Emerging Role of miR-21-5p in Neuron–Glia Dysregulation and Exosome Transfer Using Multiple Models of Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Ethics
2.2. Organotypic Hippocampal Cultures
2.3. Human Neuroblastoma Cell Culture
2.4. Human Neuroblastoma Live-Cell Staining and Transplantation
2.5. MCI Patients, Clinical Assessment and CSF Collection
2.6. RNA Isolation and Purification from CSF
2.7. Culture and Differentiation of Patient iPSCs
2.8. Primitive Hematopoietic Differentiation of iPSCs into Microglia
2.9. Neural Induction of iPSCs and Sphere Formation
2.9.1. Isolation and Maturation of Neurons
2.9.2. Differentiation, Maturation and Immunostimulation of Astrocytes
2.10. RNA Extraction and RT-qPCR
2.11. Immunocytochemistry and Immunohistochemistry
2.12. Confocal Microscopy and Post-Acquisition Analysis
2.13. Exosome Isolation
2.14. Evaluation of the Phagocytic Ability of Microglia
2.15. Determination of Synaptic Puncta in Neurons
2.16. Protein Extraction and Western Blot
2.17. Soluble Cytokine Determination
2.18. Statistical Analysis
3. Results
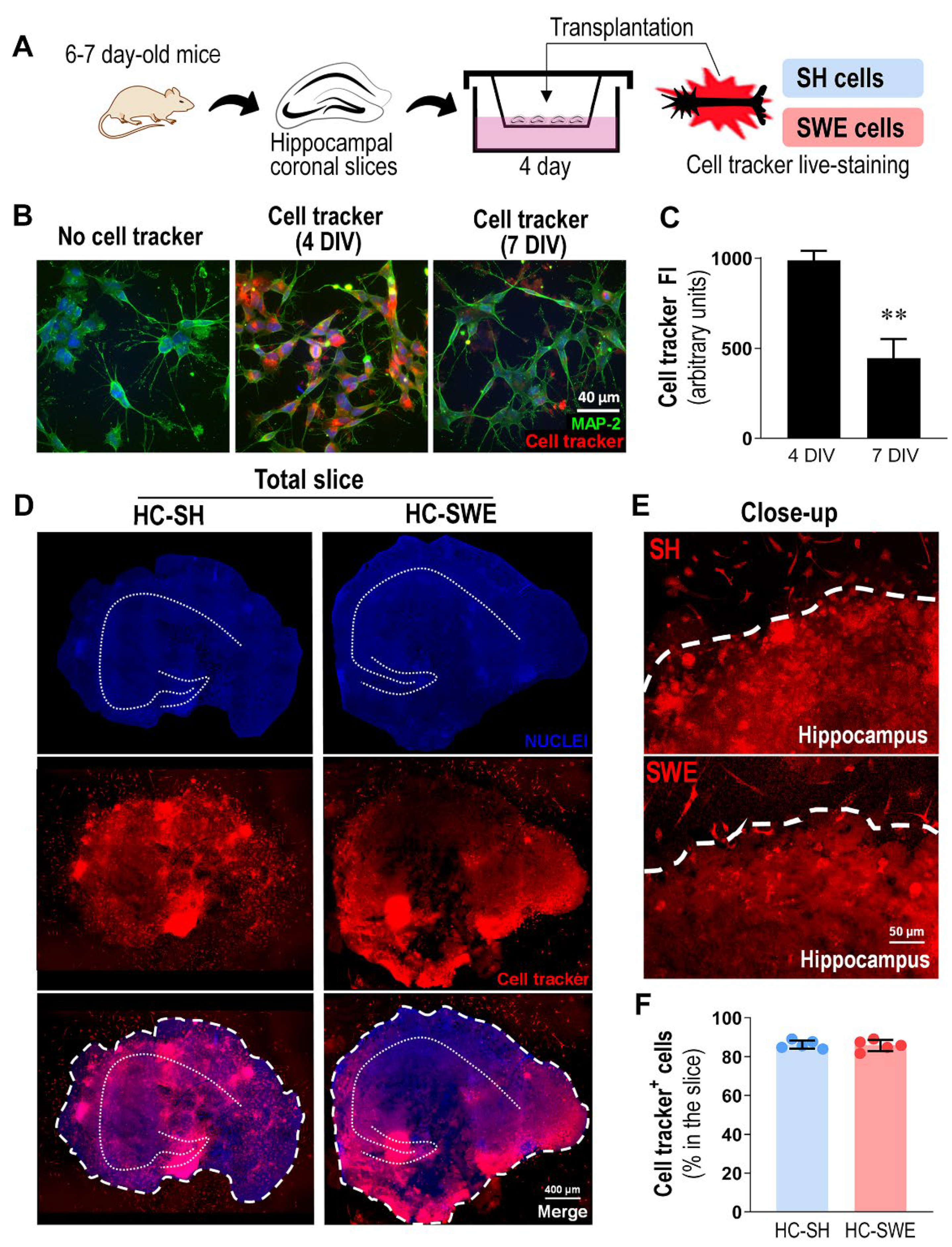
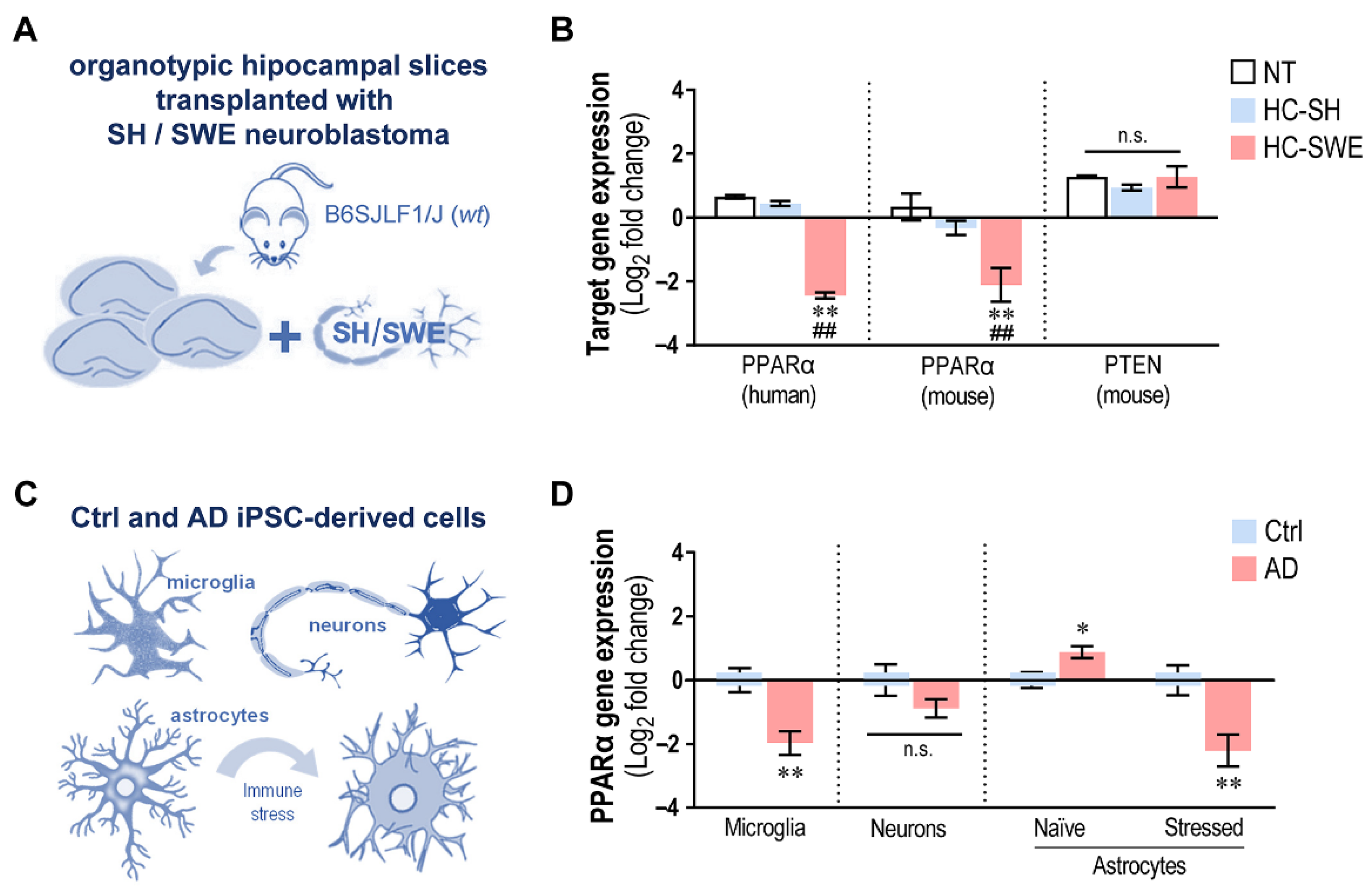
3.1. SH and SWE Human Neuroblastoma Cells Were Successfully Transplanted into 4 DIV Organotypic Hippocampal Cultures from WT Mice
3.2. Hippocampal Slices Transplanted with SWE Cells Show Microglial Activation and Astrocyte Reactivity, Together with miR-21 Overexpression
3.3. miR-21 Upregulation in the CSF Discriminates MCI-AD from MCI-Ctrl Patients
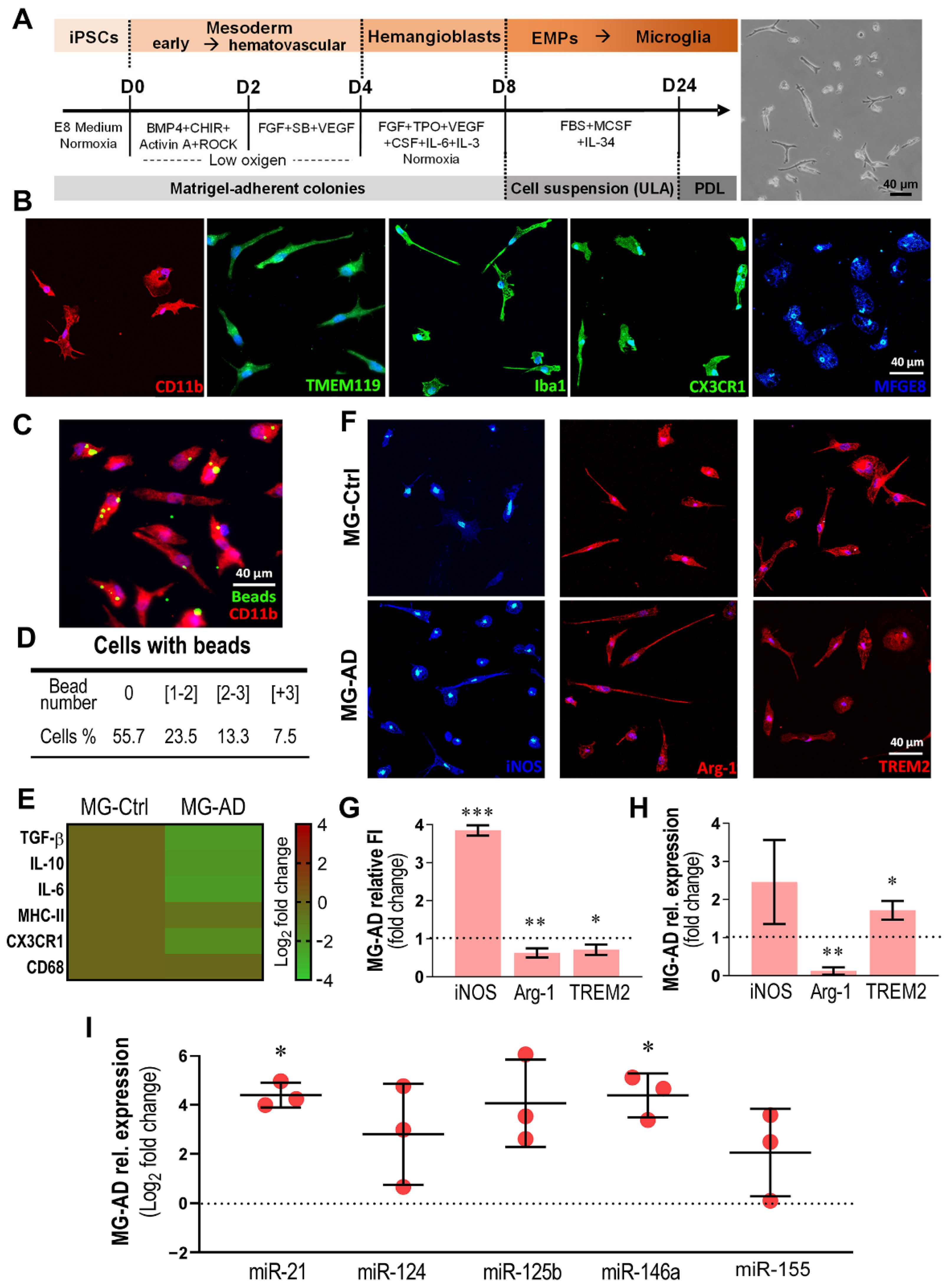
3.4. MG-AD Cells Show Dysregulated Immunoreactive Markers and Upregulated miR-146a and miR-21
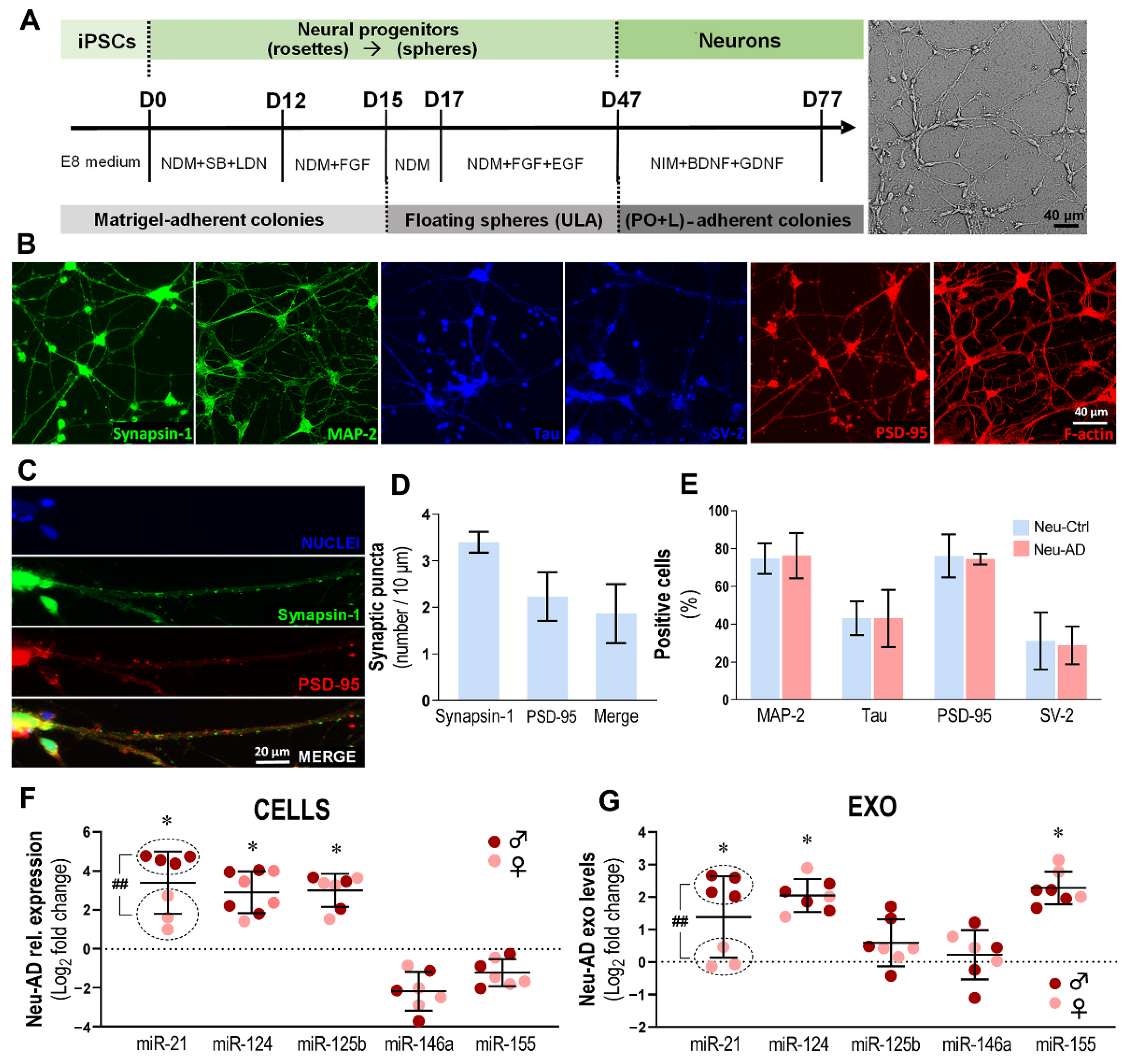
3.5. Neu-AD Cells Exhibit Upregulated Intracellular and Exosomal miR-21 and miR-124, Together with miR-125b in Cells and miR-155 Only in Exosomes, and Sex-Biased miR-21 Expression
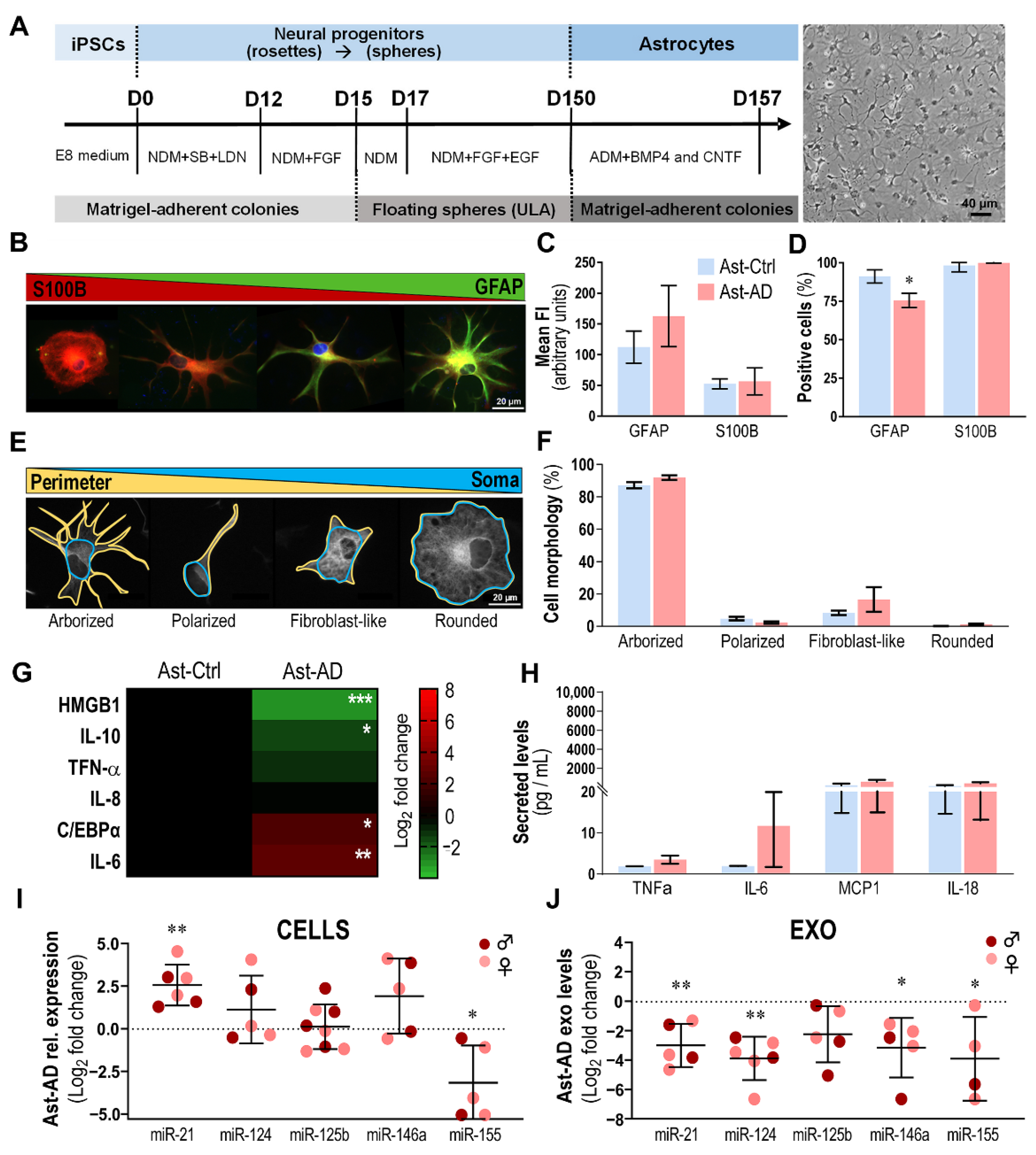
3.6. Ast-AD Cells Show Phenotypic Diversity, GFAP+ Cell Deficiency, Altered Inflammatory Gene/miRNA Expression and Contrasting Cellular/Exosomal miR-21 Profile
3.7. Immunostimulation Drives S100B Imbalance and Morphological Changes in Ast-AD
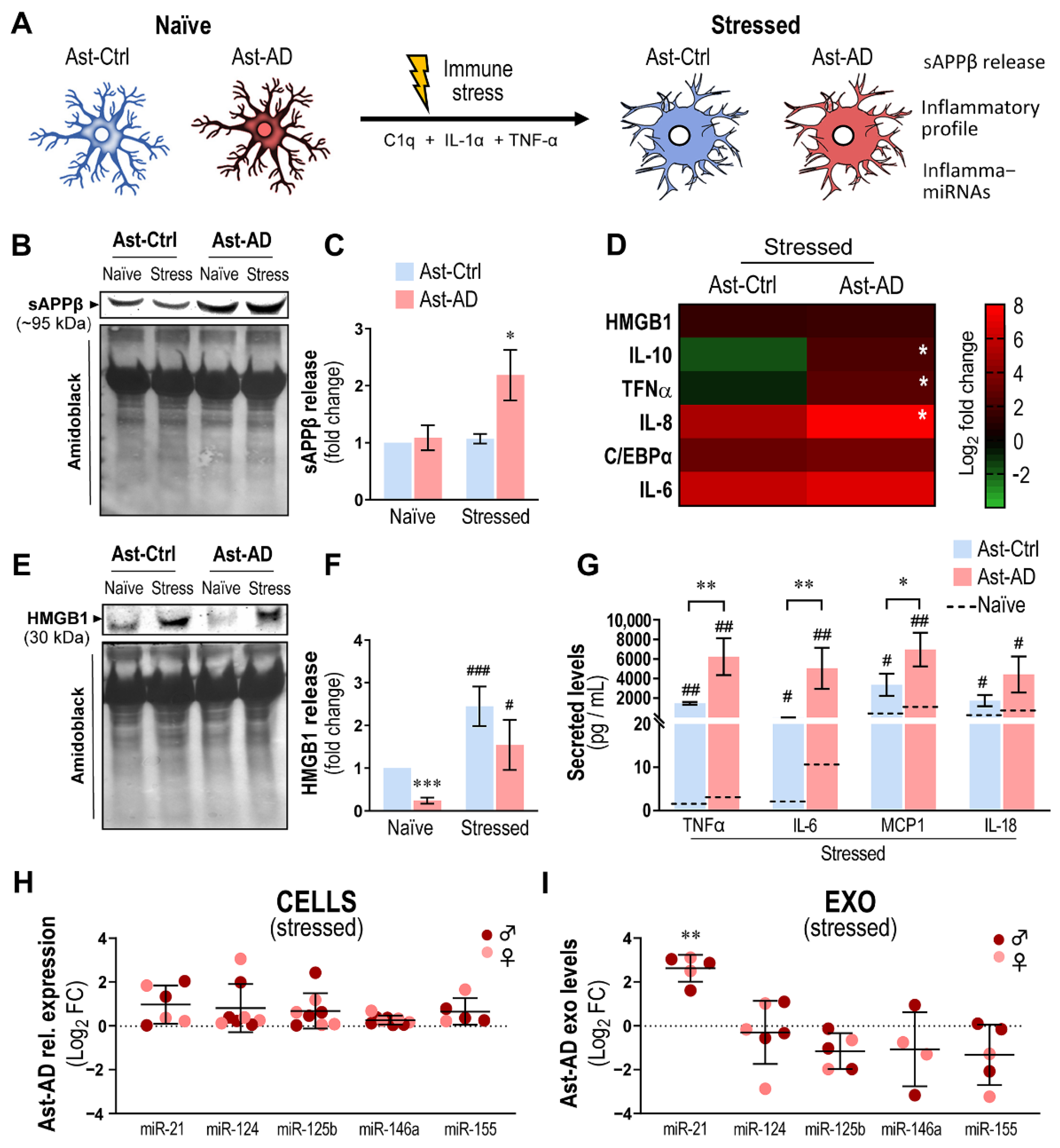
3.8. Immunostimulated Ast-AD Cells Show Increased Inflammatory Gene Expression, Exosomal Enrichment in miR-21 and Release of sAPPβ and Cytokines
3.9. miR-21 Upregulation Suppresses its PPARα Target in SWE-Transplanted Hippocampal Slices and in AD iPSC-Derived Cells Depending on the Concurrent Dysregulated miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Hayek, Y.H.; Wiley, R.E.; Khoury, C.P.; Daya, R.P.; Ballard, C.; Evans, A.R.; Karran, M.; Molinuevo, J.L.; Norton, M.; Atri, A. Tip of the Iceberg: Assessing the Global Socioeconomic Costs of Alzheimer’s Disease and Related Dementias and Strategic Implications for Stakeholders. J. Alzheimer’s Dis. 2019, 70, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Schott, J.M.; Aisen, P.S.; Cummings, J.L.; Howard, R.J.; Fox, N.C. Unsuccessful Trials of Therapies for Alzheimer’s Disease. Lancet 2019, 393, 29. [Google Scholar] [CrossRef]
- Cummings, J.; Lee, G.; Nahed, P.; Zadeh, M.E.; Kambar, N.; Zhong, K.; Fonseca, J.; Taghva, K. Alzheimer’s Disease Drug Development Pipeline: 2022. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2022, 8, e12295. [Google Scholar] [CrossRef] [PubMed]
- Markesbery, W.R. Neuropathologic Alterations in Mild Cognitive Impairment: A Review. J. Alzheimer’s Dis. 2010, 19, 221–228. [Google Scholar] [CrossRef]
- Albert, M.S.; DeKosky, S.T.; Dickson, D.; Dubois, B.; Feldman, H.H.; Fox, N.C.; Gamst, A.; Holtzman, D.M.; Jagust, W.J.; Petersen, R.C.; et al. The Diagnosis of Mild Cognitive Impairment Due to Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 270–279. [Google Scholar] [CrossRef]
- Zou, K.; Abdullah, M.; Michikawa, M. Current Biomarkers for Alzheimer’s Disease: From CSF to Blood. J. Pers. Med. 2020, 10, 85. [Google Scholar] [CrossRef]
- Klyucherev, T.O.; Olszewski, P.; Shalimova, A.A.; Chubarev, V.N.; Tarasov, V.V.; Attwood, M.M.; Syvänen, S.; Schiöth, H.B. Advances in the Development of New Biomarkers for Alzheimer’s Disease. Transl. Neurodegener. 2022, 11, 25. [Google Scholar] [CrossRef]
- Miller, M.B.; Huang, A.Y.; Kim, J.; Zhou, Z.; Kirkham, S.L.; Maury, E.A.; Ziegenfuss, J.S.; Reed, H.C.; Neil, J.E.; Rento, L.; et al. Somatic Genomic Changes in Single Alzheimer’s Disease Neurons. Nature 2022, 604, 714–722. [Google Scholar] [CrossRef]
- De Strooper, B.; Karran, E. The Cellular Phase of Alzheimer’s Disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef]
- Preman, P.; Alfonso-Triguero, M.; Alberdi, E.; Verkhratsky, A.; Arranz, A.M. Astrocytes in Alzheimer’s Disease: Pathological Significance and Molecular Pathways. Cells 2021, 10, 540. [Google Scholar] [CrossRef]
- González-Reyes, R.E.; Nava-Mesa, M.O.; Vargas-Sánchez, K.; Ariza-Salamanca, D.; Mora-Muñoz, L. Involvement of Astrocytes in Alzheimer’s Disease from a Neuroinflammatory and Oxidative Stress Perspective. Front. Mol. Neurosci. 2017, 10, 427. [Google Scholar] [CrossRef] [PubMed]
- Scimemi, A.; Meabon, J.S.; Woltjer, R.L.; Sullivan, J.M.; Diamond, J.S.; Cook, D.G. Amyloid- 1-42 Slows Clearance of Synaptically Released Glutamate by Mislocalizing Astrocytic GLT-1. J. Neurosci. 2013, 33, 5312–5318. [Google Scholar] [CrossRef] [PubMed]
- Olabarria, M.; Noristani, H.N.; Verkhratsky, A.; Rodríguez, J.J. Concomitant Astroglial Atrophy and Astrogliosis in a Triple Transgenic Animal Model of Alzheimer’s Disease. Glia 2010, 58, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s Disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Bamberger, M.E.; Harris, M.E.; McDonald, D.R.; Husemann, J.; Landreth, G.E. A Cell Surface Receptor Complex for Fibrillar β-Amyloid Mediates Microglial Activation. J. Neurosci. 2003, 23, 2665–2674. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s Disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Hickman, S.E.; Allison, E.K.; El Khoury, J. Microglial Dysfunction and Defective -Amyloid Clearance Pathways in Aging Alzheimer’s Disease Mice. J. Neurosci. 2008, 28, 8354–8360. [Google Scholar] [CrossRef]
- Minter, M.R.; Taylor, J.M.; Crack, P.J. The Contribution of Neuroinflammation to Amyloid Toxicity in Alzheimer’s Disease. J. Neurochem. 2016, 136, 457–474. [Google Scholar] [CrossRef]
- Drummond, E.; Wisniewski, T. Alzheimer’s Disease: Experimental Models and Reality. Acta Neuropathol. 2017, 133, 155–175. [Google Scholar] [CrossRef]
- Abreu, C.M.; Gama, L.; Krasemann, S.; Chesnut, M.; Odwin-Dacosta, S.; Hogberg, H.T.; Hartung, T.; Pamies, D. Microglia Increase Inflammatory Responses in IPSC-Derived Human BrainSpheres. Front. Microbiol. 2018, 9, 2766. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. IPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Brownjohn, P.W.; Smith, J.; Solanki, R.; Lohmann, E.; Houlden, H.; Hardy, J.; Dietmann, S.; Livesey, F.J. Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem. Cell Rep. 2018, 10, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Valadez-Barba, V.; Cota-Coronado, A.; Hernández-Pérez, O.R.; Lugo-Fabres, P.H.; Padilla-Camberos, E.; Díaz, N.F.; Díaz-Martínez, N.E. IPSC for Modeling Neurodegenerative Disorders. Regen. Ther. 2020, 15, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiao, B.; Zhou, M.; Zhou, T.; Shen, L. Modeling Alzheimer’s Disease with Induced Pluripotent Stem Cells: Current Challenges and Future Concerns. Stem Cells Int. 2016, 2016, 7828049. [Google Scholar] [CrossRef]
- Garcia, G.; Pinto, S.; Cunha, M.; Fernandes, A.; Koistinaho, J.; Brites, D. Neuronal Dynamics and MiRNA Signaling Differ between SH-SY5Y APPSwe and PSEN1 Mutant IPSC-Derived AD Models upon Modulation with MiR-124 Mimic and Inhibitor. Cells 2021, 10, 2424. [Google Scholar] [CrossRef]
- Konttinen, H.; Cabral-da-Silva, M.E.C.; Ohtonen, S.; Wojciechowski, S.; Shakirzyanova, A.; Caligola, S.; Giugno, R.; Ishchenko, Y.; Hernández, D.; Fazaludeen, M.F.; et al. PSEN1ΔE9, APPswe, and APOE4 Confer Disparate Phenotypes in Human IPSC-Derived Microglia. Stem Cell Rep. 2019, 13, 669–683. [Google Scholar] [CrossRef]
- Oksanen, M.; Petersen, A.J.; Naumenko, N.; Puttonen, K.; Lehtonen, Š.; Olivé, M.G.; Shakirzyanova, A.; Leskelä, S.; Sarajärvi, T.; Viitanen, M.; et al. PSEN1 Mutant IPSC-Derived Model Reveals Severe Astrocyte Pathology in Alzheimer’s Disease. Stem Cell Rep. 2017, 9, 1885–1897. [Google Scholar] [CrossRef]
- Jones, V.C.; Atkinson-Dell, R.; Verkhratsky, A.; Mohamet, L. Aberrant IPSC-Derived Human Astrocytes in Alzheimer’s Disease. Cell Death Dis. 2017, 8, e2696. [Google Scholar] [CrossRef]
- Zhang, Z.; Almeida, S.; Lu, Y.; Nishimura, A.L.; Peng, L.; Sun, D.; Wu, B.; Karydas, A.M.; Tartaglia, M.C.; Fong, J.C.; et al. Downregulation of MicroRNA-9 in IPSC-Derived Neurons of FTD/ALS Patients with TDP-43 Mutations. PLoS ONE 2013, 8, 76055. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, H.; Si, Y.; Wu, N.; Cao, H.; Mei, B.; Meng, B. MiR-125b Promotes Tau Phosphorylation by Targeting the Neural Cell Adhesion Molecule in Neuropathological Progression. Neurobiol. Aging 2019, 73, 41–49. [Google Scholar] [CrossRef]
- Kozuka, T.; Omori, Y.; Watanabe, S.; Tarusawa, E.; Yamamoto, H.; Chaya, T.; Furuhashi, M.; Morita, M.; Sato, T.; Hirose, S.; et al. MiR-124 Dosage Regulates Prefrontal Cortex Function by Dopaminergic Modulation. Sci. Rep. 2019, 9, 3445. [Google Scholar] [CrossRef] [PubMed]
- Schonrock, N.; Ke, Y.D.; Humphreys, D.; Staufenbiel, M.; Ittner, L.M.; Preiss, T.; Götz, J. Neuronal MicroRNA Deregulation in Response to Alzheimer’s Disease Amyloid-β. PLoS ONE 2010, 5, e11070. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. MiR-21: A Non-specific Biomarker of All Maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Bian, Z. MicroRNA-21 Is a Versatile Regulator and Potential Treatment Target in Central Nervous System Disorders. Front. Mol. Neurosci. 2022, 15, 5. [Google Scholar] [CrossRef]
- Feng, M.-G.; Liu, C.-F.; Chen, L.; Feng, W.-B.; Liu, M.; Hai, H.; Lu, J.-M. MiR-21 Attenuates Apoptosis-Triggered by Amyloid-β via Modulating PDCD4/ PI3K/AKT/GSK-3β Pathway in SH-SY5Y Cells. Biomed. Pharmacother. 2018, 101, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of Extracellular MiRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Gaetani, S.; Sorgentoni, G.; Agarbati, S.; Laggetta, M.; Matacchione, G.; Gobbi, M.; Rossi, T.; Galeazzi, R.; Piccinini, G.; et al. Circulating Inflamma-MiRs as Potential Biomarkers of Cognitive Impairment in Patients Affected by Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 108. [Google Scholar] [CrossRef]
- Pinto, S.; Cunha, C.; Barbosa, M.; Vaz, A.R.; Brites, D. Exosomes from NSC-34 Cells Transfected with HSOD1-G93A Are Enriched in Mir-124 and Drive Alterations in Microglia Phenotype. Front. Neurosci. 2017, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.; Ribeiro, A.R.; Monteiro, M.; Garcia, G.; Vaz, A.R.; Brites, D.; Rita, A.; Brites, D.; Vaz, A.R.; Brites, D. Secretome from SH-SY5Y APPSwe Cells Trigger Time-Dependent CHME3 Microglia Activation Phenotypes, Ultimately Leading to MiR-21 Exosome Shuttling. Biochimie 2018, 155, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.; Fernandes, A.; Stein, F.; Brites, D. Protective Signature of IFNγ-Stimulated Microglia Relies on MiR-124-3p Regulation from the Secretome Released by Mutant APP Swedish Neuronal Cells. Front. Pharmacol. 2022, 13, 1273. [Google Scholar] [CrossRef]
- Barbosa, M.; Santos, M.; de Sousa, N.; Duarte-Silva, S.; Vaz, A.R.; Salgado, A.J.; Brites, D. Intrathecal Injection of the Secretome from ALS Motor Neurons Regulated for MiR-124 Expression Prevents Disease Outcomes in SOD1-G93A Mice. Biomedicines 2022, 10, 2120. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, L. Inflamma-MicroRNAs in Alzheimer’s Disease: From Disease Pathogenesis to Therapeutic Potentials. Front. Cell Neurosci. 2021, 15, 443. [Google Scholar] [CrossRef]
- Brites, D. Regulatory Function of MicroRNAs in Microglia. Glia 2020, 68, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic Reactive Astrocytes Are Induced by Activated Microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.L.; Vaz, A.R.; Diógenes, M.J.; van Rooijen, N.; Sebastião, A.M.; Fernandes, A.; Silva, R.F.M.; Brites, D. Neuritic Growth Impairment and Cell Death by Unconjugated Bilirubin Is Mediated by NO and Glutamate, Modulated by Microglia, and Prevented by Glycoursodeoxycholic Acid and Interleukin-10. Neuropharmacology 2012, 62, 2398–2408. [Google Scholar] [CrossRef]
- Hiragi, T.; Andoh, M.; Araki, T.; Shirakawa, T.; Ono, T.; Koyama, R.; Ikegaya, Y. Differentiation of Human Induced Pluripotent Stem Cell (HiPSC)-Derived Neurons in Mouse Hippocampal Slice Cultures. Front. Cell Neurosci. 2017, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-Mental State”. A Practical Method for Grading the Cognitive State of Patients for the Clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Morris, J.C. The Clinical Dementia Rating (CDR): Current Version and Scoring Rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef] [PubMed]
- Engelborghs, S.; Niemantsverdriet, E.; Struyfs, H.; Blennow, K.; Brouns, R.; Comabella, M.; Dujmovic, I.; Flier, W.; Frölich, L.; Galimberti, D.; et al. Consensus Guidelines for Lumbar Puncture in Patients with Neurological Diseases. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2017, 8, 111–126. [Google Scholar] [CrossRef]
- Teunissen, C.E.; Tumani, H.; Engelborghs, S.; Mollenhauer, B. Biobanking of CSF: International Standardization to Optimize Biomarker Development. Clin. Biochem. 2014, 47, 288–292. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly Efficient Neural Conversion of Human ES and IPS Cells by Dual Inhibition of SMAD Signaling. Nat. Biotechnol 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Krencik, R.; Zhang, S.-C. Directed Differentiation of Functional Astroglial Subtypes from Human Pluripotent Stem Cells. Nat. Protoc. 2011, 6, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Method. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Caldeira, C.; Oliveira, A.F.; Cunha, C.; Vaz, A.R.; Falcão, A.S.; Falcão, A.S.; Fernandes, A.; Brites, D.; Falcão, A.S.; Fernandes, A.; et al. Microglia Change from a Reactive to an Age-like Phenotype with the Time in Culture. Front. Cell. Neurosci. 2014, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Kouroupi, G.; Taoufik, E.; Vlachos, I.S.; Tsioras, K.; Antoniou, N.; Papastefanaki, F.; Chroni-Tzartou, D.; Wrasidlo, W.; Bohl, D.; Stellas, D.; et al. Defective Synaptic Connectivity and Axonal Neuropathology in a Human IPSC-Based Model of Familial Parkinson’s Disease. Proc. Natl. Acad. Sci. USA 2017, 114, E3679–E3688. [Google Scholar] [CrossRef] [PubMed]
- Vaz, A.R.; Pinto, S.; Ezequiel, C.; Cunha, C.; Carvalho, L.A.; Moreira, R.; Brites, D. Phenotypic Effects of Wild-Type and Mutant SOD1 Expression in N9 Murine Microglia at Steady State, Inflammatory and Immunomodulatory Conditions. Front. Cell. Neurosci. 2019, 13, 109. [Google Scholar] [CrossRef]
- Falcão, A.S.; Silva, R.F.M.; Vaz, A.R.; Gomes, C.; Fernandes, A.; Barateiro, A.; Tiribelli, C.; Brites, D. Cross-Talk between Neurons and Astrocytes in Response to Bilirubin: Adverse Secondary Impacts. Neurotox. Res. 2014, 26, 1–15. [Google Scholar] [CrossRef]
- Landucci, E.; Llorente, I.; Anuncibay-Soto, B.; Pellegrini-Giampietro, D.; Fernández-López, A. Using Organotypic Hippocampal Slice Cultures to Gain Insight into Mechanisms Responsible for the Neuroprotective Effects of Meloxicam: A Role for Gamma Aminobutyric and Endoplasmic Reticulum Stress. Neural. Regen. Res. 2019, 14, 65. [Google Scholar] [CrossRef]
- Zhang, T.; Ke, W.; Zhou, X.; Qian, Y.; Feng, S.; Wang, R.; Cui, G.; Tao, R.; Guo, W.; Duan, Y.; et al. Human Neural Stem Cells Reinforce Hippocampal Synaptic Network and Rescue Cognitive Deficits in a Mouse Model of Alzheimer’s Disease. Stem Cell Rep. 2019, 13, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, F.-R.; Yu, M.-Q. MicroRNA-21-5p Promotes the Inflammatory Response after Spinal Cord Injury by Targeting PLAG1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5878–5885. [Google Scholar] [CrossRef]
- Yin, Z.; Han, Z.; Hu, T.; Zhang, S.; Ge, X.; Huang, S.; Wang, L.; Yu, J.; Li, W.; Wang, Y.Y.; et al. Neuron-Derived Exosomes with High MiR-21-5p Expression Promoted Polarization of M1 Microglia in Culture. Brain Behav. Immun. 2020, 83, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Yelamanchili, S.V.; Lamberty, B.G.; Rennard, D.A.; Morsey, B.M.; Hochfelder, C.G.; Meays, B.M.; Levy, E.; Fox, H.S. MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease. PLoS Pathog. 2015, 11, e1005032. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Guo, C.; Nie, P.; Liu, Y.; Ma, J. Essential Role of MFG-E8 for Phagocytic Properties of Microglial Cells. PLoS ONE 2013, 8, e55754. [Google Scholar] [CrossRef] [PubMed]
- Fuller, A.D.; Van Eldik, L.J. MFG-E8 Regulates Microglial Phagocytosis of Apoptotic Neurons. J. Neuroimmune Pharmacol. 2008, 3, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Pomeshchik, Y.; Klementieva, O.; Gil, J.; Martinsson, I.; Hansen, M.G.; de Vries, T.; Sancho-Balsells, A.; Russ, K.; Savchenko, E.; Collin, A.; et al. Human IPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Rep. 2020, 15, 256–273. [Google Scholar] [CrossRef]
- Bhalala, O.G.; Pan, L.; Sahni, V.; McGuire, T.L.; Gruner, K.; Tourtellotte, W.G.; Kessler, J.A. MicroRNA-21 Regulates Astrocytic Response Following Spinal Cord Injury. J. Neurosci. 2012, 32, 17935–17947. [Google Scholar] [CrossRef]
- Li, H.-J.; Pan, Y.-B.; Sun, Z.-L.; Sun, Y.-Y.; Yang, X.-T.; Feng, D.-F. Inhibition of MiR-21 Ameliorates Excessive Astrocyte Activation and Promotes Axon Regeneration Following Optic Nerve Crush. Neuropharmacology 2018, 137, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.; Bellaver, B.; Souza, D.G.; Schu, G.; Fontana, I.C.; Venturin, G.T.; Greggio, S.; Fontella, F.U.; Schiavenin, M.L.; Machado, L.S.; et al. Clozapine Induces Astrocyte-Dependent FDG-PET Hypometabolism. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 2251–2264. [Google Scholar] [CrossRef]
- Carter, S.F.; Herholz, K.; Rosa-Neto, P.; Pellerin, L.; Nordberg, A.; Zimmer, E.R. Astrocyte Biomarkers in Alzheimer’s Disease. Trends Mol. Med. 2019, 25, 77–95. [Google Scholar] [CrossRef]
- Singh, D. Astrocytic and Microglial Cells as the Modulators of Neuroinflammation in Alzheimer’s Disease. J. Neuroinflamm. 2022, 19, 206. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and Microglial Activation in Alzheimer Disease: Where Do We Go from Here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Brambilla, L.; Martorana, F.; Guidotti, G.; Rossi, D. Dysregulation of Astrocytic HMGB1 Signaling in Amyotrophic Lateral Sclerosis. Front. Neurosci. 2018, 12, 622. [Google Scholar] [CrossRef]
- Chuppa, S.; Liang, M.; Liu, P.; Liu, Y.; Casati, M.C.; Cowley, A.W.; Patullo, L.; Kriegel, A.J. MicroRNA-21 Regulates Peroxisome Proliferator–Activated Receptor Alpha, a Molecular Mechanism of Cardiac Pathology in Cardiorenal Syndrome Type 4. Kidney Int. 2018, 93, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Orellana, F.; Leroy, T.; Ribeiro, F.; Kreis, A.; Leroy, K.; Lalloyer, F.; Baugé, E.; Staels, B.; Duyckaerts, C.; Brion, J.-P.; et al. Regulation of PPARα by APP in Alzheimer Disease Affects the Pharmacological Modulation of Synaptic Activity. JCI Insight 2021, 6, e150099. [Google Scholar] [CrossRef] [PubMed]
- Raha, S.; Ghosh, A.; Dutta, D.; Patel, D.R.; Pahan, K. Activation of PPARα Enhances Astroglial Uptake and Degradation of β-Amyloid. Sci. Signal 2021, 14, eabg4747. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Chen, D.; Chen, N. The Regulation of MicroRNAs in Alzheimer’s Disease. Front. Neurol. 2020, 11, 288. [Google Scholar] [CrossRef]
- Walgrave, H.; Zhou, L.; de Strooper, B.; Salta, E. The Promise of MicroRNA-Based Therapies in Alzheimer’s Disease: Challenges and Perspectives. Mol. Neurodegener. 2021, 16, 76. [Google Scholar] [CrossRef]
- Wei, W.; Wang, Z.-Y.; Ma, L.-N.; Zhang, T.-T.; Cao, Y.; Li, H. MicroRNAs in Alzheimer’s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 2020, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Pascoal, T.A.; Benedet, A.L.; Ashton, N.J.; Kang, M.S.; Therriault, J.; Chamoun, M.; Savard, M.; Lussier, F.Z.; Tissot, C.; Karikari, T.K.; et al. Microglial Activation and Tau Propagate Jointly across Braak Stages. Nat. Med. 2021, 27, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Gentile, G.; Morello, G.; La Cognata, V.; Guarnaccia, M.; Conforti, F.L.; Cavallaro, S. Dysregulated MiRNAs as Biomarkers and Therapeutical Targets in Neurodegenerative Diseases. J. Pers. Med. 2022, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fan, M.; Zheng, Q.; Hao, S.; Yang, L.; Xia, Q.; Qi, C.; Ge, J. MicroRNAs in Alzheimer’s Disease: Potential Diagnostic Markers and Therapeutic Targets. Biomed. Pharmacother. 2022, 148, 112681. [Google Scholar] [CrossRef]
- Vaz, A.R.; Vizinha, D.; Morais, H.; Colaço, A.R.; Loch-Neckel, G.; Barbosa, M.; Brites, D. Overexpression of MiR-124 in Motor Neurons Plays a Key Role in ALS Pathological Processes. Int. J. Mol. Sci. 2021, 22, 6128. [Google Scholar] [CrossRef]
- Barbosa, M.; Gomes, C.; Sequeira, C.; Gonçalves-Ribeiro, J.; Pina, C.C.; Carvalho, L.A.; Moreira, R.; Vaz, S.H.; Vaz, A.R.; Brites, D. Recovery of Depleted MiR-146a in ALS Cortical Astrocytes Reverts Cell Aberrancies and Prevents Paracrine Pathogenicity on Microglia and Motor Neurons. Front. Cell Dev. Biol. 2021, 9, 930. [Google Scholar] [CrossRef]
- Gomes, C.; Sequeira, C.; Likhite, S.; Dennys, C.N.; Kolb, S.J.; Shaw, P.J.; Vaz, A.R.; Kaspar, B.K.; Meyer, K.; Brites, D. Neurotoxic Astrocytes Directly Converted from Sporadic and Familial ALS Patient Fibroblasts Reveal Signature Diversities and MiR-146a Theragnostic Potential in Specific Subtypes. Cells 2022, 11, 1186. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Wood, A.; Bowlby, M. Brain Slices as Models for Neurodegenerative Disease and Screening Platforms to Identify Novel Therapeutics. Curr. Neuropharmacol. 2007, 5, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Gerace, E.; Landucci, E.; Bani, D.; Moroni, F.; Mannaioni, G.; Pellegrini-Giampietro, D.E. Glutamate Receptor-Mediated Neurotoxicity in a Model of Ethanol Dependence and Withdrawal in Rat Organotypic Hippocampal Slice Cultures. Front. Neurosci. 2019, 12, 1053. [Google Scholar] [CrossRef]
- Foidl, B.M.; Humpel, C. Differential Hyperphosphorylation of Tau-S199, -T231 and -S396 in Organotypic Brain Slices of Alzheimer Mice. A Model to Study Early Tau Hyperphosphorylation Using Okadaic Acid. Front. Aging Neurosci. 2018, 10, 113. [Google Scholar] [CrossRef]
- Jang, S.; Kim, H.; Kim, H.; Lee, S.K.; Kim, E.W.; Namkoong, K.; Kim, E. Long-Term Culture of Organotypic Hippocampal Slice from Old 3xTg-AD Mouse: An Ex Vivo Model of Alzheimer’s Disease. Psychiatry Investig. 2018, 15, 205–213. [Google Scholar] [CrossRef]
- Delbridge, A.R.D.; Huh, D.; Brickelmaier, M.; Burns, J.C.; Roberts, C.; Challa, R.; Raymond, N.; Cullen, P.; Carlile, T.M.; Ennis, K.A.; et al. Organotypic Brain Slice Culture Microglia Exhibit Molecular Similarity to Acutely-Isolated Adult Microglia and Provide a Platform to Study Neuroinflammation. Front. Cell Neurosci. 2020, 14, 444. [Google Scholar] [CrossRef]
- Perez-Nievas, B.G.; Serrano-Pozo, A. Deciphering the Astrocyte Reaction in Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 114. [Google Scholar] [CrossRef]
- Pahrudin Arrozi, A.; Shukri, S.N.S.; Wan Ngah, W.Z.; Mohd Yusof, Y.A.; Ahmad Damanhuri, M.H.; Makpol, S. Evaluation of the Expression of Amyloid Precursor Protein and the Ratio of Secreted Amyloid Beta 42 to Amyloid Beta 40 in SH-SY5Y Cells Stably Transfected with Wild-Type, Single-Mutant and Double-Mutant Forms of the APP Gene for the Study of Alzheimer’s Disease Pathology. Appl. Biochem. Biotechnol. 2017, 183, 853–866. [Google Scholar] [CrossRef]
- Lopez-Suarez, L.; Awabdh, S.A.; Coumoul, X.; Chauvet, C. The SH-SY5Y Human Neuroblastoma Cell Line, a Relevant in Vitro Cell Model for Investigating Neurotoxicology in Human: Focus on Organic Pollutants. Neurotoxicology 2022, 92, 131–155. [Google Scholar] [CrossRef]
- Penney, J.; Ralvenius, W.T.; Tsai, L.-H. Modeling Alzheimer’s Disease with IPSC-Derived Brain Cells. Mol. Psychiatry 2020, 25, 148–167. [Google Scholar] [CrossRef]
- Kovalevich, J.; Langford, D. Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: Totowa, NJ, USA, 2013; Volume 1078, pp. 9–21. [Google Scholar]
- Xicoy, H.; Wieringa, B.; Martens, G.J.M. The SH-SY5Y Cell Line in Parkinson’s Disease Research: A Systematic Review. Mol. Neurodegener. 2017, 12, 10. [Google Scholar] [CrossRef]
- Munõz-San Martín, M.; Reverter, G.; Robles-Cedenõ, R.; Buxò, M.; Ortega, F.J.; Gómez, I.; Tomàs-Roig, J.; Celarain, N.; Villar, L.M.; Perkal, H.; et al. Analysis of MiRNA Signatures in CSF Identifies Upregulation of MiR-21 and MiR-146a/b in Patients with Multiple Sclerosis and Active Lesions. J. Neuroinflamm. 2019, 16, 220. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory Study on MicroRNA Profiles from Plasma-Derived Extracellular Vesicles in Alzheimer’s Disease and Dementia with Lewy Bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef]
- Sørensen, S.S.; Nygaard, A.-B.; Christensen, T. MiRNA Expression Profiles in Cerebrospinal Fluid and Blood of Patients with Alzheimer’s Disease and Other Types of Dementia—An Exploratory Study. Transl. Neurodegener. 2016, 5, 6. [Google Scholar] [CrossRef]
- De Leeuw, S.; Tackenberg, C. Alzheimer’s in a Dish—Induced Pluripotent Stem Cell-Based Disease Modeling. Transl. Neurodegener. 2019, 8, 21. [Google Scholar] [CrossRef]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; Van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing Sporadic and Familial Alzheimer’s Disease Using Induced Pluripotent Stem Cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef]
- Yang, J.; Li, S.; He, X.-B.; Cheng, C.; Le, W. Induced Pluripotent Stem Cells in Alzheimer’s Disease: Applications for Disease Modeling and Cell-Replacement Therapy. Mol. Neurodegener. 2016, 11, 39. [Google Scholar] [CrossRef]
- Tomov, M.L.; O’Neil, A.; Abbasi, H.S.; Cimini, B.A.; Carpenter, A.E.; Rubin, L.L.; Bathe, M. Resolving Cell State in IPSC-Derived Human Neural Samples with Multiplexed Fluorescence Imaging. Commun. Biol. 2021, 4, 786. [Google Scholar] [CrossRef]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.-H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient Derivation of Microglia-like Cells from Human Pluripotent Stem Cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef]
- Reich, M.; Paris, I.; Ebeling, M.; Dahm, N.; Schweitzer, C.; Reinhardt, D.; Schmucki, R.; Prasad, M.; Köchl, F.; Leist, M.; et al. Alzheimer’s Risk Gene TREM2 Determines Functional Properties of New Type of Human IPSC-Derived Microglia. Front. Immunol. 2021, 11, 3918. [Google Scholar] [CrossRef]
- Ma, C.; Hunt, J.B.; Selenica, M.-L.B.; Sanneh, A.; Sandusky-Beltran, L.A.; Watler, M.; Daas, R.; Kovalenko, A.; Liang, H.; Placides, D.; et al. Arginase 1 Insufficiency Precipitates Amyloid-β Deposition and Hastens Behavioral Impairment in a Mouse Model of Amyloidosis. Front. Immunol. 2021, 11, 3376. [Google Scholar] [CrossRef]
- Combs, C.K.; Karlo, J.C.; Kao, S.-C.; Landreth, G.E. β-Amyloid Stimulation of Microglia and Monocytes Results in TNFα-Dependent Expression of Inducible Nitric Oxide Synthase and Neuronal Apoptosis. J. Neurosci. 2001, 21, 1179–1188. [Google Scholar] [CrossRef]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Arginase 1+ Microglia Reduce Aβ Plaque Deposition during IL-1β-Dependent Neuroinflammation. J. Neuroinflamm. 2015, 12, 203. [Google Scholar] [CrossRef]
- Lue, L.F.; Schmitz, C.; Walker, D.G. What Happens to Microglial TREM2 in Alzheimer’s Disease: Immunoregulatory Turned into Immunopathogenic? Neuroscience 2015, 302, 138–150. [Google Scholar] [CrossRef]
- Liang, C.; Zou, T.; Zhang, M.; Fan, W.; Zhang, T.; Jiang, Y.; Cai, Y.; Chen, F.; Chen, X.; Sun, Y.; et al. MicroRNA-146a Switches Microglial Phenotypes to Resist the Pathological Processes and Cognitive Degradation of Alzheimer’s Disease. Theranostics 2021, 11, 4103–4121. [Google Scholar] [CrossRef]
- Sheedy, F.J. Turning 21: Induction of MiR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef]
- Kondo, T.; Asai, M.; Tsukita, K.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Imamura, K.; Egawa, N.; Yahata, N.; Okita, K.; et al. Modeling Alzheimer’s Disease with IPSCs Reveals Stress Phenotypes Associated with Intracellular Aβ and Differential Drug Responsiveness. Cell Stem Cell 2013, 12, 487–496. [Google Scholar] [CrossRef]
- Gouder, L.; Vitrac, A.; Goubran-Botros, H.; Danckaert, A.; Tinevez, J.-Y.; André-Leroux, G.; Atanasova, E.; Lemière, N.; Biton, A.; Leblond, C.S.; et al. Altered Spinogenesis in IPSC-Derived Cortical Neurons from Patients with Autism Carrying de Novo SHANK3 Mutations. Sci. Rep. 2019, 9, 94. [Google Scholar] [CrossRef]
- Ochalek, A.; Mihalik, B.; Avci, H.X.; Chandrasekaran, A.; Téglási, A.; Bock, I.; Giudice, M.L.; Táncos, Z.; Molnár, K.; László, L.; et al. Neurons Derived from Sporadic Alzheimer’s Disease IPSCs Reveal Elevated TAU Hyperphosphorylation, Increased Amyloid Levels, and GSK3B Activation. Alzheimer’s Res. Ther. 2017, 9, 90. [Google Scholar] [CrossRef]
- Cornacchia, D.; Studer, L. Back and Forth in Time: Directing Age in IPSC-Derived Lineages. Brain Res. 2017, 1656, 14–26. [Google Scholar] [CrossRef]
- Zhang, T.; Ni, S.; Luo, Z.; Lang, Y.; Hu, J.; Lu, H. The Protective Effect of MicroRNA-21 in Neurons after Spinal Cord Injury. Spin. Cord 2019, 57, 141. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, S.; Ding, Y.; Nong, L.; Li, H.; Gao, G.; Zhou, D.; Xu, N. MicroRNA-21 Promotes Neurite Outgrowth by Regulating PDCD4 in a Rat Model of Spinal Cord Injury. Mol. Med. Rep. 2017, 16, 2522–2528. [Google Scholar] [CrossRef]
- Hao, P.; Waxman, D.J. Functional Roles of Sex-Biased, Growth Hormone–Regulated MicroRNAs MiR-1948 and MiR-802 in Young Adult Mouse Liver. Endocrinology 2018, 159, 1377–1392. [Google Scholar] [CrossRef]
- Acosta, C.; Anderson, H.D.; Anderson, C.M. Astrocyte Dysfunction in Alzheimer Disease. J. Neurosci. Res. 2017, 95, 2430–2447. [Google Scholar] [CrossRef]
- Konttinen, H.; Gureviciene, I.; Oksanen, M.; Grubman, A.; Loppi, S.; Huuskonen, M.T.; Korhonen, P.; Lampinen, R.; Keuters, M.; Belaya, I.; et al. PPARβ/δ-Agonist GW0742 Ameliorates Dysfunction in Fatty Acid Oxidation in PSEN1ΔE9 Astrocytes. Glia 2018, 67, 146–159. [Google Scholar] [CrossRef]
- Gaikwad, S.; Puangmalai, N.; Bittar, A.; Montalbano, M.; Garcia, S.; McAllen, S.; Bhatt, N.; Sonawane, M.; Sengupta, U.; Kayed, R. Tau Oligomer Induced HMGB1 Release Contributes to Cellular Senescence and Neuropathology Linked to Alzheimer’s Disease and Frontotemporal Dementia. Cell Rep. 2021, 36, 109419. [Google Scholar] [CrossRef]
- Gomes, C.; Sequeira, C.; Barbosa, M.; Cunha, C.; Vaz, A.R.; Brites, D. Astrocyte Regional Diversity in ALS Includes Distinct Aberrant Phenotypes with Common and Causal Pathological Processes. Exp. Cell Res. 2020, 395, 112209. [Google Scholar] [CrossRef]
- Szpakowski, P.; Ksiazek-Winiarek, D.; Turniak-Kusy, M.; Pacan, I.; Glabinski, A. Human Primary Astrocytes Differently Respond to Pro- and Anti-Inflammatory Stimuli. Biomedicines 2022, 10, 1769. [Google Scholar] [CrossRef] [PubMed]
- Madhyastha, R.; Madhyastha, H.; Nurrahmah, Q.I.; Purbasari, B.; Maruyama, M.; Nakajima, Y. MicroRNA 21 Elicits a Pro-Inflammatory Response in Macrophages, with Exosomes Functioning as Delivery Vehicles. Inflammation 2021, 44, 1274–1287. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Wetzel, I.; Marriott, I.; Dréau, D.; D’Avanzo, C.; Kim, D.Y.; Tanzi, R.E.; Cho, H. A 3D Human Triculture System Modeling Neurodegeneration and Neuroinflammation in Alzheimer’s Disease. Nat. Neurosci. 2018, 21, 941–951. [Google Scholar] [CrossRef]
- Adamowicz, M.; Kempinska-Podhorodecka, A.; Abramczyk, J.; Banales, J.M.; Milkiewicz, P.; Milkiewicz, M. Suppression of Hepatic PPARα in Primary Biliary Cholangitis Is Modulated by MiR-155. Cells 2022, 11, 2880. [Google Scholar] [CrossRef]
- Ramanan, S.; Kooshki, M.; Zhao, W.; Hsu, F.-C.; Robbins, M.E. PPARα Ligands Inhibit Radiation-Induced Microglial Inflammatory Responses by Negatively Regulating NF-ΚB and AP-1 Pathways. Free Radic. Biol. Med. 2008, 45, 1695–1704. [Google Scholar] [CrossRef]
- Roy, A.; Jana, M.; Kundu, M.; Corbett, G.T.; Rangaswamy, S.B.; Mishra, R.K.; Luan, C.-H.; Gonzalez, F.J.; Pahan, K. HMG-CoA Reductase Inhibitors Bind to PPARα to Upregulate Neurotrophin Expression in the Brain and Improve Memory in Mice. Cell Metab. 2015, 22, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Wójtowicz, S.; Strosznajder, A.K.; Jeżyna, M.; Strosznajder, J.B. The Novel Role of PPAR Alpha in the Brain: Promising Target in Therapy of Alzheimer’s Disease and Other Neurodegenerative Disorders. Neurochem. Res. 2020, 45, 972–988. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Luo, Z.; Volinia, S.; Rassenti, L.Z.; Kipps, T.J.; Croce, C.M. The Down-Regulation of MiR-125b in Chronic Lymphocytic Leukemias Leads to Metabolic Adaptation of Cells to a Transformed State. Blood 2012, 120, 2631–2638. [Google Scholar] [CrossRef]
- Xie, W.; Qianqian, Y.; Yadong, Z.; Yi, Z.; Ronghan, W.; Wenzhao, L. Knockdown of MicroRNA-21 Promotes Neurological Recovery After Acute Spinal Cord Injury. Neurochem. Res. 2018, 48, 1641–1649. [Google Scholar] [CrossRef]
- Chen, C.-H.; Hsu, S.-Y.; Chiu, C.-C.; Leu, S. MicroRNA-21 Mediates the Protective Effect of Cardiomyocyte-Derived Conditioned Medium on Ameliorating Myocardial Infarction in Rats. Cells 2019, 8, 935. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic. Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Balint, V.; Stanisavljevic Ninkovic, D.; Anastasov, N.; Lazic, S.; Kovacevic-Grujicic, N.; Stevanovic, M.; Lazic, A. Inhibition of MiR-21 Promotes Cellular Senescence in NT2-Derived Astrocytes. Biochemistry 2021, 86, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chen, Z.; Du, H.; Liu, R.; Wang, W.; Li, H.; Ning, B. Silencing MiR-21 Induces Polarization of Astrocytes to the A2 Phenotype and Improves the Formation of Synapses by Targeting Glypican 6 via the Signal Transducer and Activator of Transcription-3 Pathway after Acute Ischemic Spinal Cord Injury. FASEB J. 2019, 33, 10859–10871. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, W.; Wang, S.; Xie, W.; Li, H.; Ning, B. MicroRNA-21 Regulates Astrocytic Reaction Post-Acute Phase of Spinal Cord Injury through Modulating TGF-β Signaling. Aging 2018, 10, 1474. [Google Scholar] [CrossRef] [PubMed]
- López-Leal, R.; Díaz-Viraqué, F.; Catalán, R.J.; Saquel, C.; Enright, A.; Iraola, G.; Court, F.A. Schwann Cell Reprogramming into Repair Cells Increases MiRNA-21 Expression in Exosomes Promoting Axonal Growth. J. Cell Sci. 2020, 133, jcs239004. [Google Scholar] [CrossRef]

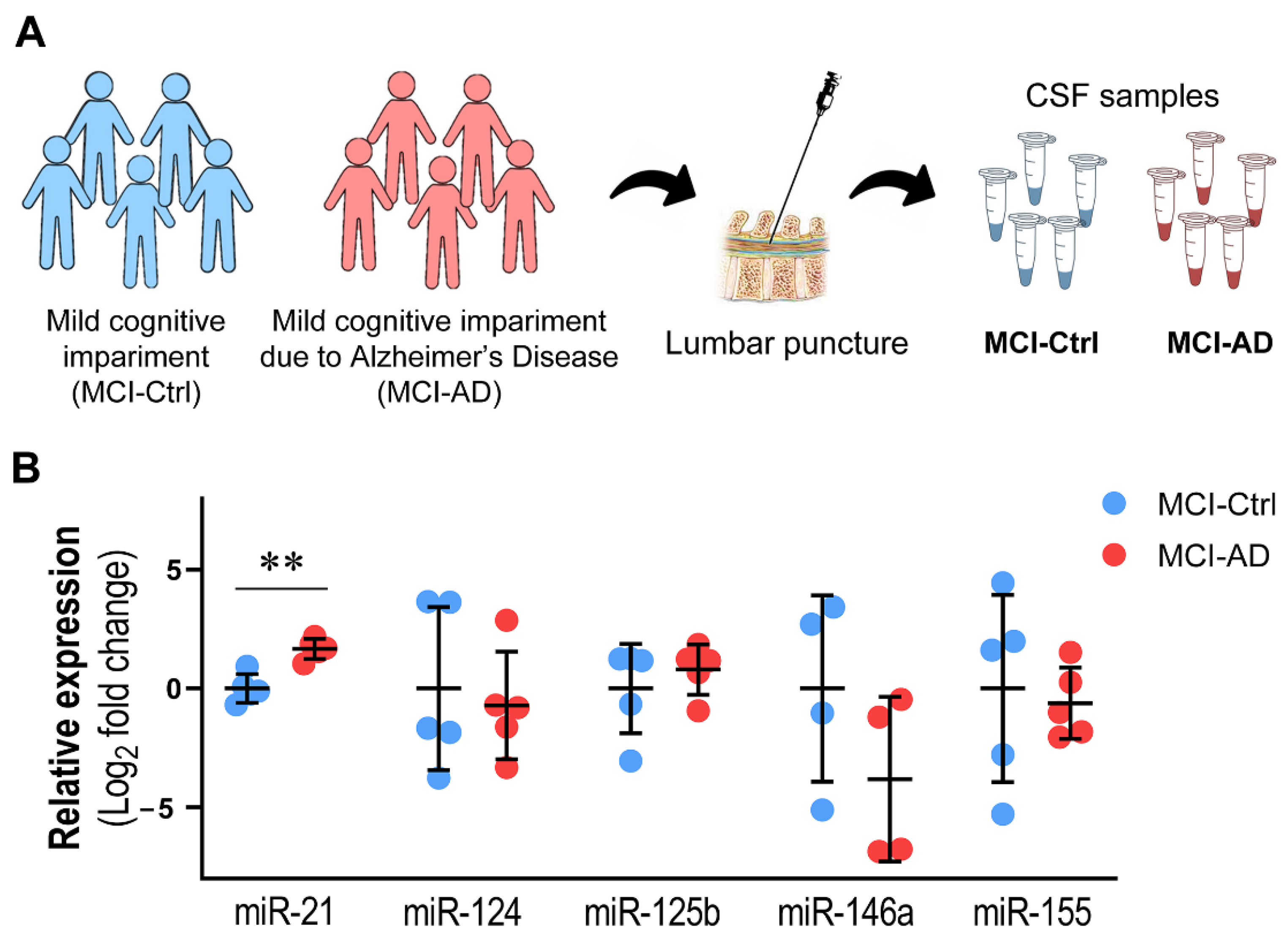

| Name | Patient | Sex | Age | MMSE Score | CDR Scale | PiB-PET | CSF tTau (pg/mL) | CSF pTau (pg/mL) | CSF Aβ42 (pg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| MCI-Ctrl | LIS-105 | F | 65 y | 25 | 0.5 | − | 214 | 37 | 1070 |
| LIS-106 | F | 78 y | ND | 0.5 | − | 280 | 46 | 1190 | |
| LIS-107 | F | 73 y | 28 | 0.5 | − | 296 | 46 | 1010 | |
| LIS-113 | M | 57 y | 27 | 0.5 | − | 194 | 29 | 617 | |
| LIS-115 | M | 60 y | 22 | 0.5 | − | 126 | 19 | 696 | |
| MCI-AD | LIS-096 | M | 58 y | 28 | 0.5 | + | 1100 | 140 | 450 |
| LIS-097 | F | 74 y | 27 | 0.5 | + | 566 | 75 | 555 | |
| LIS-102 | F | 71 y | 27 | 0.5 | + | 1140 | 111 | 274 | |
| LIS-103 | M | 72 y | ND | 0.5 | + | 539 | 59 | 494 | |
| LIS-104 | M | 72 y | ND | 0.5 | + | 186 | 30 | 463 |
| Patient | Suffix | Diff. | Sex | Age at Biopsy | Mutation Genotype | APOE Genotype | Health Status | Sample Origin | Reprogr. Method | Karyotype | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl1 | Ctrl | Neuron Astrocyte | F | Adult | - | ε3/ε3 | Healthy | Skin biopsy | SeV 1.0 | 46XX Normal | [27] |
| Ctrl3 | Ctrl | Microglia | F | 44 y | - | ε3/ε3 | Healthy | Skin biopsy | SeV 1.0 | 46XX Normal | [26] |
| AD2 | AD | Neuron Astrocyte | M | 48 y | PSEN1ΔE9 | ε3/ε3 | EOAD | Skin biopsy | SeV 2.0 | 46XY Normal | [27] |
| AD3 | AD | Microglia Neuron Astrocyte | F | 47 y | PSEN1ΔE9 | ε3/ε3 | Pre-symptomatic AD | Skin biopsy | SeV 2.0 | 46XX Normal | [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, G.; Pinto, S.; Ferreira, S.; Lopes, D.; Serrador, M.J.; Fernandes, A.; Vaz, A.R.; Mendonça, A.d.; Edenhofer, F.; Malm, T.; et al. Emerging Role of miR-21-5p in Neuron–Glia Dysregulation and Exosome Transfer Using Multiple Models of Alzheimer’s Disease. Cells 2022, 11, 3377. https://doi.org/10.3390/cells11213377
Garcia G, Pinto S, Ferreira S, Lopes D, Serrador MJ, Fernandes A, Vaz AR, Mendonça Ad, Edenhofer F, Malm T, et al. Emerging Role of miR-21-5p in Neuron–Glia Dysregulation and Exosome Transfer Using Multiple Models of Alzheimer’s Disease. Cells. 2022; 11(21):3377. https://doi.org/10.3390/cells11213377
Chicago/Turabian StyleGarcia, Gonçalo, Sara Pinto, Sofia Ferreira, Daniela Lopes, Maria João Serrador, Adelaide Fernandes, Ana Rita Vaz, Alexandre de Mendonça, Frank Edenhofer, Tarja Malm, and et al. 2022. "Emerging Role of miR-21-5p in Neuron–Glia Dysregulation and Exosome Transfer Using Multiple Models of Alzheimer’s Disease" Cells 11, no. 21: 3377. https://doi.org/10.3390/cells11213377
APA StyleGarcia, G., Pinto, S., Ferreira, S., Lopes, D., Serrador, M. J., Fernandes, A., Vaz, A. R., Mendonça, A. d., Edenhofer, F., Malm, T., Koistinaho, J., & Brites, D. (2022). Emerging Role of miR-21-5p in Neuron–Glia Dysregulation and Exosome Transfer Using Multiple Models of Alzheimer’s Disease. Cells, 11(21), 3377. https://doi.org/10.3390/cells11213377













