from sALS patients [297] |
| SOD1 | Binds copper and zinc ions, responsible for freeing superoxide radicals from cells [312] | Exos from SOD1G93A mice model SVZ-derived NSCs, differentiated into G93A neuronal cells | ADSC-exos alleviated aggregation of cytosolic SOD1 in G93A ALS mice isolated neuronal cells [318] |
| MVs and Exos from sALS patients’ plasma | Protein levels are higher in ALS patients’ plasma-derived Exos than MVs [81] | ||
| TDP-43 and pTDP-43 | RNA regulation (transcriptional regulation, alternative splicing and mRNA stabilization) [325] | Exos from ALS patients CSF | TDP-43 accumulation [326] |
| MVs and Exos from sALS patients’ plasma | Protein levels are higher in ALS patients’ plasma-derived MVs than Exos [81] | ||
| TNF-R2 | Proinflammatory proteins activation [327] | EVs from ALS patients’ CSF | Upregulation in serum of ALS patients. TNF-R2 knocking down in ALS mouse model results in motor neuron protection [298] |
| UBA1 | Involved in ubiquitination of proteins for degradation by the UPS [328] | EVs from ALS patients’ CSF (C9orf72 mutation) | Upregulation in ALS patients’ CSF-derived EVs [298] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afonso, G.J.M.; Cavaleiro, C.; Valero, J.; Mota, S.I.; Ferreiro, E. Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives. Cells 2023, 12, 1763. https://doi.org/10.3390/cells12131763
Afonso GJM, Cavaleiro C, Valero J, Mota SI, Ferreiro E. Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives. Cells. 2023; 12(13):1763. https://doi.org/10.3390/cells12131763
Chicago/Turabian StyleAfonso, Gonçalo J. M., Carla Cavaleiro, Jorge Valero, Sandra I. Mota, and Elisabete Ferreiro. 2023. "Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives" Cells 12, no. 13: 1763. https://doi.org/10.3390/cells12131763
APA StyleAfonso, G. J. M., Cavaleiro, C., Valero, J., Mota, S. I., & Ferreiro, E. (2023). Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives. Cells, 12(13), 1763. https://doi.org/10.3390/cells12131763








